Abstract
A lysine histidine transporter (LHT) cDNA was isolated and characterized from the roots of Panax ginseng, designated PgLHT. The cDNA is 1,865 bp with an open reading frame that codes for a protein with 449 amino acids and a calculated molecular mass of 50.6 kDa with a predicted isoelectric point of 8.87. Hydropathy analysis shows that PgLHT is an integral membrane protein with 9 putative membrane-spanning domains. Multiple sequence alignments show that PgLHT shares a high homology with other plant LHTs. The expression profile of the gene was investigated by real-time quantitative polymerase chain reaction during various chemical treatments. PgLHT was up-regulated in the presence of abscisic acid, salicylic acid, methyl jasmonate, NaCl, and amino acids. To further explore the function of PgLHT gene, full-length cDNA of PgLHT was introduced into P. ginseng by Agrobacterium rhizogenes A4. The overexpression of PgLHT in the hairy roots led to an obviously increase of biomass compared to the controls, and after addition of the amino acids, the overexpressed-PgLHT hairy roots grew more rapidly than untreated controls during early stage of the culture cycle. The results suggested that the PgLHT isolated from ginseng might have role in the environmental stresses and growth response.
Keywords: Panax ginseng, LHT gene, Abiotic stresses, Real-time quantitative polymerase chain reaction
INTRODUCTION
Amino acids play central roles both as building blocks of proteins and as intermediates in metabolism. They are needed for cellular structure, growth, and development of plants. Amino acid uptake into cells and cellular compartments depends on membrane transporters and amino acid transporters have been identified in many organisms including bacteria, animals, and plants [1,2]. Amino acid transport is involved in all processes associated with resource allocation during plant growth and development [3,4]. Therefore, identifying various amino acid transporter genes that participate in resource allocation is essential for understanding this fundamental process.
Amino acid transporters in higher plants are classified into two superfamilies: the amino acid transporter (ATF) and the amino acid-polyamine-choline facilitator (APC) [2,4]. Members of the ATF superfamily have been shown to have 9 to 11 membrane-spanning domains, whereas members of the APC superfamily contain 12 or 14 putative transmembrane domains [5,6]. The ATF superfamily contains the amino acid permease (AAP), lysine- and histidine-specific transporter (LHT), proline transporter, γ-aminobutyric acid transporter, aromatic and neutral amino acid transporter, and auxin transporter [3].
AAP1 was the first isolated by functional complementation [7,8] and it was subsequently shown to be part of a small gene family of related transporters that are differentiated by substrate specificity and expression patterns [9,10]. Since then many AAPs were separated and all Arabidopsis AAPs (AtAAPs) have been identified as H+-coupled amino acid uptake systems [9,11]. AAPs have been suggested to be involved in a number of physiological processes in plants including amino acid uptake from the soil [12].
However, much less is known about the LHTs, a family of 10 members was found in Arabidopsis. AtLHT1 was originally described as a lysine and histidine selective transporter [6], but other studies with AtLHT1 and AtLHT2 suggest that LHTs preferentially transport neutral and acidic amino acids with high affinity [11,13]. Although AtLHT1 was previously reported to be a broad-spectrum amino acid transporter [11], these abundant amino acids are possibly the main physiological substrates of AtLHT1. In recent investigations demonstrated that AtLHT1 is a negative modulator of disease resistance and that its substrate, Gln, when integrated with the SA pathway, plays pivotal roles in plant defense responses [14]. In this study, a putative LHT gene was isolated and characterized from the roots of Panax ginseng. Sequence comparison showed that this ginseng gene may belong to the LHT subfamily and may be a plasma membrane-located protein, as indicated by the existence of its 9 transmembrane helices. Isolation and functional analysis of the PgLHT gene from P. ginseng will help to improve our knowledge of amino acid metabolism and accumulation in this important medicinal plant.
MATERIALS AND METHODS
Plant materials
Ginseng (P. ginseng Meyer) was collected from Fusong County, Jinlin Province, China. Four-year-old fresh ginseng roots were used for gene cloning and hairy roots induction. Ginseng stems, roots, leaves, and hairy roots were used for gene expression analysis.
LHT gene cloning and sequence analysis
Total RNA was isolated from roots of 4-year-old ginseng plants. cDNA was synthesized using BD SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s protocol. All polymerase chain reaction (PCR) products were cloned into the pGEM-T-Easy vector (Promega, Madison, WI, USA) and sequenced. To amplify the core fragments, a 729 bp fragment cDNA was amplified using two degenerate primers: 5′-TGGCAAATGGTKGADATGCATGA-3′ as forward primer, 5′-WATCTGGTADCTWCCDATDACATG-3′ as reverse primer. PCR reaction was performed in a 50 μL reaction volume containing 5 μL of 10×Ex Taq Buffer (Mg2+ Plus), 0.5 μL of dNTP (10 mM), 1 μL of each primer (10 μM), 41.25 μL of ddH2O, 0.25 μL (1 U) of Ex Taq polymerase (Takara, Ohtsu, Japan), and 1 μL of cDNA template. The PCR conditions were: 94℃ for 30 s, 65℃ for 30 s, 72℃ for 60 s (10 cycles, scaledown of 1℃ per cycle); 94℃ for 30 s, 56℃ for 60 s, 72℃ for 60 s (25 cycles); 72℃ for 10 min.
