Summary
Microarray analysis of the genome of Lactobacillus acidophilus identified a number of operons that were differentially expressed in response to carbohydrate source or constitutively expressed regardless of carbohydrate source. These included operons implicated in the transport and catabolism of fructooligosaccharides (FOS), lactose (lac), trehalose (tre) and genes directing glycolysis. Analysis of these operons identified a number of putative promoter and repressor elements, which were used to construct a series of expression vectors for use in lactobacilli, based on the broad host range pWV01 replicon. A β‐glucuronidase (GusA3) reporter gene was cloned into each vector to characterize expression from each promoter. GUS reporter assays showed FOS, lac and tre based vectors to be highly inducible by their specific carbohydrate and repressed by glucose. Additionally, a construct based on the phosphoglycerate mutase (pgm) promoter was constitutively highly expressed. To demonstrate the potential utility of these vectors, we constructed a plasmid for the overexpression of the oxalate degradation pathway (Frc and Oxc) of L. acidophilus NCFM. This construct was able to improve oxalate degradation by L. gasseri ATCC 33323 and compliment a L. acidophilus oxalate‐deficient mutant. Development of these expression vectors could support several novel applications, including the expression of enzymes, proteins, vaccines and biotherapeutics by intestinal lactobacilli.
Introduction
Lactobacilli are members of the lactic acid bacteria, a functional group related by formation of lactic acid as the primary product of carbohydrate metabolism. Lactobacilli have long been considered beneficial, occupying important niches in the gastrointestinal (GI) tracts of humans and animals. Select lactobacilli are increasingly recognized as modulators of health, gaining interest as microbes used as health‐promoting functional food ingredients, and as delivery vectors for vaccines and biotherapeutics (Wells et al., 1996).
There continues to be great interest in the development of genetic tools for production of proteins and enzymes from lactic acid bacteria. Several gene expression systems for lactobaclli have been developed. The widely used nisin‐controlled expression (NICE) system, originally developed for use in Lactococcus lactis (de Ruyter et al., 1996), has been adapted for use in lactobacilli (Kleerebezem et al., 1997; Wu et al., 2006). Expression systems based on control by other bacteriocins (Axelsson et al., 2003; Mathiesen et al., 2004) and lactose (Gosalbes et al., 2001) are also available.
Lactobacillus acidophilus NCFM is a probiotic culture widely used in nutritional supplements, dairy products and infant formulas. The availability of the L. acidophilus genome sequence (Altermann et al., 2005), gene expression profiling (Azcarate‐Peril et al., 2005; 2006; Barrangou et al., 2006) and functional genomic studies (Russell and Klaenhammer, 2001a;Barrangou et al., 2003; Duong et al., 2006) has provided considerable insight into the physiology of this organism and established a technical basis that can be used to improve both the fermentation and probiotic functionalities of this organism.
In this study, we exploit the genome sequence, gene expression profiling and functional genomic data to construct a series of expression vectors and analyse their properties using a β‐glucuronidase (GusA3) reporter protein. Additionally, one vector encoding a strong constitutive promoter was employed for overexpression of the L. acidophilus oxalate‐degradation operon and complementation of a deletion mutation therein (Azcarate‐Peril et al., 2005).
Results
Promoter selection
Previous global gene expression analysis found a number of genes involved in carbohydrate transport and metabolism to be differentially expressed during growth on various carbohydrates (Barrangou et al., 2006). Also, a number of genes were identified as being highly constitutively expressed (Fig. 1). Putative promoters (Fig. 2) from the FOS (PFOS), lac (Plac) and tre (Ptre) operons (Fig. 3) were selected for use in the construction of carbohydrate‐inducible vectors, while the promoter for pgm (Ppgm) was selected for use in the construction of a constitutive expression vector.
Figure 1.
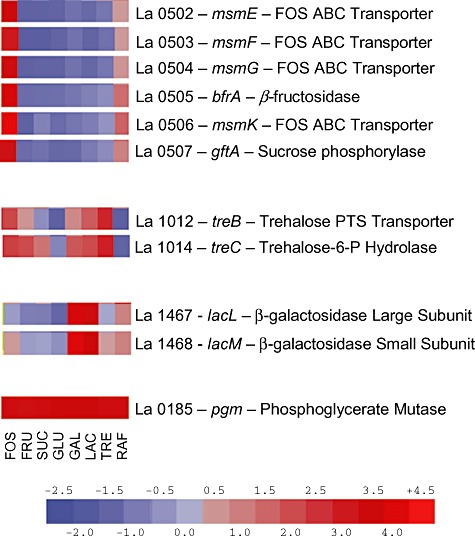
Microarray expression data for select genes and operons. Gene expression of FOS, lac and tre operons and the pgm gene in L. acidophilus grown on fructooligsaccharides (FOS), fructose (FRU), sucrose (SUC), glucose (GLU), galactose (GAL), lactose (LAC), trehalose (TRE) and raffinose (RAF) is shown colormetrically. Scale represents least squares means of overall gene expression level.
Figure 2.
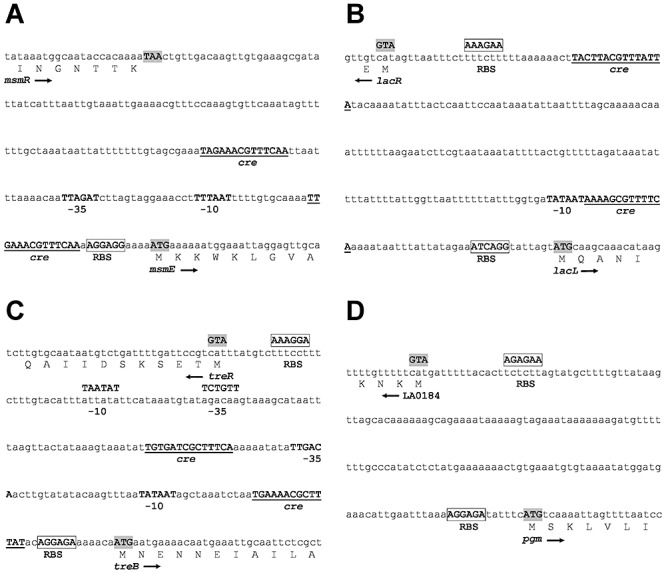
Nucleotide sequence of select intergenic regions. Native promoter regions for (A) FOS, (B) lac and (C) tre operons and (D) pgm. The consensus catabolite response element (cre) sequence WTGNAANCGNWNNCW (Barrangou et al., 2003) was used to identify cre‐like sequences in the promoter regions. Putative –35 and –10 regions are in bold; putative cre‐like sequences are underlined; putative ribosome binding sites (RBS) are boxed; and putative translation start sites are in shaded text.
