Summary
Nisin is the prototypical example of the lantibiotic family of antimicrobial peptides and has been employed as a food preservative for over half a century. It has also attracted attention due to its potency against a number of multidrug‐resistant clinical pathogens. Nisin A is the originally isolated form of Nisin and a further five natural variants have been described which differ by up to 10 amino acids (of 34 in total in Nisin A). Nisins A, Z, F and Q are produced by Lactococcus lactis, while Nisins U and U2 are produced by Streptococcus sp. In this study we bioengineered the nisA gene of a Nisin A producer to generate genes encoding Nisins Z, F, Q, U and U2. We determined that while active Nisin Z, F and Q can be produced against this genetic background, active forms of Nisin U and U2 are not generated. Minimum inhibitory concentration studies with Nisin A, Z, F and Q variants against a series of different clinically significant pathogens establish differences in specific activities against selected targets. Nisin F was most impressive, being the most active, or one of the most active, against the MRSA strain ST 525, EC 676, EC 725, VISA 22900, VISA 22781, hVISA 35197, Staphylococcus aureus 8325‐4 and L. lactis HP. Nisin Z was most active against ST 299, hVISA 32683 and, together with Nisin F, HP but had contrastingly poor activity against ST 525, EC 676 and 8325‐4. Nisin F, Q and A exhibited similar potency against VISA 22900. This was the only target against which Nisin Q and Nisin A were among the most active variants.
Introduction
Nisin is an antimicrobial peptide which belongs to the lantibiotic class of bacteriocins (Class I) and has been employed as a food preservative in over 50 countries (Cotter et al., 2005a). Lantibiotics are characterized by the presence of unusual amino acid residues, including the lanthionines which give these peptides their name (Guder et al., 2000; Chatterjee et al., 2005; Cotter et al., 2005a; Willey and van der Donk, 2007). Lantibiotics, including nisin, can have multiple mechanisms of action facilitated through the binding of lipid II and insertion into bacterial membranes (Brotz et al., 1998; Breukink et al., 1999; Hasper et al., 2006). While Nisin A was first discovered in 1928 in fermenting milk cultures (Rogers and Whittier, 1928), five other naturally occurring variants of Nisin have been described. These are Nisin Z, F and Q, which like Nisin A are produced by Lactococcus lactis, and Nisin U and U2 which are produced by Streptococcus sp. There also exist a number of other more distantly related peptides within the Nisin subgroup of lantibiotics, such as subtilin (Jansen and Hirschmann, 1944), ericin A and ericin S (Stein et al., 2002; Piper et al., 2009a), but which are not regarded as Nisin variants. Nisin Z, first isolated from L. lactis NIZO 22186 from a dairy product, differs from Nisin A by just one amino acid in the final active peptide, His27Asn (Mulders et al., 1991) (Fig. 1). Nisin F is produced by L. lactis F10 isolated from a freshwater catfish in South Africa, and it differs from Nisin A with respect to two amino acid residues, His27Asn (as in Nisin Z) and Ile30Val (de Kwaadsteniet et al., 2008) (Fig. 1). Nisin Q, produced by L. lactis strain 61‐14 isolated from a river in Japan, contains both of the substitutions observed in Nisin F as well as two additional variations, Ala15Val and Met21Leu (Fukao et al., 2008) (Fig. 1). Two more distantly related peptides, Nisin U and U2, are produced by Streptococcus uberis 42 (Nisin U) and Streptococcus agalactiae D536 (Nisin U2), and differ from Nisin A by 9 and 10 amino acids respectively (Wirawan et al., 2006) (Fig. 1). These peptides are also three amino acid residues shorter than