To isolate the LHT full-length cDNA of P. ginseng, 5′ RACE and 3′ RACE were carried out using the SMART RACE amplification kit (Clontech) with 5′ RACE primer (5′-CTCGGGTGTTGAAGGGATTGTTGC-3′) and 3′ RACE primer (5′-GTCCACTTTGTC CTCTCCCATCTCC-3′) respectively. PCR reaction systems were used in the kit-recommended 50 μL PCR reaction. The PCR conditions were: 94℃ for 4 min; 94℃ for 30 s, 72℃ for 90 s, 5 cycles; 94℃ for 30 s, 70℃ for 30 s, 72℃ for 2 min, 5 cycles; 94℃ for 30 s, 68℃ for 30 s, 72℃ for 2 min, 20 cycles.
The open reading frame (ORF) was analyzed using an ORF finder tool from the web (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Multiple sequence alignments were performed using BLAST and ClustalX program [15,16]. Phylogenetic analysis of deduced amino acid alignments was performed using the neighbor-joining method with the MEGA 4.1 program [17]. Bootstrap analysis with 1,000 replicates was used to assess the strength of nodes in the tree. Prediction of transmembrane domains was generated using the TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0) and TMpred server (http://www.ch.embnet.org/ software/TMPRED_form.html).
Chemical treatment of hairy roots
To investigate the response of PgLHT gene to various chemical treatments, two weeks cultured hairy roots (transformed by Agrobacterium rhizogenes A4 without any foreign DNA) were used. For chemical treatment, hairy roots were maintained in Murashige and Skoog (MS) media containing indicated concentrations of chemicals; 20 μM abscisic acid (ABA), 200 μM salicylic acid (SA), 100 μM methyl jasmonate (MeJA), 50 mM NaCl, and 36 μM L-amino acids (Thr, Ser, Asn, Glu, Gln, Pro, Gly, Ala, Val, Cys, Met, Leu, Tyr, Phe, Lys, His, Arg, Asp 2 μM each). The hairy root samples were collected after post-treatment. The treated hairy roots were immediately frozen in liquid nitrogen and stored at -70℃ until required.
Real-time quantitative polymerase chain reaction analysis
The gene transcript levels of different tissues and hairy roots of ginseng were quantified by real-time quantitative PCR (qRT-PCR) using the SYBR Green stains on the Mastercycler ep realplex2 detection system (Eppendorf, Hamburg, Germany). Total RNA was extracted from ginseng samples using EZNA Plant RNA kit (Omega Biotek, Doraville, GA, USA) according to the manufacturer's instructions. Reverse transcription reactions were performed using RevertAid First Strand cDNA Synthesis kit (Fermentas, Hanover, MD, USA) according to the supplier's manual. A 25-μL reaction mixture contained 12.5 μL of 2×SYBR Premix Ex Taq II (Takara), 1 μL each of 10 μM forward and reverse primer and a suitable amount of cDNA. The PCR conditions were: 95℃ for 30 s; 40 cycles of 95℃ for 10 s, 60℃ for 30 s. At the end of the amplification cycle, a melting analysis was conducted to verify the specificity of the reaction. This was carried out by heating the amplification products from 60℃ to 95℃ at 0.5℃/10 s. Post qRT-PCR calculations to analyze relative gene expression were performed according to the 2-ΔΔCt method as described [18]. The data for the genes expression are shown after normalization with the β-actin internal control (GenBank accession no. AY907207). All qRT-PCR reactions were performed in triplicate. Specific primers for PgLHT were 5′-CTGGGAGATGTAGCGTTTG C-3′ as forward, 5′-GGTGTTGAAGGGATTGTTGC-3′ as reverse; and β-actin were 5′-TGC CCCAGAAGAGCACCCTGT-3′ as forward, 5′-AGCATACAGGGAAAGATCGGCTTGA-3′ as reverse.
Vector construction and plant transformation
The PgLHT cDNA was amplified by specific primers (5′-CGGGATCCCGCACCATGG TTTCCCAAGCTCAGAC-3′ as forward and 5′-CGCGAGCTCGGCTCCTTTGCTGCTTAT GAGAA-3′ as reverse) from P. ginseng and was digested with BamH I and Sac I to yield a 1,350 bp fragment containing the complete open reading frame. This fragment was then ligated into the same restriction site of pBI121 vector which contains a CaMV35S promoter, a nopaline synthase (NOS) terminator, and a kanamycin resistance marker. The recombinant binary vector pBI121 was subsequently confirmed by DNA sequence analysis. The pBI121 vector harboring PgLHT construct was introduced into A. rhizogenes A4 by a conventional method [19].