Figure 3.
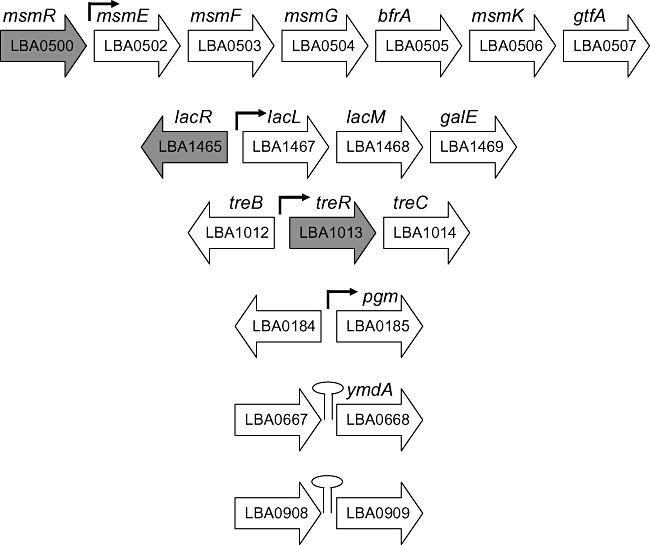
Layouts of selected operons and genes. Open reading frames (ORFs) are represented by arrows with ORF number inside the arrow and gene names above. Transcriptional regulators are shown in grey; promoters are shown as black arrows; and Rho‐independent terminators shown as stem loops. ORF representations are not shown to scale.
Terminator selection
Lactobacillus acidophilus NCFM sequences that could potentially function as Rho‐independent terminators were identified by dyad symmetry analysis using Clone Manager (SciEd, Durham, NC, USA) and TransTerm (Ermolaeva et al., 2000). Two putative terminators, Term908 and Term667, were selected for use in this series of expression vectors (Table 1). Terminators were selected based upon the following criteria: low Gibbs free energy (Δ°G) (Term908: Δ°G = −24.4 kcal mol−1; Term667: Δ°G = −23.5 kcal mol−1), location within a ‘head‐to‐tail’ intergenic region (Fig. 3) and, TransTerm confidence score (100 for both). Additionally, both terminators were predicted to function as bidirectional terminators by TransTerm. Transcription arrays from Barrangou and colleagues (2006) also showed cessation of expression of downstream genes.
Table 1.
Transcriptional terminators.
| Terminator | Δ°G (kcal mol−1) | Sequence |
|---|---|---|
| Term 908 | −24.4 |  |
| Term 667 | −23.5 |  |
Plasmid construction
The vectors constructed in this study are shown in Fig. 4. Terminators were cloned into pTRK846, sequentially. Transformants were screened and selected in order maintain the native ‘head‐to‐tail’ orientation from L. acidophilus to prevent transcription into the expression cassette. The resulting construct, pTRK847, was used as the base for this series of expression vectors into which regulatory elements for the FOS (pTRK848), lac (pTRK849), tre (pTRK850) operons and pgm (pTRK882) were cloned. Plasmid construction was confirmed by sequencing. The Lactobacillus gusA3 gene from pTRK782 (Callanan et al., 2007) was directionally cloned into each vector to create plasmids pTRK888 (pFOS), pTRK889 (pLAC), pTRK890 (pTRE) and pTRK892 (pPGM) for expression analysis (Table 2).
Figure 4.
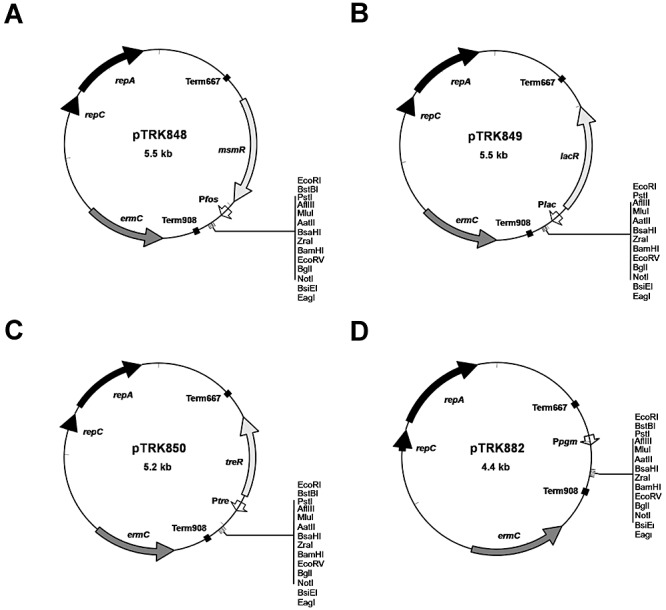
Construction of expression vectors. Restriction maps for expression vectors. Details of construction are given in the text. Black arrows, replication determinants; dark grey arrows, antibiotic resistance markers (ermC: erythromycin resistance maker); black boxes, transcriptional terminators; light grey arrows, transcriptional regulators (msmR: FOS repressor; treR: trehalose repressor; lacR: lactose repressor); white arrows, promoters (Pfos: FOS promoter; Plac: lac promoter; Ptre: tre promoter; Ppgm: pgm promoter). All restriction sites shown are unique.