the other Nisin variants (Rollema et al., 1996) (Fig. 1). In addition to the genes which encode these structural peptides (and their accompanying leaders), Nisin production also requires the presence of genes required for transport (nisT; Nisin A designations are employed), leader cleavage (nisP), post‐translational modification (nisB and nisC), regulation (nisRK) and immunity (self‐protection; nisI and nisEFG) (Guder et al., 2000). While these genes are present in producers of all variants sequenced (the Nisin F and U2 gene clusters have not been sequenced), they can differ in gene order, as is the case with Nisin U (Wirawan et al., 2006) (Fig. 2). The percentage similarity between the individual biosynthetic, regulatory and immunity proteins produced also varies (Fig. 2), with the Nisin U genes being most distantly related to those associated with Nisin A. In recent years it has been shown that Nisin biosynthetic proteins can be harnessed, both in vivo and in vitro. This technology has facilitated the incorporation of lantibiotic‐associated post‐translational modifications into a range of unrelated peptides (Kuipers et al., 2006; Kluskens et al., 2009; Kuipers et al., 2009; Majchrzykiewicz et al., 2010; for review see Moll et al., 2010). In addition to the long established use of Nisin as a food preservative, the high potency and multiple mechanisms of action have also been the focus of studies designed around using Nisin in clinical or veterinary applications against drug‐resistant pathogens. The efficacy of Nisin A, and to a much lesser extent Nisins Z and F, against methicillin‐resistant Staphylococcus aureus (MRSA), vancomycin‐resistant enterococci (VRE) and (heterogeneous) vancomycin intermediate S. aureus[(h)VISA] has been tested (Severina et al., 1998; Giacometti et al., 2000; Morency et al., 2001; Brumfitt et al., 2002; Kuwano et al., 2005; de Kwaadsteniet et al., 2008; De Kwaadsteniet et al., 2009; Piper et al., 2009b). In this study, we use a bioengineering approach to convert a Nisin A‐producing strain into a producer of Nisins F, Q and Z. This bank of bioengineered strains provided us with the opportunity to directly compare the antimicrobial activity of these four natural Nisin variants.
Figure 1.
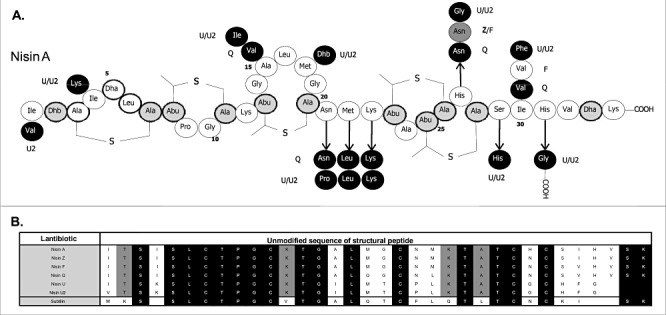
Comparison of natural Nisin variants. A. Structure of Nisin A with modified residues indicated by grey circles/black text. Residues in Nisin F, Q, Z, U and U2 that differ from Nisin A are indicated by a white circle/grey text (Nisin F), dark grey circle/white text (Nisin Q), a dark grey circle/black text (Nisin Z) and black circles/white text (Nisin U/U2). B. Alignment of unmodified Nisin variant and, for comparison purposes, subtilin propeptides. Residues that are conserved across all peptides are indicated by a black box and white text. Those conserved among the Nisin variants are indicated by a grey box and black text.
Figure 2.
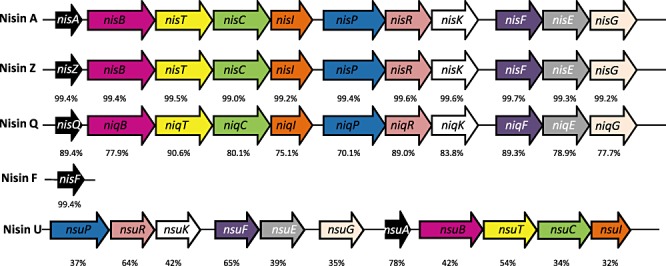
Nisin gene clusters. Analysis of the various Nisin loci illustrates that Nisin A, Z and Q all contain 11 open reading frames (ORFs) arranged in an identical manner on the chromosome, i.e. nisA(Z)(Q) BTCIPRKFEG (Engelke et al., 1994; Immonen et al., 1995; Yoneyama et al., 2008). Nisin U also has 11 ORFs; however, they are arranged differently within the cluster (Wirawan et al., 2006). Homologous genes are represented by arrows of the same colour (the Nisin F and Nisin U2 gene cluster sequences are unavailable). Numbers underneath arrows show the % identity of the gene relative to its Nisin A‐associated counterpart [pairwise alignment of protein sequences was carried out using Needleman–Wunsch global algorithms accessed via the European Bioinformatics (EBI) web server].