The 4-year-old fresh ginseng roots were used to induce hairy roots. Ginseng roots were sterilized and cultured as previously described methods [20]. Sterilized roots were sliced into 3-5 mm-thick discs. Root discs were wounded with a sterilized needle loaded with A. rhizogenes A4 containing pBI121 binary vector. After 3 days, the root discs were transferred onto the MS medium containing 100 mg/L kanamycin and 400 mg/L cefotaxime for selection of transformed roots. After cultivation of 30 to 50 days, the hairy roots were picked out and transferred on fresh medium with cefotaxime and kanamycin for several generations until the bacteria were eliminated. Hairy roots were transformed by A. rhizogenes A4 harboring empty pBI121 vector used as controls.
Analysis of transgenic hairy roots
RT-PCR and qRT-PCR were done to determine the transgenic hairy roots. RNA extraction and reverse transcription reaction were performed by following the method used as before. In the RT-PCR, specific primers for PgLHT were (5′-GCCTCTGTCCACTTTG TCCTCT-3′) as forward and (5′-TCTCGGGTGTTGAAGGGATTGT-3′) as reverse; and β-actin (GenBank accession no. AY907207) were 5′-CGTGATCTTACAGATAGCTTCA TGA-3′ as forward, 5′-AG AGAAGCTAAGATTGATCCTCC-3′ as reverse. PCR reaction was performed in a 50 μL reaction volume containing 5 μL of 10×Taq buffer (Mg2+ free), 3 μL of MgCl2 (25 mM), 4 μL of dNTP (2.5 mM each), 2 μL of each primer (10 μM), 32.75 μL of ddH2O, 0.25 μL (1 U) of Taq polymerase (Takara) and 1 μL of cDNA template. The amplifications were carried out under the following conditions: 95℃ for 30 s; 30 cycles of 95℃ for 30 s, 58℃ for 30 s, 72℃ for 40 s; 72℃ for 10 min.
Analysis of growth pattern
To evaluate the effects of overexpression of PgLHT on the growth of hairy roots, the overexpression-PgLHT lines and control lines were cultured in liquid MS medium with amino acids or not, harvested the hairy roots and the media was collected at the indicated time respectively. The harvested hairy roots were rinsed with sterile dH2O, dried on filter paper and weighed securely. The media were filtered and amino acids of the media were assayed using the L-amino acid quantitation kit (BioVision, Milpitas, CA, USA) according to the manufacturer’s instructions. A total 50 μL reaction mix containing: 46 μL L-amino acid assay buffer, 2 μL L-amino acid probe, 2 μL L-amino acid enzyme mix. And then 50 μL of the reaction mix was added to each well containing the test samples, incubated 30 minutes at 37℃ and assayed at 570 nm. All samples were corrected background by subtracting the value of the 0 μM amino acid controls. Amino acid concentration was calculated according to the formula of the manufacturer’s instructions.
RESULTS AND DISCUSSION
Isolation and amino acid sequence analysis of PgLHT
In order to isolate full-length of ginseng LHT gene, a pair of degenerative primers was designed according to the most conservative region of a known lys- and his-specific amino acid transporter from Arabidopsis and other plants. A 729 bp fragment was cloned successfully. 3′RACE and 5′RACE were carried out using the corresponding gene specific primers designed in the light of the fragment which was cloned by degenerate primers. This gene is named PgLHT with a GenBank accession number JX896444. The PgLHT cDNA is 1,865 base pairs (bp) long and have 5′-UTR (129 bp) and 3′-UTR (386 bp) sequences at both ends. The ORF encodes for a protein of 449 amino acids with a predicted molecular weight of 50.6 kDa with a predicted isoelectric point of 8.87.
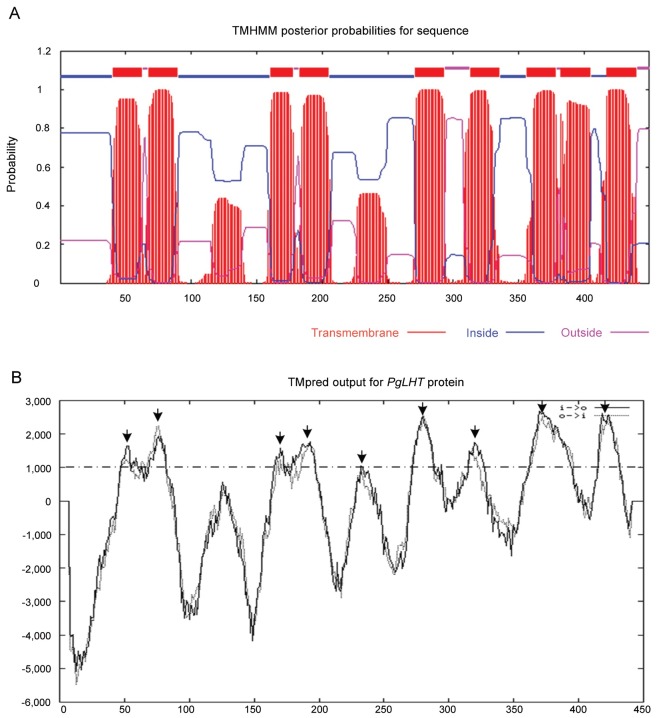
The model for the location of the trans-membrane helices predicted by TMHMM 2.0 is shown in Fig. 1A. Hydropathy analysis shows that PgLHT is an integral membrane protein with 9 putative membrane-spanning domains, and amino acid residues are located at 41-63, 68-90, 161-178, 183-205, 271-293, 313-335, 356-378, 382-404, and 417-439 respectively. We also analyzed the structure by using TMpred software which revealed also the possible existence of 9 membrane-spanning regions, located at 44-63, 66-87, 161-177, 183-202, 221-242, 270-290, 309-327, 361-380, and 417-435, respectively, the putative membrane- spanning regions are marked by arrows (Fig. 1B). These domains are consistent with its plant LHT family [2].