Table 2.
Bacterial strains and plasmids.
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| L. acidophilus | ||
| NCK 56 | NCFM, human intestinal isolate | Barefoot and Klaenhammer (1983) |
| NCK 1825 | NCFM w/ pTRK888 | This study |
| NCK 1826 | NCFM w/ pTRK889 | This study |
| NCK 1827 | NCFM w/ pTRK890 | This study |
| NCK 1829 | NCFM w/ pTRK892 | This study |
| NCK 1728 | NCFM, Δfrc (Lba0395) | Azcarate‐Peril et al. (2006) |
| NCK 1889 | NCFM w/ pTRK928 | This study |
| NCK 1895 | NCFM w/ pTRK882 | This study |
| NCK 1897 | NCK 1728 w/ pTRK928 | This study |
| NCK 1899 | NCK 1728 w/ pTRK882 | This study |
| L. gasseri | ||
| ATCC 33323 | Neotype | American Type Culture Collection |
| NCK 1935 | ATCC 33323 w/ pTRK888 | This study |
| NCK 1936 | ATCC 33323 w/ pTRK889 | This study |
| NCK 1937 | ATCC 33323 w/ pTRK890 | This study |
| NCK 1938 | ATCC 33323 w/ pTRK892 | This study |
| NCK 99 | ADH, Human intestinal isolate | Kleeman and Klaenhammer (1982) |
| NCK 1967 | ADH w/ pTRK928 | This study |
| NCK 1968 | ADH w/ pTRK882 | This study |
| NCK 1969 | ATCC 33323 w/ pTRK928 | This study |
| NCK 1970 | ATCC 33323 w/ pTRK882 | This study |
| E. coli | ||
| MC 1061 | Strr, E. coli transformation host | Casadaban and Cohen (1980) |
| NCK 1751 | MC 1061 w/ pTRK846 | This study |
| NCK 1752 | MC 1061 w/ pTRK847 | This study |
| NCK 1753 | MC 1061 w/ pTRK848 | This study |
| NCK 1754 | MC 1061 w/ pTRK849 | This study |
| NCK 1755 | MC 1061 w/ pTRK850 | This study |
| NCK 1814 | MC 1061 w/ pTRK882 | This study |
| Plasmids | ||
| pGK12 | ori (pWV01), Emr, Cmr, Gram‐positive shuttle vector | Kok et al. (1984) |
| pORI28 | ori (pWV01), Emr, source of MCS | Law et al. (1995) |
| pTRK782 | Source of gusA3 reporter gene | Callanan et al. (2007) |
| pTRK846 | Emr, Δcat derivative of pGK12 with MCS from pORI28 | This study |
| pTRK847 | Emr,pTRK846 with Term 908 and Term 667 | This study |
| pTRK848 | 1.4‐kb msmR/PFOS PCR fragment cloned into pTRK847 | This study |
| pTRK849 | 1.4‐kb lacR/PlacPCR fragment cloned into pTRK847 | This study |
| pTRK850 | 1.4‐kb treR/Ptre PCR fragment cloned into pTRK847 | This study |
| pTRK882 | 350‐bp Ppgm PCR fragment cloned into pTRK847 | This study |
| pTRK888 | gusA3 cloned into pTRK848 | This study |
| pTRK889 | gusA3 cloned into pTRK849 | This study |
| pTRK890 | gusA3 cloned into pTRK850 | This study |
| pTRK892 | gusA3 cloned into pTRK882 | This study |
| pTRK928 | 7.6 kb, pTRK882 with 3.1 kb PCR product from primers OXF/OXR | This study |
GUS activity
The GUS assays were performed using L. acidophilus NCFM and Lactobacillus gasseri ATCC 33323 transformants that carried the GUS reporter constructs. A time‐course experiment was performed using cultures of L. acidophilus NCK 1825, harbouring the FOS‐inducible GUS reporter construct, pTRK888. GUS activity of cell‐free extracts (CFEs) made from 10 ml aliquots of culture taken at 0, 1, 2 and 3 h post induction was determined (Fig. 5). Cultures induced with FOS had a 100‐fold greater GUS activity at 1, 2 and 3 h post induction as compared with cultures exposed to a non‐inducing carbohydrate, fructose, and a repressive carbohydrate, glucose. No GUS activity was detected at 0 h post induction, indicating that GUS expression was undetectable in MRS medium. Since no significant increase in specific activity was detected at 2 and 3 h post induction by FOS, GUS activity for the remaining carbohydrate inducible and constitutive expressing constructs was assayed at 1 h post induction.
Figure 5.
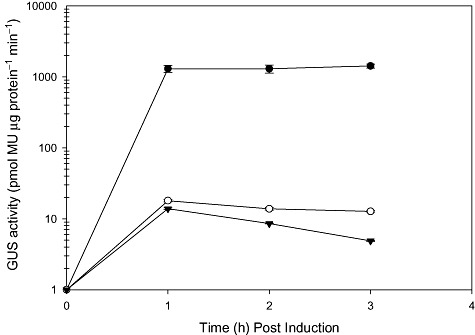
Time‐course of induction. CFEs of L. acidophilus harbouring pTRK888 (PFOS) were assayed for GUS activity at 0, 1, 2 and 3 h post induction. Cultures were induced in (●) FOS, (o) fructose or (▾) glucose. Error bars represent the standard error of the means (SEM) for three independent experiments.
The GUS activity of cultures of L. acidophilus and L. gasseri harbouring pTRK888, pTRK889 and pTRK890 exposed to an inducing carbohydrate, fructose and glucose was determined at 1 h post induction. The GUS activity was greatest in each strain exposed to its specific inducing carbohydrate. In each case, a detectable level of basal expression from the inducible promoters was observed in cells exposed to glucose (lowest) or fructose (intermediate) (Fig. 6). In L. acidophilus, induction of GUS activity by the inducing carbohydrates was 18‐fold higher in pTRK888, 50‐fold higher in pTRK889, and 14‐fold higher in pTRK890 as compared with glucose. In L. gasseri GUS activity was induced 16‐fold in pTRK888, fourfold in pTRK889 and fivefold in pTRK890. Additionally, when specifically induced, GUS activity was found to be at least 10‐fold higher in CFEs from L. gasseri than in CFEs from L. acidophilus.