Results
Bioengineering of nisA
Lactococcus lactis NZ9800 is a non‐producing derivative of the Nisin A‐producing strain L. lactis NZ9700 (Kuipers et al., 1995) in which there is a deletion within the nisA structural gene. Production of Nisin A by NZ9800 can be restored through the introduction of nisA in trans on a vector such as pDF05, i.e. pCI372‐nisA (Field et al., 2008). Here a number of new pCI372 derivatives were generated in the hope of providing a source of the peptides corresponding to the natural Nisin variants Nisin F, Q, Z, U and U2, while also determining if the Nisin A production machinery can, without manipulation, successfully be employed to produce these variants. As Nisin Z and Nisin F differ from Nisin A by one and two amino acids, respectively, a PCR‐based strategy was employed to alter the relevant codons in the pCI372‐nisA. However, as Nisin Q, U and U2 differ more dramatically from Nisin A, an alternative gene synthesis‐based strategy was employed to create the relevant genes. The synthesized genes were designed such that they deviated from the nisA sequence by as little as possible, including the retention of the NisA leader. In all instances, the newly generated pCI372 derivatives, i.e. pCI372nisF, pCI372nisQ, pCI372nisZ, pCI372nisU and pCI372nisU2, were sequenced to confirm the integrity of the newly created/introduced genes and each construct was introduced into L. lactis strain NZ9800.
Production of Nisin variants
Preliminary antimicrobial assays (deferred antagonism assays) were performed with L. lactis NZ9800 containing the various pCI372 constructs. NZ9800 containing pCI372nisA has previously been shown to produce active Nisin A (Field et al., 2008), but we also observed that strains containing pCI372nisF, pCI372nisQ and pCI372nisZ all inhibited the L. lactis HP indicator strain (Fig. 3). Thus deferred antagonism bioactivity assays against L. lactis HP established that these strains produced an active antimicrobial. However, strains containing pCI372nisU or pCI372nisU2 did not inhibit this indicator, or any other of the indicator strains employed (data not shown) indicating that these strains did not produce a bacteriocin. Colony mass spectrometry detected the production of Nisins A, F, Q and Z from the relevant strains (Fig. 3); no peaks were detected which would correspond to Nisin U and U2. Additional investigations were carried out to investigate the basis for the apparent non‐production of these peptides, which could be as a consequence of an inability to auto‐induce their own production (Eichenbaum et al., 1998; Bryan et al., 2000) or the inability of the biosynthetic machinery to modify the bioengineered peptide or cleave the associated leader. These investigations employed mass spectrometric analysis of lysates of cells with and without induction with Nisin A‐containing supernatant. However, peptides of mass corresponding to those of U and U2 (modified, unmodified, with or without leader) were again not detected (data not shown). Thus the basis for the non‐production of these peptides is not apparent. We anticipate that the use of alternate expression systems, which differ more dramatically from that in a wild‐type cell (such as those used to introduce lanthionine residues into non‐nisin peptides (Rink et al., 2005; Kuipers et al., 2006; Rink et al., 2007; Kluskens et al., 2009; Kuipers et al., 2009; Majchrzykiewicz et al., 2010)), could be employed to generate Nisin U and U2. However, the inability of the Nisin A machinery in its natural form (i.e. not expressed from vectors) to generate Nisin U or Nisin U2 suggests that the classification of these peptides as true Nisin variants is debatable. It could be argued that these peptides, which unlike the other Nisin variants are not produced naturally by lactococci, should instead be regarded as members of the extended Nisin‐like peptide subgroup of lantibiotics (Cotter et al., 2005b; Piper et al., 2009a), as is the case for subtilin (Bacillus subtilis) (Jansen and Hirschmann, 1944), ericin A/S (B. subtilis) (Stein et al., 2002), epidermin (Staphylococcus epidermidis) (Schnell et al., 1988), pep 5 (S. epidermidis) (Kaletta and Entian, 1989) and gallidermin (Staphylococcus gallinarum) (Kellner et al., 1988).
