Abstract
Metoprolol, a commonly prescribed beta-blocker, is primarily metabolized by cytochrome P450 2D6 (CYP2D6), an enzyme with substantial genetic heterogeneity. Several smaller studies have shown that metoprolol pharmacokinetics is influenced by CYP2D6 genotype and metabolizer phenotype. To increase robustness of metoprolol pharmacokinetic estimates, a systematic review and meta-analysis of pharmacokinetic studies that administered a single oral dose of immediate release metoprolol was performed. Pooled analysis (n= 264) demonstrated differences in peak plasma metoprolol concentration, area under the concentration-time curve, elimination half-life, and apparent oral clearance that were 2.3-, 4.9-, 2.3-, and 5.9-fold between extensive and poor metabolizers, respectively, and 5.3-, 13-, 2.6-, and 15-fold between ultra-rapid and poor metabolizers (all p<0.001). Enantiomer-specific analysis revealed genotype-dependent enantio-selective metabolism, with nearly 40% greater R- vs S-metoprolol metabolism in ultra-rapid and extensive metabolizers. This study demonstrates a marked effect of CYP2D6 metabolizer phenotype on metoprolol pharmacokinetics and confirms enantiomer specific metabolism of metoprolol.
Keywords: metoprolol, CYP2D6, poor metabolizer, pharmacokinetics, pharmacogenetics, pharmacogenomics, meta-analysis
INTRODUCTION
Metoprolol is one of the most commonly used beta-blockers in medicine and primarily used in the treatment of hypertension and heart failure. The cytochrome P450 2D6 (CYP2D6) enzyme is predominantly involved (approximately 70-80%) in the hepatic metabolism of metoprolol. (1, 2) Since the first description of a CYP2D6 sparteine-debrisoquin polymorphism in the mid-1970’s, substantial genetic heterogeneity has been reported with close to 100 different polymorphisms identified. (3-6) Some CYP2D6 polymorphisms render the enzyme completely inactive while others do not modify the activity of the enzyme. Based on these CYP2D6 gene variants, four metabolizer phenotypes are used to characterize drug metabolism via CYP2D6 in vivo: ultra-rapid metabolizer (UM), extensive metabolizer (EM), intermediate metabolizer (IM) and poor metabolizer (PM) phenotype. As a result of these metabolizer phenotypes, the plasma concentration of metoprolol available for target effects may range from subtherapeutic levels in the UM group to supratherapeutic and potentially toxic concentrations in the PM group, increasing the probability of adverse effects such as hypotension and bradycardia. (7, 8)
Pharmacokinetic studies often have small sample sizes that can result in less precise parameter estimates. Systematic reviews and meta-analyses have been developed to provide more robust estimates by pooling available data from several studies. Therefore, a pooled analysis of available pharmacokinetic data was performed in order to present a more accurate estimate of CYP2D6 metabolizer phenotype-dependent effects on metoprolol pharmacokinetics.
RESULTS
The systematic review of the literature identified 13 independent studies that met the pre-defined search criteria (6, 9-21), with a total sample size of 264 participants (Figure 1). Two major analyses were performed: one for studies quantifying racemic metoprolol pharmacokinetics (n=172) and another for studies that distinguished between R- and S-metoprolol enantiomers (n=128). One study (n=36) provided data that were used in both analyses.
Figure 1. PRISMA study selection flow diagram.
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram depicting the steps undertaken in the systematic review prior to meta-analysis, including the number of records identified, included and excluded. Detailed reasons for exclusion can be found in Supplemental Table 3.