Fig. 1. Results of transmembrane helices prediction by TMHMM and TMPred. (A) The result of transmembrane helices prediction by TMHMM Server ver. 2.0. Red bars indicate transmembrane domains, blue lines indicate intracellular loops and magenta lines indicate extracellular loops. (B) Hydrophobicity plot of PgLHT obtained using TMPred. The ordinate represents the TMPred score divided by 1,000. Positive scores suggest hydrophobic region. The putative membrane-spanning regions are marked by arrows.
Homology analysis
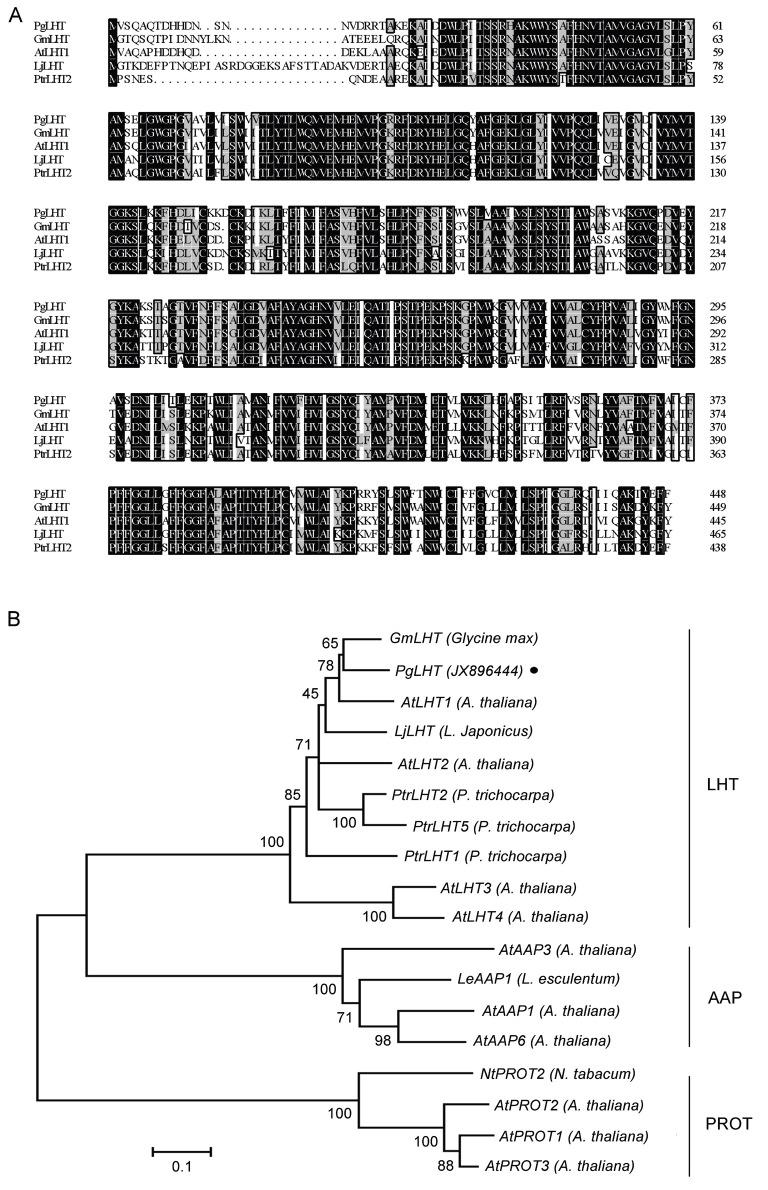
An amino acid sequence comparison using BLASTP revealed the high degree of similarity between the predicted ginseng LHT transporter and other LHT transporters from plants. The alignment using ClustalX is shown in Fig. 2A. Sequence analysis showed high identities with other species such as Glycine max (GmLHT, XM003547627, 83.41% identity), Arabidopsis thaliana (AtLHT1, NM180778, 79.65% identity), Lotus japonicus (LjLHT, JF705953, 72.46% identity), Populus trichocarpa (XM002315593, 69.23% identity). Phylogenetic analysis was performed based on LHTs amino acid sequences with the MEGA 4.1 program. PgLHT shares the highest homology with the LHT from G. max (Fig. 2B).