Figure 6.
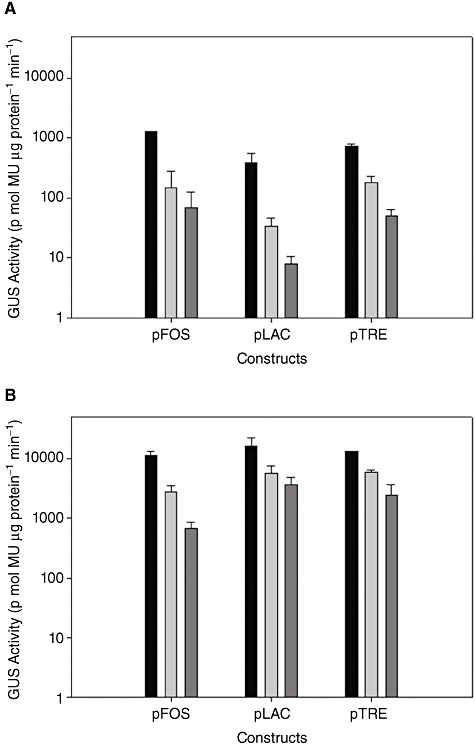
β ‐Glucurondiase activity of inducible vectors. GUS activity of (A) L. acidophilus and (B) L. gasseri harbouring pTRK888 (pFOS), pTRK889 (pLAC) and pTRK890 (pTRE) in inducing carbohydrate (black), fructose (light grey) and glucose (dark grey) at 1 h post induction. Inducing carbohydrates were as follows: pTRK888, FOS; pTRK889, lactose; pTRK890, trehalose. Error bars represent the SEM for three independent experiments.
The GUS activity of Lactobacillus cultures harbouring pTRK892 (Ppgm) when exposed to different carbohydrates was also determined (Fig. 7). The GUS activity was found to be highly expressed across all conditions in both organisms with levels of expression approximately 10‐fold over the levels observed with the induced constructs. There was no significant difference in GUS activity between carbohydrates. GUS activity was found to be threefold to 16‐fold higher in CFEs from L. gasseri than in CFEs from L. acidophilus.
Figure 7.
β ‐Glucurondiase activity of constitutive vector. GUS activity of (A) L. acidophilus and (B) L. gasseri harbouring pTRK892 at 1 h post induction. Cultures were incubated in SSM + 1% carbohydrate or MRS. Error bars represent the SEM for three independent experiments.
Analysis of oxalate degradation in recombinant Lactobacillus
In order to determine the effect of overexpressing the L. acidophilus NCFM oxalate operon on oxalate degradation by lactobacilli, pTRK928 (Fig. 8) was transformed into L. acidophilus NCFM (NCK 56), an L. acidophilus frc deletion mutant (NCK 1728), L. gasseri ATCC 33323 and L. gasseri ADH. The empty vector, pTRK882, was transformed into these strains to serve as controls. The ability of L. acidophilus (Fig. 9A) and L. gasseri (Fig. 9B) cultures to degrade oxalate over 96‐hours was determined. Oxalate degradation in the L. acidophilus strains was 5–52% over the 96‐hour time‐course. The frc deletion mutant strain harbouring the control plasmid (NCK 1899) degraded only 5% of the oxalate, while the frc deletion harbouring pTRK928 (NCK 1897) degraded 48%. Additionally, there was no significant difference in oxalate degradation by L. acidophilus NCFM harbouring pTRK928 or the empty vector. While expression of the oxalate operon using pTRK928 showed no significant improvement in the oxalate degradation ability of L. acidophilus NCFM, pTRK928 was able to complement the deletion mutation, improving oxalate degradation in the frc mutant from 5% to 48%. Oxalate degradation in the L. gasseri strains was 20–45% over 96 h with the L. gasseri ATCC 33323‐derived strains degrading 10–25% greater amounts of oxalate than the L. gasseri ADH‐derived strains. No significant difference in oxalate degradation was measured between the L. gasseri ADH‐derived strains, indicating that pTRK928 was unable to confer improved oxalate degrading ability on this particular strain. However, L. gasseri ATCC 33323 harbouring pTRK928 (NCK 1969) degraded 15% more oxalate than its control (NCK 1970), indicating that pTRK928 was able to improve oxalate degrading ability of the parent strain.
Figure 8.
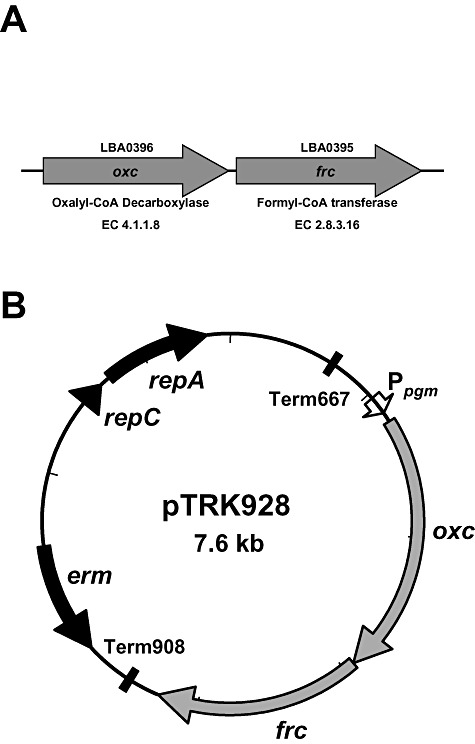
Schematic of oxalate operon and plasmid. A. oxc and frc from Lactobacillus acidophilus NCFM. B. Oxalate operon expression construct, pTRK928. Promoter, Ppgm,is shown in white; oxc and frc are shown in light grey; erythromycin resistance marker, erm, is shown in dark grey; replication determinants are shown in black; terminators are shown as black boxes.