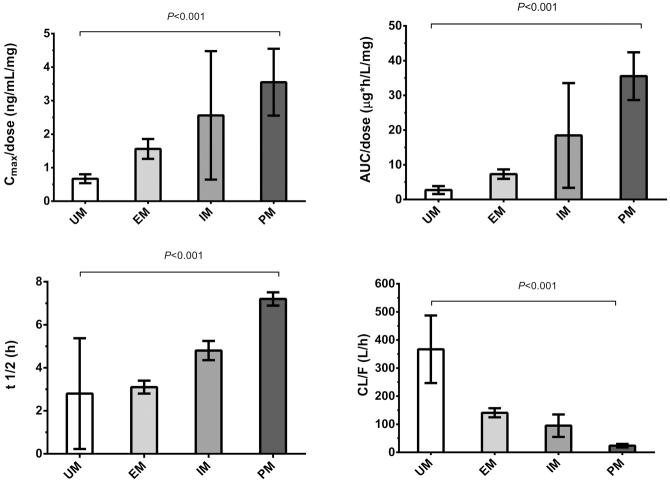
A meta-analysis of the pooled racemic data demonstrated a clear effect in all queried pharmacokinetic (PK) parameters. Comparing both extreme phenotypes, ultra-rapid metabolizers (UM) and poor metabolizers (PM), a 5.3-fold difference in peak dose-normalized plasma metoprolol concentration (C-max/dose; 90% CI 3.9 – 6.9-fold, p<0.001), a 13-fold difference in dose-normalized area under the plasma concentration vs time curve (AUC/dose; 90% CI 9.4 – 19.7-fold, p<0.001), a 2.6-fold difference in elimination half-life (t1/2; 90% CI 2.1 – 3.2-fold, p<0.001) and a 15-fold difference in apparent oral clearance (CL/F; 90% CI 14.0 – 16.6-fold, p<0.001) was observed (see Table 1 and Figure 2). Between extensive metabolizers (EM, i.e., the majority of the population and the reference group) and poor metabolizers a 2.3-fold difference in Cmax/dose (90% CI 2.2 – 2.4-fold, p<0.001), 4.9-fold difference in AUC/dose (90% CI 4.7 – 5.0-fold, p<0.001), 2.3-fold difference in t1/2 (90%CI 2.3 – 2.3-fold, p<0.001) and a 5.9-fold difference CL/F (90% CI 5.6 – 6.1-fold, p<0.001) was found. Between EM and UM, a 2.3-fold difference in Cmax/dose (90% CI 1.9 – 2.9-fold, p=0.11), 2.7-fold difference in AUC/dose (90% CI 1.9 – 4.0-fold, p=0.17), 1.1-fold difference in t1/2 (90% CI 0.7 – 3.6-fold, p=0.94) and a 2.6-fold difference CL/F (90% CI 1.9 – 3.3-fold, p<0.001) was found.
Table 1. Pooled analysis of metoprolol pharmacokinetics stratified by CYP2D6 phenotype.
| n | Cmax/dose (ng/mL/mg) |
AUC/dose (μg*h/L/mg) |
t 1/2 (h) | CL/F (L/h) | |
|---|---|---|---|---|---|
| UM | 12 | 0.67 [0.55 – 0.79] | 2.73 [1.71 – 3.75] | 2.8 [0.5 – 5.1] | 367 [259 – 474] |
| EM | 122 | 1.56 [1.25 – 1.86] | 7.31 [5.96 – 8.66] | 3.1 [2.8 – 3.4] | 141 [127 – 157] |
| IM | 11 | 2.56 [0.88 – 4.25] | 18.46 [5.19 – 31.73] | 4.8 [4.5 – 5.2] | 95 [60 -130] |
| PM | 27 | 3.55 [2.60 – 4.50] | 35.53 [28.97 – 42.09] | 7.2 [7.0 – 7.5] | 24 [17 – 30] |
Data presented as mean [95% confidence interval]. UM- ultra-rapid metabolizer, EM- extensive metabolizer, IM- intermediate metabolizer, PM- poor metabolizer, Cmax/dose- peak metoprolol concentration divided by the dose of metoprolol given, AUC/dose- area under the curve divided by the dose of metoprolol given, t½ - half-life, CL/F- oral clearance
Figure 2. A-D. Effects of CYP2D6 metabolizer phenotype on selected pharmacokinetic parameters.