Fig. 2. Sequence homology and phylogenetic analysis of PgLHT compared to other lysine histidine transporter (LHT) proteins. (A) Comparison of the putative amino acid sequences of PgLHT and those of LHT genes from the following plants: Glycine max (GmLHT, XM003547627), Arabidopsis thaliana (AtLHT1, NM180778), Lotus japonicus (LjLHT, JF705953), Populus trichocarpa (PtrLHT2, XM002315593). (B) Phylogenetic analysis of the amino acid sequences of PgLHT and the other plant LHTs. Phylogenetic tree of plant LHTs distances between each clone and group are calculated with the program Mega 4.1 using the neighbor-joining method. The scale bar (number of substitutions/site) corresponds to the relative branch length, where a scale of 0.1 represents 10% change. AAP, amino acid permease; PROT, proline transporter.
Tissue-specific expression patterns of PgLHT
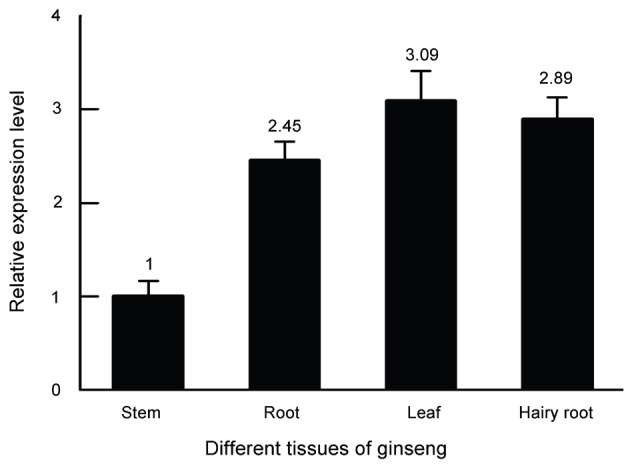
The qRT-PCR analysis was performed to investigate the patterns of PgLHT gene expression in different tissues of P. ginseng. As illustrated in Fig. 3, the PgLHT transcript was constitutive expressed in roots, stems, leaves, and hairy roots, the highest level was found in leaves, significantly lower expression was found in stems.
Fig. 3. Relative mRNA expression levels of PgLHT in different tissues of ginseng. The error bars represent the standard error of means of three independent replicates.
In Arabidopsis, amino acid transporter genes have also various expression patterns in different tissues such as AAP3 is exclusively expressed in roots and AAP4 mainly in leaves, stems, and flowers whereas AAP5 is found in all tissues. ProT1 was found to be expressed in all organs of Arabidopsis, but highest levels were found in roots, stems, and flowers [21].
Expression profiles of PgLHT in response to chemical treatments
Hormone-induced plant growth regulators like ABA, SA, or jasmonic acid (JA) differentially activate stressed-related genes expression [22]. As shown in Fig. 4, PgLHT was upregulated at different degree to various hormone treatments. Under ABA treatment, the PgLHT expression increased steadily and gradually reached a higher level at 24 h (5.5-fold), which can be explained by the role of ABA. Recent studies suggested that ABA-mediated signaling also plays an important part in plant responses to environmental stresses and plant pathogens [23].
Fig. 4. Relative mRNA expression levels of PgLHT at various time points after chemical treatments. Abscisic acid (ABA), 20 μM; salicylic acid (SA), 200 μM; methyl jasmonate (MeJA), 100 μM; NaCl, 50 mM; amino acids (AAs), 36 μM. The error bars represent the standard error of means of three independent replicates.
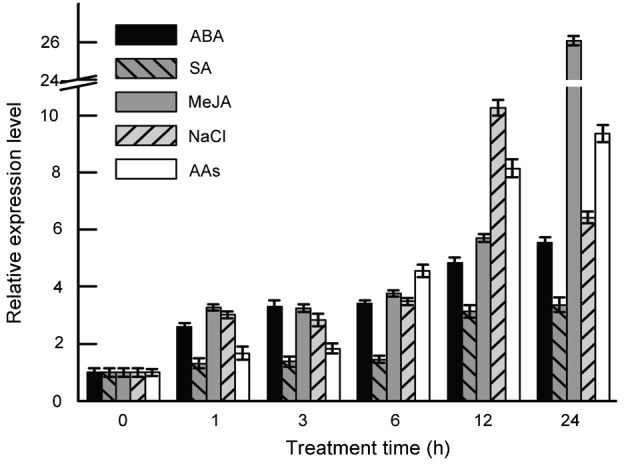
In the case of SA treatment, PgLHT expression maintained a lower level and reached higher point at 12 h with 3.14-fold and then sustained as steady level. AtLHT1, an important amino acid transporter, initially defined as a lys- and his-specific transporter [6], was later reported to transport a broad spectrum of amino acids in a heterologous yeast expression system [12]. It was also observed that AtLHT1 expression was induced within 1 h of SA treatment. It is demonstrated that AtLHT1 negatively modulates defense responses by interacting with the SA-dependent pathway [14]. The results imply that PgLHT may be mediating the defense responses in SA-dependent way in ginseng.