Figure 9.
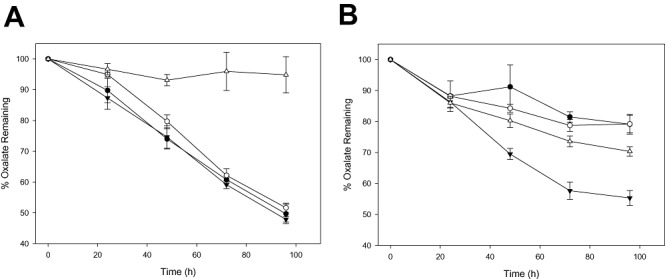
Oxalate degradation by Lactobacillus cultures. Cultures were consecutively transferred in MRS containing 0.05% oxalate then transferred to MRS containing 0.1% oxalate. Cultures were assayed for oxalate at 0, 24, 48, 72 and 96 h after the final transfer. A. Oxalate degrading activity of L. acidophilus cultures: (●) L. acidophilus NCFM + pTRK928 (NCK 1889), (○) L. acidophilus NCFM + pTRK882 (NCK 1895), (▾) L. acidophilus frc deletion + pTRK928 (NCK 1897), (▵) L. acidophilus frc deletion + pTRK882 (NCK 1899). B. Oxalate degrading activity of L. gasseri cultures: (●) L. gasseri ADH + pTRK928 (NCK 1967), (○) L. gasseri ADH + pTRK882 (NCK 1968), (▾) L. gasseri ATCC 33323 + pTRK928 (NCK 1969), (▵) L. gasseri ATCC 33323 + pTRK882 (NCK 1970). Error bars are standard deviations of means from triplicate measurements of three independent experiments.
Discussion
Gene expression analysis in L. acidophilus using microarrays identified a number of operons related to carbohydrate metabolism whose expression was tightly regulated by carbohydrates (Barrangou et al., 2006). Specifically, genes within these operons were found to be highly induced when the organism was grown on a specific carbohydrate and to be very highly repressed when grown on glucose. Based on these data and previous functional characterization using gene insertion knockouts, the FOS (Barrangou et al., 2003), lac (Russell and Klaenhammer, 2001a) and tre (Duong et al., 2006) operons were selected to serve as the basis for our carbohydrate‐inducible vectors.
β‐Glucuronidase reporter assays show the inducible vectors to be inducible and regulated by carbohydrates in both L. acidophilus and L. gasseri. In all strains, the inducing carbohydrates elicited the highest GUS activity, while glucose repressed GUS activity by a factor of 4 or more. GUS activity of pTRK890 (Ptre) showed the least difference in activity between trehalose and glucose in both organisms. This was in agreement with the microarray expression data in L. acidophilus and is most likely due to trehalose present in yeast extract used to prepare media. GUS activity of Lactobacillus cultures harbouring pTRK892 (Ppgm) was not found to be significantly different when exposed to MRS or different carbohydrates but, in all cases, was constitutively highly expressed at over 10‐fold over levels observed with inducible vectors. The results showed that the expression systems maintain regulation of expression similar to the native genes positioned in the L. acidophilus genome. Modified Lactobacillus GusA3 enzyme was previously shown to offer superior performance and reproducibility in acidifying conditions over native Lactobacillus GusA enzyme (Callanan et al., 2007).
Additional benefits may be gained by using these specific carbohydrates. Fructooligosaccharides are prebiotic compounds thought to promote the growth of probiotic microorganisms in the GI tract (Gibson and Roberfroid, 1995). Fructooligosaccharides remain undigested by the GI tract and are available to the lactobacilli in the lower GI tract. By using a FOS‐inducible expression vector, it may be possible to target the expression of desired proteins in the lower GI tract, where the undigested FOS are more prevalent. This may be useful for the use of lactobacilli as live vaccine or biotherapeutic delivery vectors. Trehalose is a glucose disaccharide commonly used as a cryoprotectant. It has been shown to confer protection from freezing, heat and desiccation stress. This conferred protection may benefit the end‐user of a trehalose‐inducible expression system. Lactose is a very inexpensive and readily available carbohydrate and the primary carbohydrate source available in milk, potentially facilitating the use of lactose‐inducible vectors in dairy applications.
Previous efforts at developing a FOS‐inducible expression vector in our group found expression from a pGK12‐derived vector using only the FOS promoter to be very leaky (Miller et al., 2004). This was possibly due to repressor titration (Williams et al., 1998) or transcriptional readthrough from promoters outside the expression cassette (Gibson et al., 1987). In order to avoid this, the respective repressors from each operon and Rho‐independent terminators were included in the vectors constructed for this study.
Gosalbes and colleagues (2001) previously described an expression vector based on the lactose operon in Lactobacillus casei ATCC 393. This vector was based on pIAβ5, containing a p15A replicon from pACYC184, and functioned only in L. casei. The expression systems described herein are based on the pGK12 shuttle replicon and contain determinants, which allow replication in both Escherichia coli and most lactic acid bacteria (Kok et al., 1984). This makes it possible to build constructs in E. coli for eventual transformation into lactobacilli and, likely, other lactic acid bacteria. While we have only tested this system in L. acidophilus and L. gasseri, we expect that this series of vectors will be useful in the wide host range available to pGK12.