A Peak dose-normalized plasma metoprolol concentration (Cmax/dose); B Dose-normalized area under the plasma concentration (AUC/dose); C Elimination half-life (t1/2); D Apparent oral clearance (CL/F) The difference between ultra-rapid metabolizers and poor metabolizers in all four parameters is highly statistically significant (all p<0.001). Error bars represent standard deviations.
Overall, the observed heterogeneity between the studies, as indicated by the I2 statistics (ranges from 0% to 100%; the lower the number, the lower the degree of heterogeneity), was high and ranged between 80% and 95%, In addition to the gene-dose effect noted in the analysis of racemic metoprolol, a stereoselective metabolism of metoprolol that becomes more pronounced with an increasing number of functional alleles (http://www.cypalleles.ki.se/cyp2d6.htm) was observed in the pooled enantiomer-specific analysis (see Table 2). As a result of genotype-dependent metabolism of the S-enantiomer, peak S-metoprolol concentrations and AUC are approximately 40% higher than those of R-metoprolol in the UM and EM groups (approximate S/R ratios: 1.5 and 1.5 for UM; 1.3 and 1.5 in EM, respectively) and are approximately 20% higher for the IM group (approximate S/R ratios: 1.1 and 1.3 in IM, respectively). The preference for the metabolism of R-metoprolol is also seen in the different fold-changes between the UM and PM phenotypes regarding different pharmacokinetic parameters (5.3 vs. 3.8-fold difference in peak metoprolol concentration, 14.5 vs. 9.5-fold difference in AUC and 30.4 vs. 22.4-fold difference in CL/F for R vs. S-enantiomer, respectively).
Table 2. Pooled analysis of R- and S-metoprolol pharmacokinetics stratified by CYP2D6 phenotype.
| n | Cmax/dose (ng/mL/mg) | AUC/dose (ug*h/L/mg) | t 1/2 (h) | CL/F (L/h) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| R | S | R | S | R | S | R | S | ||
| UM | 12 | 0.32 [0.24–0.40] |
0.47 [0.33-0.61] |
1.27 [0.86-1.68] |
1.90 [1.34-2.46] |
ND | ND | 547 [393-701] |
380 [272-488] |
| EM | 90 | 0.66 [0.50-0.81] |
0.89 [0.69-1.09] |
2.94 [2.31-3.58] |
4.27 [3.53-5.01] |
3.1 [2.7-3.6] |
3.3 [2.7-3.8] |
170 [131-208] |
104 [91-117] |
| IM | 12 | 1.09 [0.89-1.29] |
1.24 [1.04-1.44] |
7.61 [5.54-9.68] |
9.59 [7.31-11.87] |
4.6 [3.9-5.3] |
5.2 [4.5-5.9] |
61 [46-75] |
48 [36-60] |
| PM | 14 | 1.69 [122-216] |
1.77 [129-225] |
18.39 [16.55-20.24] |
18.03 [16.23-19.84] |
7.8 [7.3-8.3] |
6.8 [6.3-7.3] |
18 [9-27] |
17 [4-31] |
Data presented as mean [95% confidence interval]. UM- ultra-rapid metabolizer, EM- extensive metabolizer, IM- intermediate metabolizer, PM- poor metabolizer, Cmax/dose- peak metoprolol concentration divided by the dose of metoprolol given, AUC/dose- area under the curve divided by the dose of metoprolol given, t½ - half-life, CL/F- oral clearance, ND- not done
Because the extensive metabolizer group has been broadly defined as having “at least one” active allele, an analysis was performed to determine if a semi-quantitative dose effect could be observed amongst different allele combinations.(13, 15) After assigning a score of 1 for each fully active allele, 0.5 for alleles with decreased activity, and 0 for inactive alleles/deletions, a clear trend towards decreased metabolism with lower semi-quantitative dose was noted with regards to peak metoprolol concentration and AUC (see Table 3).