PgLHT expression level increased immediately after MeJA treatment and increased slowly until 12 h and reached 26.05-fold at 24 h. As well-known, MeJA is a lipid-based hormone signals that regulate a wide range of processes in plants, ranging from controlling plant insect defense, wounding responses, development to secondary metabolite synthesis [24]. In general, genes that were markedly up-regulated by JA or MeJA, were not SA-induced and vice versa, consistent with the known antagonistic effect of JA/MeJA and SA on gene expression [25]. As an exception to this rule, PgLHT was up-regulated by MeJA as well as SA.
At a salinity stress, PgLHT expression level increased gradually to 10.27-fold at 12 h and then decreased rapidly at 24 h. High salinity reduces plant growth by affecting the plant’s osmotic or ionic homeostasis [26]. Salinity stress in soil or water is one of the major abiotic stresses especially in arid and semi-arid regions and can severely limit plant growth and yield. ProT1 and ProT2 are widely expressed proline-specific amino acid transporters in Arabidopsis, they respond differentially to changes in water and salinity stress. ProT2 expression was strongly induced under water or salinity stress [21], implying that PgLHT are similar to ProT2 and play an important role in nitrogen distribution during water stress.
A mixture of 18 amino acids was added to the culture of the hairy roots to investigate the expression pattern of PgLHT. After addition the amino acids, PgLHT expression increased steadily to a higher level (9.36 fold) at 24 h. Recent studies of Arabidopsis have identified several transporters as being important for amino acid uptake, and the transporters are regulated by its substrates of amino acids [27], PgLHT was upregulated in a similar way after addition the amino acids. Therefore, there may be a strong relationship between PgLHT expression and uptake of amino acid in ginseng.
Overexpression analysis of PgLHT in ginseng hairy roots
P. ginseng root was transformed using A. rhizogenes A4 with the PgLHT. Several independent transgenic lines were established by selection on kanamycin. Kanamycin resistant hairy roots were confirmed by RT-PCR and qRT-PCR analysis (Fig. 5).
Fig. 5. Real-time (RT)- polymerase chain reaction (PCR) and real-time quantitative (qRT)-PCR analysis of PgLHT transcripts in control (empty vector; C-6 and C-9) and overexpression-PgLHT (A-4, A-7, A-12, and A-16) hairy root lines harvested at 15 day after subculture of the culture cycle. (A) RT-PCR. (B) qRT-PCR. The error bars represent the standard error of means of three independent replicates.
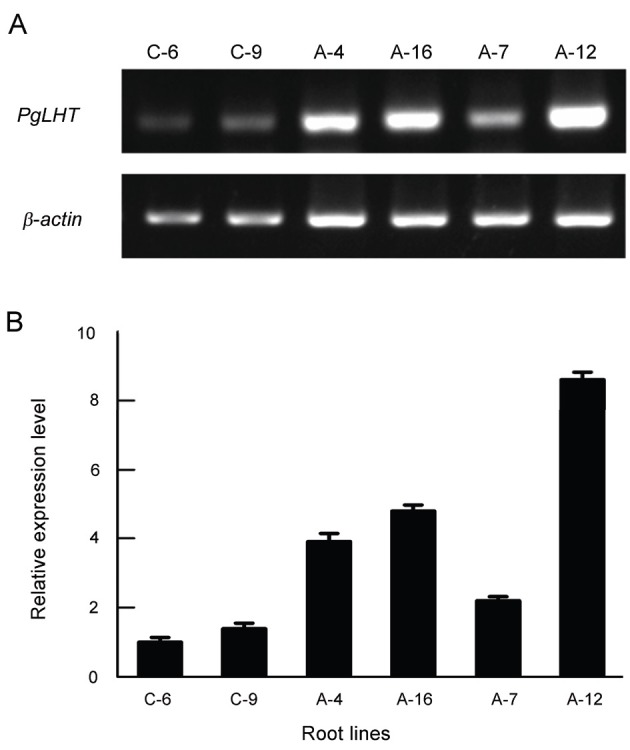
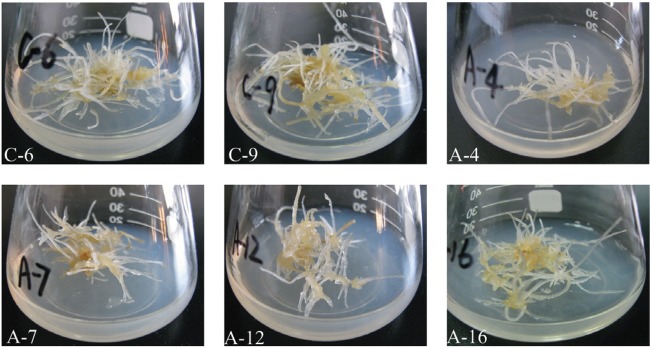
The expression levels of all overexpression-PgLHT lines tested, except A-7, are much higher than the average of controls (transformed with empty vector) (Fig. 5). The transformed hairy roots were indicated in Fig. 6. Two control hairy root lines and four overexpression-PgLHT lines were adapted to liquid MS medium with 5% sucrose and maintained for more than six generations before further analysis.