Figure 3.
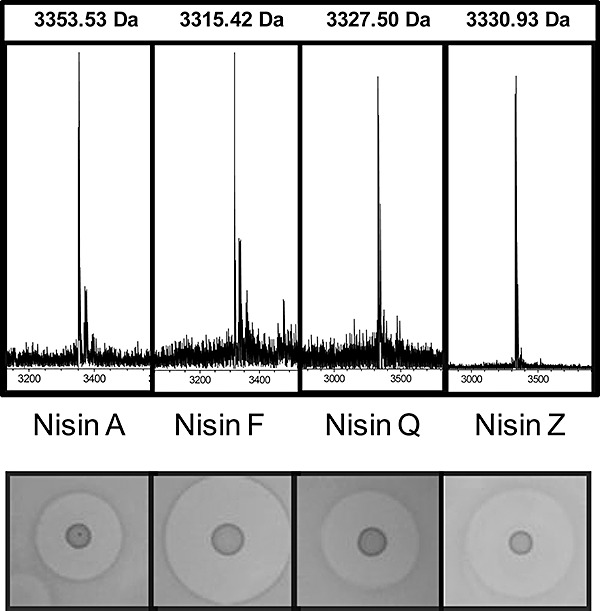
The masses of Nisin A, F, Q and Z conform with those predicted previously i.e. 3353.53 Da (Zendo et al., 2003) respectively. Bioactivity studies, in the form of deferred antagonism assays using 3 µl of spots of the respective overnight culture, were carried out with all four producers using L. lactis HP as an indicator.
Comparison of the antimicrobial activity of Nisin A, F, Q and Z
The creation of strains of NZ9800 capable of producing Nisins A, F, Q and Z, provided us with an opportunity to compare the antimicrobial activity of equimolar concentrations of purified forms of these peptides in broth against a specific set of indicators. In addition to L. lactis HP, we chose S. aureus or Enterococcus sp. isolates, including many clinical isolates. These were ST 299 (MRSA), ST 525 (MRSA), EC 676 (VRE), EC 725 (VRE), VISA 22900, VISA 22781, hVISA 35197, hVISA 32683 and the laboratory strains S. aureus Newman and 8325‐4. It was apparent from these studies that the Nisin variants differ in their specific activities against selected targets (Table 1). Nisin F was most impressive, being the most active, or one of the most active, against the MRSA strain ST 525, EC 676, EC 725, VISA 22900, VISA 22781, hVISA 35197, S. aureus 8325‐4 and L. lactis HP. Nisin Z was most active against ST 299, hVISA 32683 and, together with Nisin F, HP but had contrastingly poor activity against ST 525, EC 676 and 8325‐4. Nisin F, Q and A exhibited similar potency against VISA 22900. This was the only target against which Nisin Q and Nisin A were among the most active variants.
Table 1.
MIC values of Nisin A, F, Q and Z against representative targets.
| Nisin A (mg l−1) | Nisin F (mg l−1) | Nisin Q (mg l−1) | Nisin Z (mg l−1) | |
|---|---|---|---|---|
| ST 299 (MRSA) | 5.2 | 5.1 | 10.3 | 2.6 |
| ST 525 (MRSA) | 10.4 | 1.2 | 2.5 | 20.8 |
| EC 676 (VRE) | 20.9 | 5.1 | 10.3 | > 83.2 |
| EC 725 (VRE) | 10.4 | 2.5 | 10.3 | 5.2 |
| VISA 22900 | 41.9 | 41.4 | 41.5 | > 83.2 |
| VISA 22781 | > 83.8 | 82.8 | > 83.1 | > 83.2 |
| hVISA 35197 | 10.4 | 2.5 | 5.1 | 5.2 |
| hVISA 32683 | 2.6 | 2.5 | 5.1 | 1.3 |
| Staphylococcus aureus Newman | 10.4 | 5.1 | 5.1 | 20.8 |
| Staphylococcus aureus 8325‐4 | 20.9 | 5.1 | 10.3 | 41.6 |
| Lactococcus lactis HP | 0.13 | 0.064 | 5.1 | 0.066 |
The standard deviation in all cases is 0 reflecting identical triplicate results.