Table 3. Extensive metabolizer semiquantitative gene dose sub-analysis of metoprolol pharmacokinetics stratified by CYP2D6 phenotype.
| n | Cmax/dose (ng/mL/mg) |
AUC/dose (ug*h/L/mg) |
t 1/2 (h) | CL/F (L/h) | |
|---|---|---|---|---|---|
| Two fully active alleles (SQD=2) | 25 | 1.14 [0.94-1.33] | 5.39 [4.28-6.49] | 3.4 [2.7-4.2] | 168 [136-200] |
|
One fully active allele, one
semiactive allele (SQD=1.5) |
7 | 2.18 [1.65-2.71] | 9.96 [7.58-12.34] | 3.2 [2.2-4.2] | ND |
|
One fully active allele, one
inactive allele (SQD =1) |
1 | 3.32 | 34.17 | 8.2 | ND |
Data presented as mean [95% confidence interval]. SQD- Semiquantitative gene dose, Cmax/dose- peak metoprolol concentration divided by the dose of metoprolol given, AUC/dose- area under the curve divided by the dose of metoprolol given, t½ - half-life, CL/F- oral clearance, ND- not done
Lastly, data regarding the influence of CYP2D6 metabolizer phenotype on the clinical effects of metoprolol were extracted. In these studies, exercise-induced heart rates of healthy volunteers were determined before and after administration of metoprolol. Twenty participants from one study (17) were excluded, as the data appeared duplicated from an earlier study.(16) A greater change in heart rate was observed in PM, an effect that continued virtually unchanged for more than 12 hours while in EM the hemodynamic effects of metoprolol markedly decreased (see Table 4). This effect was also noted in the 12-hour area under the effect curve.
Table 4. Effect of metabolizer phenotype on changes in exercise induced heart rate.
| Study | N | % chg HR (1.5 h.) |
% chg HR (12 h.) |
12 h. Heart rate AUEC |
|---|---|---|---|---|
| Hamelin | EM- 10 | 21% | 5% +/− 7% | ND |
| PM- 6 | 24% | 14% +/− 2% | ND | |
| Hemeryck | EM- 8 | ND | ND | 203 +/− 75 (%*h) |
|
Sharma,
2005 |
EM- 16 | 24% | 8% | 275 +/− 63 (beats*h/min) |
| PM- 4 | 31% | 26% | 423 +/− 85 (beats*h/min) |
Data presented as mean +/− standard deviation where applicable. EM- extensive metabolizer, PM- poor metabolizer, % chg HR- percent change in heart rate, AUEC- area under the effect curve, ND- not done
DISCUSSION
This meta-analysis provides a more robust evidence of the effects of different CYP2D6 metabolizer phenotypes on metoprolol pharmacokinetics A clear gene-dose effect was observed regarding metoprolol pharmacokinetics, whereby metabolism of metoprolol was proportional to the number of active CYP2D6 alleles present, as evident in the four PK parameters measured (peak metoprolol concentration [Cmax], area under the curve [AUC], half-life [t1/2] and oral clearance [CL/F]). In addition, CYP2D6 exhibits a preference towards metabolism of the R-enantiomer of metoprolol. The magnitude of this effect is dependent upon the degree of metabolism exerted by the enzyme: poor metabolizers show no preference while the difference between the enantiomers increases gradually through IM, EM and UM phenotypes. Lastly, this pooled systematic analysis shows that these pharmacokinetic differences influence clinical effects of metoprolol such as heart rate and blood pressure.
With varying degrees of statistical significance, each of the studies included in this meta-analysis demonstrated a gene-dose effect of CYP2D6 metabolizer phenotype on metoprolol pharmacokinetics. Consequently, the pooled analysis also demonstrates this effect, however, with greater statistical robustness given the substantially larger sample size of 264 participants while the largest individual study only consisted of 36 participants. In addition, no individual study presented pharmacokinetic data for all four metabolizer groups. Two studies presented oral clearance (13) and AUC(15) as a semiquantitative gene dose to provide a better understanding of individual allele-specific effects. The pooled analysis, however, now allows for a direct comparison of four PK parameters stratified by four major metabolizer phenotypes, with an additional semiquantitative gene dose subanalysis of the extensive metabolizer group.