Fig. 6. Hairy roots induced from roots of Panax ginseng. Hairy roots of controls (C-6 and C-9) and overexpressionPgLHT lines (A-4, A-7, A12, and A-16) cultivated on solid Murashige and Skoog medium at 26℃ at 15 d after subculture of the culture cycle. All lines presented the typical traits of hairy roots and had no morphological difference.
The hairy root lines harvested midway through a culture cycle of 4 wk were subjected to qRT-PCR analysis again. The stable overexpression-PgLHT lines (A-12, A-16) were obtained for the further investigation.
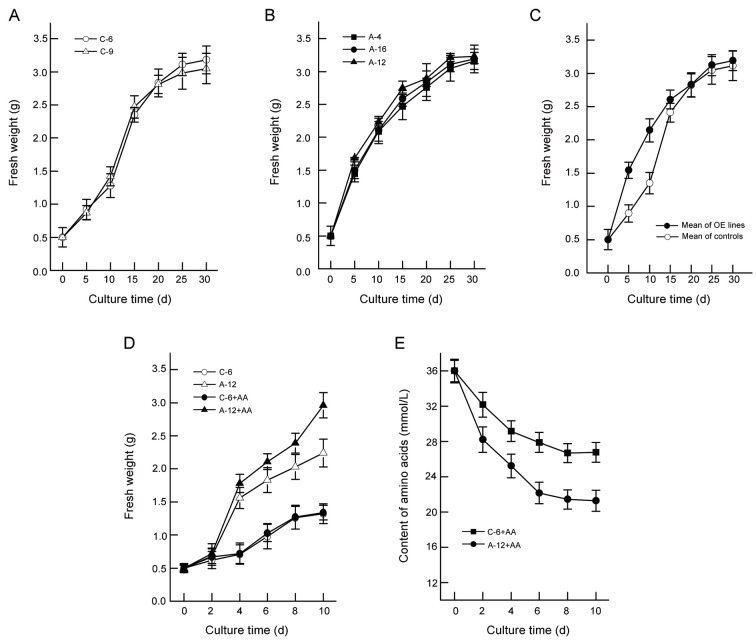
In liquid MS medium, the overexpression-PgLHT lines showed a faster growth than control lines until 15 d after subculture, as determined by Student’s t-test (p<0.05) between the average of overexpression-PgLHT lines and control lines, but there was no obvious difference in the final biomass (Fig. 7A-C).
Fig. 7. Growth curves (A-D) and amino acids analysis (E) of hairy root lines. (A) Control lines. (B) Overexpression-PgLHT lines. (C) Average of controls and overexpression-PgLHT (OE) lines. (D) The growth curves of control (C-6) and overexpression-PgLHT line (A-12) after addition of the amino acids (AAs). (E) The amino acids content in the media of control (C-6) and overexpression-PgLHT line (A-12) after addition of amino acids (36 μM, AAs). The error bars represent the standard error of means of three independent replicates. The initial inoculum for all lines weighed 0.5 g.
This implied that overexpression-PgLHT had positive effects on hairy root growth during early days of the culture cycle. To further analyze the effect on the initial stage of overexpression-PgLHT line, the growth curves until 10 days after subculture were studied. As shown in Fig. 7D, there was an obvious increase of overexpression-PgLHT line (A-12) than control line (C-6) and biomass of overexpression-PgLHT hairy root line increased with the addition of amino acids (Fig. 7D, A-12+AA) in a time-dependent manner up to 2.96 g fresh weigh at 10 d, there was a 1.32-fold increase from the values without amino acids (A-12). Both overexpression-PgLHT line and control line caused amino acids content to a obvious decrease (2 to 6 d after subculture) in the media after exogenous addition of amino acids, but its was maintained at a steady level from 8 d after subculture. Amino acids content in overexpression-PgLHT lines are obviously lower than its control lines (Fig. 7E). Comparison of Fig. 7A-D, we presume that PgLHT may participate in the uptake of amino acids or other nutrients from the media.
In this study, overexpression-PgLHT lines of P. ginseng did not show any morphological differences (Fig. 6) when compared to controls. However, overexpression of PgLHT led to rapid growth of these hairy roots during early stage of the culture cycle. In Arabidopsis plants, LHT1 is crucial for Gln uptake at low concentrations and the magnitude of the increase in the uptake rate for Gln was similar to the magnitude of the increase in the growth response of plants overexpressing LHT1[27]. Hence, we can speculate that PgLHT plays an important role between amino acid uptake and growth of hairy roots in the initial culture.