Absorption of dietary oxalate in the colon contributes to hyperoxaluria (Balaji and Menon, 1997) and increases the risk of urinary calcium oxalate stone formation. Because humans lack enzymes for the breakdown of oxalate, degradation by intestinal bacteria is important to maintaining the oxalate homeostasis (Stewart et al., 2004). The use of oxalate degrading lactic acid bacteria has been demonstrated as a potential biotherapeutic route for the treatment of oxalate‐related disorders in humans (Campieri et al., 2001; Lieske et al., 2005). Lactobacillus acidophilus and L. gasseri have been previously identified as oxalate degrading probiotic lactobacilli (Azcarate‐Peril et al., 2006; Lewanika et al., 2007). To demonstrate the utility of constructs developed in this study, we investigated the possibility of improving the oxalate degrading activity of these organisms by overexpression of the L. acidophilus oxalate gene cluster. In this study the promoter for the constitutively highly expressed gene pgm was used in pTRK928 to drive expression of the oxalate operon. Plasmid pTRK928 was able significantly improve the oxalate degradation activity of L. gasseri ATCC 33323, suggesting that this strategy may be effective in other strains or may be used to add oxalate degrading activity to organisms unable to degrade oxalate. Additionally, the overexpression of oxc and frc using pTRK928 was able to return the oxalate degradation activity of the L. acidophilus frc deletion mutant (NCK 1724) to the level of the wild‐type parent strain. This illustrates the potential use of the pTRK882 expression system in complementation studies.
In this work, a series of expression vectors designed for use in lactobacilli are described. The GUS reporter gene results showed the carbohydrate‐inducible expression systems to be highly inducible by their respective carbohydrates, while being significantly repressed by glucose in both L. acidophilus and L. gasseri. Additionally, the constitutive expression vector was found to produce relatively high amounts of GUS activity across all conditions. Although only tested in L. acidophilus and L. gasseri, we expect these vectors to have the same broad host range as pGK12. These expression vectors provide alternative and useful tools for overexpression of desired proteins and enzymes in lactobacilli and potentially other members of the lactic acid bacteria. Additionally, we have shown the potential of these vectors for use in genetic complementation studies and also for expression of biotherapeutic proteins (Mohamadzadeh et al., 2009). Additional work is now in progress to exploit these new vectors for improvement of probiotic functionalities and in vivo applications.
Experimental procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. Liquid cultures of E. coli were propagated at 37°C in Luria–Bertani broth (Difco, Detroit, MI, USA) with shaking, while Lactobacillus cultures were propagated statically at 37°C in deMan, Rogosa and Sharpe (MRS) broth (Difco). Solid media was prepared with the addition of 1.5% agar (%w/v) (Difco). When appropriate, erythromycin (Fisher Scientific, Fair Lawn, NJ, USA) was added as follows: 150 µg ml−1 for E. coli and 5 µg ml−1 for Lactobacillus.
For induction experiments, Lactobacillus cultures were grown in MRS, harvested and resuspended in semi‐synthetic medium (SSM) (Barrangou et al., 2003) supplemented with 1% (w/v) of the appropriate carbohydrate. The carbohydrates added to the SSM include glucose (Fisher), fructose (Sigma, St Louis, MO, USA), Raftilose P95 (FOS) (Orafti, Tienen, Belgium), lactose (Sigma) and trehalose (Sigma).
DNA isolation and transformation
Lactobacillus acidophilus genomic DNA was isolated according to the method of Walker and Klaenhammer (1994). Escherichia coli plasmid DNA was isolated using the QIAprep Spin Miniprep (Qiagen, Valencia, CA, USA) kit. All DNA manipulations were performed according to standard procedures (Ausubel et al., 2001). Electrocompetent E. coli MC‐1061 cells were prepared and transformed according to standard protocols (Ausubel et al., 2001). Electrocompetent Lactobacillus cells were prepared as described by Walker et al. (Luchansky et al., 1991; Walker et al., 1996). Electrotransformation was performed using a Gene‐Pulser (Bio‐Rad, Hercules, CA, USA).
Construction of expression vectors
Plasmids used or constructed appear in Table 2. Plasmids were constructed using standard molecular cloning techniques (Ausubel et al., 2001). Restriction enzymes, T4 Ligase, taq DNA polymerase and expand high fidelity polymerase chain reaction (PCR) system polymerase were purchased from Roche Molecular Biochemicals (Indianapolis, IN, USA). DNA fragments were purified from agarose gels using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Orange, CA, USA). All PCRs were performed according to the manufacturer's instructions. PCR primers were designed using Clone Manager 9 (SciEd) and synthesized by Integrated DNA Technologies, Incorporated (Coralville, IA, USA) with restriction sites designed at the 5′ end of the primers to facilitate cloning when appropriate (Table 3). PCR products were purified using the QIAquick PCR purification kit (Qiagen). DNA sequencing was performed by Davis Sequencing (Davis, CA, USA).
Table 3.
PCR primers.
| Primer | Sequence (5′– 3′)* | Application |
|---|---|---|
| TD_pGK12Xba | ATGCTCTAGATTCAGGAATTGTCAGATAGG | Inverse PCR of pGK12, XbaI site |
| TD_pGK12Bgl | ATGCAGATCTACTAATGGGTGCTTTAGTTG | Inverse PCR of pGK12, BglII site |
| TD_Term908F | ATGCTCTAGATAATTCTATTGCTGAAACTG | Cloning of Term908 |
| TD_Term908R | ATGCTCTAGAAGCTTATACACTGATAATAAC | Cloning of Term908 |
| TD_Term667F | ATGCAGATCTTATTAAGATTACCGTTATCC | Cloning of Term667 |
| TD_Term667R | ATGCAGATCTCTAATAAATGGCGTAATTG | Cloning of Term667 |
| TD_PFOSE | ATGCGAATTCATTACTACAGCCAGTTAGTG | Amplification of PFOS/msmR fragment |
| TD_PFOSN | ATGCCCATGGTTGATGAATAAGGTGAAGAAAG | Amplification of PFOS/msmR fragment |
| TD_PLACE | ATGCGAATTCTTATGTTTGCTTGCATAC | Amplification of Plac/lacR fragment |
| TD_PLACN | ATGCCCATGGATACTCTCGATACTTTATTAG | Amplification of Plac/lacR fragment |
| TD_PTREE | ATGCGAATTCCACTTATTGGTGCAACAAC | Amplification of Ptre/treR fragment |
| TD_PTREN | ATGCCCATGGATCGCCATTTGAATCATAG | Amplification of Ptre/treR fragment |
| TD_PLA0185E | ATGCGAATTCAGCCTTCTTAGCTTCTTCAAC | Amplification of Ppgm fragment |
| TD_PLA0185N | ATGCCCATGGTGCGACAAGTAATAAACTAAAC | Amplification of Ppgm fragment |
| TD_OXF | ATGCGAATTCTCGTGGTTGATACATCACTC | Amplification of Lba 0395‐Lba0396 |
| TD_OXR | ATGCGCGGCCGCTTGGGTTCAGTCATTATGAC | Amplification of Lba 0395‐Lba0396 |
Restriction enzyme sites appear underlined.