The results of this study have potential implications for clinical practice. First, additional evidence supporting the validity of CYP2D6 genotyping was demonstrated by a clear correlation between genotype, metabolizer phenotype and metoprolol pharmacokinetics. Genotyping patients before initiating therapy with metoprolol would allow for the identification of PM and UM, thus potentially avoiding adverse events such as hypotension, syncope and bradycardia in the former or a lack of effect in the latter. Armed with this foreknowledge, adjustment of the initial metoprolol dose or the use of alternative drugs which are not major substrates for CYP2D6 may be considered. Alternative beta-blockers that do not depend on CYP2D6 metabolism are bisoprolol, a lipophilic drug that has been successfully tested in heart failure (22), or atenolol which is hydrophilic but has fewer positive endpoint studies. Carvedilol, a newer “third-generation” beta-blocker with unique vasodilating properties may also be a good alternative, although carvedilol undergoes some CYP2D6-dependent metabolism. Carvedilol has been shown in several studies (e.g. the COMET trial) to be superior to metoprolol in heart failure treatment. (23, 24) Given the significant effects the CYP2D6 phenotype has on metoprolol pharmacokinetics as well as the substantial prevalence of CYP2D6 ultra-rapid and poor metabolizers (combined prevalence probably >10% population), it may be possible that some of the observed differences in clinical trials comparing metoprolol to other beta-blockers may have been due to underlying CYP2D6 genotype and metabolizer phenotype. Furthermore, in the future it may be possible to target and adjust metoprolol dosing according to the CYP2D6 genotype/phenotype.
While this study observed a robust CYP2D6 gene-dose effect on metoprolol pharmacokinetics, there are several limitations. First, the results of any meta-analysis are limited by the parameters of the studies that comprise the meta-analysis. Therefore, several of the limitations inherent to the original studies are carried forward. For example, key ethnic groups were not represented in this study as only white and Asian participants were reported. The lack of diversity in this analysis could limit the generalizability of the results. Second, additional alleles or confounding genetic variation in the form of yet uncovered haplotypes could influence the conclusions drawn from the data. Lastly, the meta-analysis showed a substantial degree of heterogeneity between the pooled studies, which limits the generalizability of the findings.
In summary, this systematic review and meta-analysis provided robust evidence for the importance of CYP2D6 metabolizer phenotype in influencing plasma metoprolol pharmacokinetics and also confirmed the enantiomer-specific metabolism of metoprolol, which is dependent upon the degree of metabolism present. These results could have further implications for pharmacogenetics-oriented personalized beta-blocker therapy.
METHODS
Data Sources and Study Selection Criteria
This study followed the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines (25) A comprehensive search of the PubMed database was conducted for articles published up to October 2012 using the terms metoprolol and CYP2D6. All articles published in English that evaluated the association between CYP2D6 metabolizer phenotype and metoprolol pharmacokinetics were screened. In addition, the bibliography of all articles retrieved was scanned for the inclusion of further articles. To be included in the systematic review, the articles had to fulfill the following inclusion criteria: (1) study participants must have received a single dose of racemic, immediate acting metoprolol and did not receive any other medications (prescribed or study drugs), (2) study participants must be genotyped for CYP2D6, (3) metoprolol pharmacokinetic data were stratified by metabolizer phenotype, and (4) at least one of the four pharmacokinetic parameters (peak metoprolol concentration, area under the curve, half-life and oral clearance) must have been present. A total of153 studies were screened, of which 13 met the inclusion criteria.