In summary, we isolated PgLHT gene from ginseng roots and characterized its expression patterns in response to various chemicals. The accumulation of PgLHT transcripts was obviously induced after treatment with ABA, SA, MeJA, NaCl, and amino acids. The growth of the hairy roots was correlated with the overexpression-PgLHT which may mediate the amino acids or other nutrients uptake. The physiological function of the PgLHT is still needed to further study.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (grant no. 81071821, 81250110086, and 81101655), the Scientific Research Starting Foundation for Returned Overseas Chinese Scholars, Ministry of Education, China (grant no. 311101013), the Hunan Provincial Natural Science Foundation (grant no.12JJ3099), and the Scientific Research Fund of Hunan Provincial Education Department (11C0329). The critical reading of the manuscript by Mohammed Nuruzzaman and Sadia Afrin, Research Fellow, Molecular Biology Research Center, School of Biological Science and Technology, Central South University is greatly appreciated.
References
- 1.Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH Jr. Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 2.Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci. 2002;27:139–147. doi: 10.1016/S0968-0004(01)02054-0. [DOI] [PubMed] [Google Scholar]
- 3.Rentsch D, Schmidt S, Tegeder M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 2007;581:2281–2289. doi: 10.1016/j.febslet.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Ortiz-Lopez A, Chang H, Bush DR. Amino acid transporters in plants. Biochim Biophys Acta. 2000;1465:275–280. doi: 10.1016/S0005-2736(00)00144-9. [DOI] [PubMed] [Google Scholar]
- 5.Chang HC, Bush DR. Topology of NAT2, a prototypical example of a new family of amino acid transporters. J Biol Chem. 1997;272:30552–30557. doi: 10.1074/jbc.272.48.30552. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Bush DR. LHT1, a lysine- and histidine-specific amino acid transporter in arabidopsis. Plant Physiol. 1997;115:1127–1134. doi: 10.1104/pp.115.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frommer WB, Hummel S, Riesmeier JW. Expression cloning in yeast of a cDNA encoding a broad specificity amino acid permease from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1993;90:5944–5948. doi: 10.1073/pnas.90.13.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu LC, Chiou TJ, Chen L, Bush DR. Cloning a plant amino acid transporter by functional complementation of a yeast amino acid transport mutant. Proc Natl Acad Sci U S A. 1993;90:7441–7445. doi: 10.1073/pnas.90.16.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Mol Plant. 2010;3:997–1011. doi: 10.1093/mp/ssq047. [DOI] [PubMed] [Google Scholar]
- 10.Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem. 1995;270:16315–16320. doi: 10.1074/jbc.270.27.16315. [DOI] [PubMed] [Google Scholar]
- 11.Svennerstam H, Ganeteg U, Nasholm T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol. 2008;180:620–630. doi: 10.1111/j.1469-8137.2008.02589.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YH, Tegeder M. Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J. 2004;40:60–74. doi: 10.1111/j.1365-313X.2004.02186.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Ji Y, Bhuiyan NH, Pilot G, Selvaraj G, Zou J, Wei Y. Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell. 2010;22:3845–3863. doi: 10.1105/tpc.110.079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 18.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Zhao S, Zhang X. Antisense suppression of cycloartenol synthase results in elevated ginsenoside levels in Panax ginseng hairy roots. Plant Mol Biol Rep. 2009;27:298–304. doi: 10.1007/s11105-008-0087-7. [DOI] [Google Scholar]
- 20.Thanh NT, Murthy HN, Yu KW, Hahn EJ, Paek KY. Methyl jasmonate elicitation enhanced synthesis of ginsenoside by cell suspension cultures of Panax ginseng in 5-l balloon type bubble bioreactors. Appl Microbiol Biotechnol. 2005;67:197–201. doi: 10.1007/s00253-004-1759-3. [DOI] [PubMed] [Google Scholar]
- 21.Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YJ, Lee OR, Lee S, Kim KT, Yang DC. Isolation and characterization of a theta glutathione S-transferase gene from Panax ginseng Meyer. J Ginseng Res. 2012;36:449–460. doi: 10.5142/jgr.2012.36.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci U S A. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moons A. Transcriptional profiling of the PDR gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta. 2008;229:53–71. doi: 10.1007/s00425-008-0810-5. [DOI] [PubMed] [Google Scholar]
- 26.Chinnusamy V, Zhu J, Zhu JK. Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y) 2006;27:141–177. doi: 10.1007/0-387-25856-6_9. [DOI] [PubMed] [Google Scholar]
- 27.Svennerstam H, Jamtgard S, Ahmad I, Huss-Danell K, Nasholm T, Ganeteg U. Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. New Phytol. 2011;191:459–467. doi: 10.1111/j.1469-8137.2011.03699.x. [DOI] [PubMed] [Google Scholar]