Plasmid pTRK846 was derived from pGK12 (Kok et al., 1984) by ligating an inverse PCR product lacking the chloramphenicol resistance (Cmr) marker to the multiple cloning site (MCS) from pORI28. Plasmid pTRK847 was designed as the platform for the development of the expression vectors containing the origin of replication and erythromycin resistance (Err) marker from pGK12, an MCS and two transcriptional terminators flanking the expression region. The terminators (Table 1) were inserted by ligating PCR products from primers TD_Term908F/R and TD_Term667F/R (Table 3) into the XbaI and BglII sites of the MCS respectively. Plasmids pTRK848, pTRK849 and pTRK850 contain the promoter and repressor from the L. acidophilus FOS (LBA0500‐0507), lactose (LBA1465‐1469) and trehalose (LBA1012‐1014) operons. Also, pTRK882 contains the promoter for pgm (LBA0185) encoding phosphoglycerate mutase. These elements were obtained by PCR amplification and subsequently inserted into pTRK847 using restriction sites (see Table 2 for sources and Table 3 for primers).
To investigate expression from each promoter the gusA3 gene (Callanan et al., 2007) encoding a modified β‐glucuronidase (GusA3) was cloned into the expression region of each vector using the EcoRI and NotI sites in the MCS. In order to express the L. acidophilus oxalate gene operon (LBA0396‐LBA0395) (Fig. 8A), primers OXF and OXR (Table 3) were used to amplify a 3.2 kb DNA fragment containing the L. acidophilus oxalate gene operon. This PCR product was cloned into pTRK882 using the EcoRI and NotI restriction sites. This plasmid appears in Fig. 8B.
GUS assays
β‐Glucaronidase activity in CFEs was measured by the hydrolysis of 4‐methyl‐umbelliferyl‐β‐d‐glucuronide (MUG) (Sigma). Cultures were grown to mid‐log phase (A600 = 0.4–0.6) in MRS with 5 µg ml−1 Er, harvested by centrifugation at room temperature, washed with either MRS or SSM + 1% carbohydrate as indicated, resuspended in an equal volume of either MRS or SSM with 1% carbohydrate, and incubated at 37°C statically for 1–3 h. Cell‐free extracts were prepared as previously described (Russell and Klaenhammer, 2001b). Protein concentrations were determined using the Sigma 96‐well Assay (Sigma). For each assay, CFEs were warmed to 37°C, and 200 µl of CFE was added to 800 µl GUS buffer (100 mM sodium phosphate, 2.5 mM EDTA, 1.0 mM MUG, pH 6.0). After incubation at 37°C for 5 min, 1 ml of 0.2 M Na2CO3 was added. Activity is reported as picomoles of 4‐methylumbelliferone liberated per minute per milligram of protein.
Oxalate degradation
Oxalate degradation was measured in vitro as described previously with the following modifications (Azcarate‐Peril et al., 2006). Lactobacillus cultures were transferred three times in MRS with 0.01% (%w/v) ammonium oxalate (Fisher). Cultures were then inoculated at 1% into MRS with 0.01% ammonium oxalate and grown to OD600 = 0.5,centrifuged, and resuspended in MRS with 0.1% ammonium oxalate. Cultures were incubated statically at 37°C and samples were taken at 0, 24, 48, 72 and 96 h, centrifuged, neutralized to obtain pH values of 5–7 (according to the manufacturer's instructions) with 1 N sodium hydroxide, and stored at −20°C. The oxalate concentrations in the supernatants were determined using a diagnostic oxalate kit (Trinity Biotech, County Wicklow, Ireland).
Acknowledgments
This study was partially sponsored by the North Carolina Dairy Foundation, Danisco, Dairy Management Inc., and The Southeast Dairy Foods Research Center. Tri Duong was supported by a National Science Foundation Integrated Graduate Education and Research Traineeship in Functional Genomics and a North Carolina State University Genomics Fellowship. The authors would like to thank Rosemary Sanozky‐Dawes, Evelyn Durmaz and Erika Pfeiler for insightful discussions and technical assistance.