Genotype/Phenotype Data and Data Compilation
For each study, participants were assigned to a distinct CYP2D6 metabolizer phenotype according to previously published criteria (26-28) using either phenotypic data (i.e. debrisoquine or dextromethorphan metabolizer status) and/or genotyping data for CYP2D6. Regarding genotyping for CYP2D6, subjects with a gene duplication resulting in more than two active CYP2D6 alleles (i.e. defined as *1, *2, *33, *35) were classified as ultra-rapid metabolizers (UM), while those with at least one active allele were extensive metabolizers (EM). Subjects carrying two alleles of substrate-dependent decreased activity (i.e. *9, *10, *17, *29, *36, *41) or compound heterozygotes for one decreased activity allele in combination with a null allele (i.e. *3 to *8, *11 to *16, *19, *20, *21, *38, *40, *42, *44, *56 and *62) were termed as intermediate metabolizers (IM). A combination of two null alleles in a homozygous variant or compound heterozygous manner was classified as poor metabolizer (PM) phenotype. (13, 29, 30) The analysis focused on four common pharmacokinetic parameters: peak plasma metoprolol concentration (Cmax), area under the concentration vs time curve (AUC), elimination half-life (t1/2) and oral clearance (CL/F). Data were acquired from published tables and results, and converted to commonly reported units (Cmax(ng/mL), AUC (ng*h/mL), t1/2 (h), CL/F (L/h)) as needed. Because different doses of metoprolol were administered, Cmax and AUC were normalized by dividing by the dose given (Cmax/dose and AUC/dose).
Statistical Analysis
The statistical analysis was performed with Comprehensive Meta Analysis software, version 2.2.064 (Biostat, Englewood, NJ). Pooled pharmacokinetic parameters were calculated using a random-effects model due to the substantial heterogeneity between studies. The inverse variance method was used for weighing studies. Heterogeneity between studies was formally assessed by the I2 statistics. Non-normality of the data distribution was considered and tested for, but found to be not serious enough to warrant special treatment. Comparison of the means of pooled pharmacokinetic parameters between metabolizer phenotype groups was done by one-way ANOVA (means and 95% CI). P-values are not adjusted for multiple comparisons. The calculation for the quotient between two means and its corresponding 90% confidence interval was done using Fieller’s method (31) which was incorporated in an online calculator (http://www.graphpad.com/quickcalcs). A p-value of <0.05 was considered statistically significant. GraphPad Prism 6.0.2 (GraphPad, La Jolla, CA) was used for additional statistical analyses.
Supplementary Material
STUDY HIGHLIGHTS.
What is the current knowledge on this topic? The metabolizer phenotype of CYP2D6 has been shown to influence metoprolol pharmacokinetics in several small studies.
What question did this study address? This meta-analysis of eleven clinical trials that utilized plasma samples from healthy volunteers after a single oral dose of metoprolol addresses the impact of CYP2D6 metabolizer phenotype on metoprolol pharmacokinetics.
What this study adds to our knowledge: The pooled analysis (n= 264) demonstrated a 5.3-fold difference in peak plasma metoprolol concentration, a 13-fold difference in area under the concentration-time curve, a 2.6-fold difference in elimination half-life and a 15-fold difference in apparent oral clearance between ultra-rapid metabolizers and poor metabolizers. Enantiomer-specific analysis revealed genotype-dependent enantio-selective metabolism, with nearly 40% greater R- vs S-metoprolol metabolism in ultra-rapid and extensive metabolizers
How this might change clinical pharmacology and therapeutics: These results could have further implications in a pharmacogenetics-oriented, personalized beta-blocker therapy.
ACKNOWLEDGEMENTS
MS is supported by the German Federal Ministry of Education and Research (Virtual Liver Network grant 2318 0315755 and grant 03 IS 2061C), the Deutsche Forschungsgemeinschaft 2319 (grant SCHW 858/1-1) and the Robert-Bosch Foundation, Stuttgart, Germany. These funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The study was supported, in parts, by grants from the National Institutes of Health, Bethesda, MD (NIHK23 GM087534 to PN, K24DA00417 to EDK, and UL1RR024992 to Washington University Institute of Clinical and Translational Sciences), and the Division of Clinical and Translational Research, Department of Anesthesiology, Washington University.