References
- Altermann E., Russell W.M., Azcarate‐Peril M.A., Barrangou R., Buck B.L., McAuliffe O. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. John Wiley and Sons; 2001. [Google Scholar]
- Axelsson L., Lindstad G., Naterstad K. Development of an inducible gene expression system for Lactobacillus sakei. Lett Appl Microbiol. 2003;37:115–120. doi: 10.1046/j.1472-765x.2003.01360.x. [DOI] [PubMed] [Google Scholar]
- Azcarate‐Peril M.A., McAuliffe O., Altermann E., Lick S., Russell W.M., Klaenhammer T.R. Microarray analysis of a two‐component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl Environ Microbiol. 2005;71:5794–5804. doi: 10.1128/AEM.71.10.5794-5804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcarate‐Peril M.A., Bruno‐Barcena J.M., Hassan H.M., Klaenhammer T.R. Transcriptional and functional analysis of oxalyl‐coenzyme A (CoA) decarboxylase and formyl‐CoA transferase genes from Lactobacillus acidophilus. Appl Environ Microbiol. 2006;72:1891–1899. doi: 10.1128/AEM.72.3.1891-1899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji K.C., Menon M. Mechanism of stone formation. Urol Clin North Am. 1997;24:1–11. doi: 10.1016/s0094-0143(05)70350-5. [DOI] [PubMed] [Google Scholar]
- Barefoot S.F., Klaenhammer T.R. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983;45:1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Altermann E., Hutkins R., Cano R., Klaenhammer T.R. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc Natl Acad Sci USA. 2003;100:8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Azcarate‐Peril M.A., Duong T., Conners S.B., Kelly R.M., Klaenhammer T.R. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci USA. 2006;103:3816–3821. doi: 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callanan M.J., Russell W.M., Klaenhammer T.R. Modification of Lactobacillus beta‐glucuronidase activity by random mutagenesis. Gene. 2007;389:122–127. doi: 10.1016/j.gene.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Campieri C., Campieri M., Bertuzzi V., Swennen E., Matteuzzi D., Stefoni S. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 2001;60:1097–1105. doi: 10.1046/j.1523-1755.2001.0600031097.x. et al. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J., Cohen S.N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Duong T., Barrangou R., Russell W.M., Klaenhammer T.R. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2006;72:1218–1225. doi: 10.1128/AEM.72.2.1218-1225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M.D., Khalak H.G., White O., Smith H.O., Salzberg S.L. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- Gibson G.R., Roberfroid M.B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- Gibson T.J., Coulson A.R., Sulston J.E., Little P.F. Lorist2, a cosmid with transcriptional terminators insulating vector genes from interference by promoters within the insert: effect on DNA yield and cloned insert frequency. Gene. 1987;53:275–281. doi: 10.1016/0378-1119(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Gosalbes M.J., Perez‐Arellano I., Esteban C.D., Galan J.L., Perez‐Martinez G. Use of lac regulatory elements for gene expression in Lactobacillus casei. Lait. 2001;81:29–35. [Google Scholar]
- Kleeman E.G., Klaenhammer T.R. Adherence of Lactobacillus species to human fetal intestinal cells. J Dairy Sci. 1982;65:2063–2069. doi: 10.3168/jds.S0022-0302(82)82462-4. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Beerthuyzen M.M., Vaughan E.E., de Vos W.M., Kuipers O.P. Controlled gene expression systems for lactic acid bacteria: transferable nisin‐inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., van der Vossen J.M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Buist G., Haandrikman A., Kok J., Venema G., Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewanika T.R., Reid S.J., Abratt V.R., Macfarlane G.T., Macfarlane S. Lactobacillus gasseri Gasser AM63(T) degrades oxalate in a multistage continuous culture simulator of the human colonic microbiota. FEMS Microbiol Ecol. 2007;61:110–120. doi: 10.1111/j.1574-6941.2007.00327.x. [DOI] [PubMed] [Google Scholar]
- Lieske J.C., Goldfarb D.S., De Simone C., Regnier C. Use of a probiotic to decrease enteric hyperoxaluria. Kidney Int. 2005;68:1244–1249. doi: 10.1111/j.1523-1755.2005.00520.x. [DOI] [PubMed] [Google Scholar]
- Luchansky J.B., Tennant M.C., Klaenhammer T.R. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J Dairy Sci. 1991;74:3293–3302. doi: 10.3168/jds.S0022-0302(91)78515-9. [DOI] [PubMed] [Google Scholar]
- Mathiesen G., Sorvig E., Blatny J., Naterstad K., Axelsson L., Eijsink V.G. High‐level gene expression in Lactobacillus plantarum using a pheromone‐regulated bacteriocin promoter. Lett Appl Microbiol. 2004;39:137–143. doi: 10.1111/j.1472-765X.2004.01551.x. [DOI] [PubMed] [Google Scholar]
- Miller M., Barrangou R., Austdal I., Cargile B., Callanan M., Klaenhammer T.R. 2004. , and ) Development of a gene expression system inducible by carbohydrates in Lactobacillus acidophilus. Institute of Food Technologists Annual Meeting, Las Vegas, NV: July 1216. Abstract 99A‐27.
- Mohamadzadeh M., Duong T., Sandwick S.J., Hoover T., Klaenhammer T.R. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc Natl Acad Sci USA. 2009;106:4331–4336. doi: 10.1073/pnas.0900029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W.M., Klaenhammer T.R. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl Environ Microbiol. 2001a;67:4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W.M., Klaenhammer T.R. Identification and cloning of gusA, encoding a new beta‐glucuronidase from Lactobacillus gasseri ADH. Appl Environ Microbiol. 2001b;67:1253–1261. doi: 10.1128/AEM.67.3.1253-1261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruyter P.G., Kuipers O.P., de Vos W.M. Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.S., Duncan S.H., Cave D.R. Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol Lett. 2004;230:1–7. doi: 10.1016/S0378-1097(03)00864-4. [DOI] [PubMed] [Google Scholar]
- Walker D.C., Klaenhammer T.R. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J Bacteriol. 1994;176:5330–5340. doi: 10.1128/jb.176.17.5330-5340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.C., Aoyama K., Klaenhammer T.R. Electrotransformation of lactobacillus acidophilus group A1. FEMS Microbiol Lett. 1996;138:233–237. doi: 10.1111/j.1574-6968.1996.tb08163.x. [DOI] [PubMed] [Google Scholar]
- Wells J.M., Robinson K., Chamberlain L.M., Schofield K.M., Le Page R.W. Lactic acid bacteria as vaccine delivery vehicles. Antonie Van Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]
- Williams S.G., Cranenburgh R.M., Weiss A.M., Wrighton C.J., Sherratt D.J., Hanak J.A. Repressor titration: a novel system for selection and stable maintenance of recombinant plasmids. Nucleic Acids Res. 1998;26:2120–2124. doi: 10.1093/nar/26.9.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.M., Lin C.F., Chang Y.C., Chung T.C. Construction and characterization of nisin‐controlled expression vectors for use in Lactobacillus reuteri. Biosci Biotechnol Biochem. 2006;70:757–767. doi: 10.1271/bbb.70.757. [DOI] [PubMed] [Google Scholar]