Footnotes
CONFLICT OF INTEREST/DISCLOSURE
PN has received research support from Roche Diagnostics (Indianapolis, IN) and Express Scripts (St. Louis, MO), both unrelated to this study.
AUTHOR CONTRIBUTIONS
C.M.B., P.N. , E.D.K. , and M.S. wrote the manuscript.
C.M.B., P.N. , E.D.K. , and M.S. designed the research.
C.M.B. performed the research.
C.M.B. and P.N. analyzed the data.
REFERENCES
- (1).Lennard MS, Silas JH, Freestone S, Ramsay LE, Tucker GT, Woods HF. Oxidation phenotype--a major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–60. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- (2).Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58:521–90. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- (3).Nagele P, Liggett SB. Genetic variation, β-blockers, and perioperative myocardial infarction. Anesthesiology. 2011;115:1316–27. doi: 10.1097/ALN.0b013e3182315eb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Daly AK, et al. Nomenclature for human CYP2D6 alleles. Pharmacogenetics. 1996;6:193–201. doi: 10.1097/00008571-199606000-00001. [DOI] [PubMed] [Google Scholar]
- (5).McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark Med. 2011;5:795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacology & therapeutics. 2013 doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- (7).Rau T, et al. Impact of the CYP2D6 genotype on the clinical effects of metoprolol: a prospective longitudinal study. Clin Pharmacol Ther. 2009;85:269–72. doi: 10.1038/clpt.2008.218. [DOI] [PubMed] [Google Scholar]
- (8).Nagele P, Liggett SB. Genetic variation, beta-blockers, and perioperative myocardial infarction. Anesthesiology. 2011;115:1316–27. doi: 10.1097/ALN.0b013e3182315eb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hamelin BA, et al. Significant interaction between the nonprescription antihistamine diphenhydramine and the CYP2D6 substrate metoprolol in healthy men with high or low CYP2D6 activity. Clin Pharmacol Ther. 2000;67:466–77. doi: 10.1067/mcp.2000.106464. [DOI] [PubMed] [Google Scholar]
- (10).Hemeryck A, Lefebvre RA, De Vriendt C, Belpaire FM. Paroxetine affects metoprolol pharmacokinetics and pharmacodynamics in healthy volunteers. Clin Pharmacol Ther. 2000;67:283–91. doi: 10.1067/mcp.2000.104788. [DOI] [PubMed] [Google Scholar]
- (11).Huang J, Chuang SK, Cheng CL, Lai ML. Pharmacokinetics of metoprolol enantiomers in Chinese subjects of major CYP2D6 genotypes. Clin Pharmacol Ther. 1999;65:402–7. doi: 10.1016/S0009-9236(99)70134-7. [DOI] [PubMed] [Google Scholar]
- (12).Jin SK, et al. Influence of CYP2D6*10 on the pharmacokinetics of metoprolol in healthy Korean volunteers. J Clin Pharm Ther. 2008;33:567–73. doi: 10.1111/j.1365-2710.2008.00945.x. [DOI] [PubMed] [Google Scholar]
- (13).Kirchheiner J, et al. Impact of the ultrarapid metabolizer genotype of cytochrome P450 2D6 on metoprolol pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2004;76:302–12. doi: 10.1016/j.clpt.2004.07.002. [DOI] [PubMed] [Google Scholar]
- (14).Parker RB, Soberman JE. Effects of paroxetine on the pharmacokinetics and pharmacodynamics of immediate-release and extended-release metoprolol. Pharmacotherapy. 2011;31:630–41. doi: 10.1592/phco.31.7.630. [DOI] [PubMed] [Google Scholar]
- (15).Seeringer A, Brockmoller J, Bauer S, Kirchheiner J. Enantiospecific pharmacokinetics of metoprolol in CYP2D6 ultra-rapid metabolizers and correlation with exercise-induced heart rate. Eur J Clin Pharmacol. 2008;64:883–8. doi: 10.1007/s00228-008-0504-8. [DOI] [PubMed] [Google Scholar]
- (16).Sharma A, et al. Modulation of metoprolol pharmacokinetics and hemodynamics by diphenhydramine coadministration during exercise testing in healthy premenopausal women. J Pharmacol Exp Ther. 2005;313:1172–81. doi: 10.1124/jpet.104.081109. [DOI] [PubMed] [Google Scholar]
- (17).Sharma A, et al. Toward optimal treatment in women: the effect of sex on metoprolol-diphenhydramine interaction. J Clin Pharmacol. 2010;50:214–25. doi: 10.1177/0091270009340417. [DOI] [PubMed] [Google Scholar]
- (18).Somer M, Kallio J, Pesonen U, Pyykkö K, Huupponen R, Scheinin M. Influence of hydroxychloroquine on the bioavailability of oral metoprolol. Br J Clin Pharmacol. 2000;49:549–54. doi: 10.1046/j.1365-2125.2000.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang Y, et al. Effects of imatinib (Glivec) on the pharmacokinetics of metoprolol, a CYP2D6 substrate, in Chinese patients with chronic myelogenous leukaemia. Br J Clin Pharmacol. 2008;65:885–92. doi: 10.1111/j.1365-2125.2008.03150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Werner U, et al. Valdecoxib does not interfere with the CYP2D6 substrate metoprolol. Int J Clin Pharmacol Ther. 2006;44:397–400. doi: 10.5414/cpp44397. [DOI] [PubMed] [Google Scholar]
- (21).Werner U, Werner D, Rau T, Fromm MF, Hinz B, Brune K. Celecoxib inhibits metabolism of cytochrome P450 2D6 substrate metoprolol in humans. Clin Pharmacol Ther. 2003;74:130–7. doi: 10.1016/S0009-9236(03)00120-6. [DOI] [PubMed] [Google Scholar]
- (22).The Cardiac Insufficiency Bisoprolol Study II (CIBIS-II): a randomised trial. Lancet. 1999;353:9–13. [PubMed] [Google Scholar]
- (23).DiNicolantonio JJ, Lavie CJ, Fares H, Menezes AR, O’Keefe JH. Meta-Analysis of Carvedilol Versus Beta 1 Selective Beta-Blockers (Atenolol, Bisoprolol, Metoprolol, and Nebivolol) The American Journal of Cardiology. 2013;111:765–9. doi: 10.1016/j.amjcard.2012.11.031. [DOI] [PubMed] [Google Scholar]
- (24).Poole-Wilson PA, et al. Comparison of carvedilol and metoprolol on clinical outcomes in patients with chronic heart failure in the Carvedilol Or Metoprolol European Trial (COMET): randomised controlled trial. Lancet. 2003;362:7–13. doi: 10.1016/S0140-6736(03)13800-7. [DOI] [PubMed] [Google Scholar]
- (25).Moher D, Liberati A, Tetzlaff J, Altman DG, Group t.P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Annals of Internal Medicine. 2009;151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- (26).Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. American journal of human genetics. 1997;60:284–95. [PMC free article] [PubMed] [Google Scholar]
- (27).Griese EU, et al. Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998;8:15–26. doi: 10.1097/00008571-199802000-00003. [DOI] [PubMed] [Google Scholar]
- (28).Zanger UM, et al. Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics. 2001;11:573–85. doi: 10.1097/00008571-200110000-00004. [DOI] [PubMed] [Google Scholar]
- (29).Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- (30).Swen JJ, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89:662–73. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- (31).Fieller E. The biological standardization of Insulin. J R Statist Soc. 1940;7:S1–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.