Discussion
In this study we take advantage of a peptide bioengineering strategy, which we have employed previously to generate completely novel Nisin A derivatives (Field et al., 2008), to facilitate the creation and subsequent first direct comparison of what could be argued to be the four ‘true’ Nisin variants. The description of Nisins F, Q and Z as ‘true’ variants of Nisin A stems from our observation that Nisins U and U2 cannot be produced by a system which most closely resembles that present in natural Nisin A‐producing strains. This is despite the fact that the nisU and nisU2 genes were synthesized to incorporate the Nisin A leader and were designed to employ L. lactis codon utilization preferences and Nisin A was added exogenously to ensure that induction of the Pnis promoter was not an issue. Unfortunately, our inability to detect peptides corresponding to any form of the Nisin U and U2 peptides means that the basis for this non‐production is not known. It is not anticipated that a lack of cross‐immunity [which, in the case of lantibiotics, involves an ABC transporter (LanEFG) and/or immunity protein (LanI)] is responsible as it has previously been established that Nisin U‐, Nisin Z‐ and Nisin A‐producing strains are all resistant to both Nisin A and Nisin U (Wirawan et al., 2006). Thus presumably the non‐production of Nisin U and U2 stems from an impact on the transcription of the structural peptide‐encoding genes or a failure of one or more of the biosynthetic (NisBC), leader removal (NisP) or export (NisT) proteins to recognize the Nisin U/U2‐encoding peptides. The former possibility would most likely result due to an inability of the NisRK system to recognize and auto‐induce production to detectable levels, yet the addition of exogenous Nisin A did not rectify the problem. Were the latter alternative to be true, one might expect that unmodified or partially modified peptides would be detected. However, the expression system employed may have limited our ability to detect such peptides.
There have been only a few studies which have assessed the specific activity of more than one Nisin variant against the same indicator. Although a comparison of the minimum inhibitory concentrations (MICs) of Nisin A and Z revealed that both were equally active against Micrococcus flavus DSM 1719, Streptococccus thermophilus Rs, Clostridium tyrobutyricum BZ15, L. lactis ssp. cremoris BA3, Bacillus cereus P7 and C5 and Listeria monocytogenes 13 and 669 (de Vos et al., 1993), a more recent comparison of the activities of Nisin A, Q and Z against a range of strains showed that relative potencies did vary. Although all three were equally active against Bacillus coagulans JCM 2257, B. subtilis JCM 1465 and Micrococcus luteus IFO 12708, Nisin A and Z were the most active against Bacillus circulans JCM 2504, L. lactis ATCC 19435 and Lactobacillus sakei JCM 1157. Nisin A was the most active against Pediococcus pentosaceus JCM 5885 and Leuconostoc mesenteroides JCM 6124, Nisin A and Q were the most potent against Enterococcus faecalis JCM 5803. Nisin Q was the most active against Lactobacillus plantarum ATCC 14917 while Nisin Q and Nisin Z were more active than Nisin A against Listeria innocua ATCC 33090 (Yoneyama et al., 2008). Here, in addition to focusing to a greater extent on clinical pathogens, Nisin F was also available for direct comparison. The inclusion of Nisin F proved to be particularly important as it proved to be the most active peptide against many of the targets utilized. Our initial deferred antagonism bioactivity assays against L. lactis HP (Fig. 3) established that the Nisin F producer generated the largest zone, followed by Nisin Z, Nisin Q and lastly, Nisin A. However, it was expected that these results would not provide an accurate representation of relative specific activity given the superior diffusion properties of Nisin Z in agar (de Vos et al., 1993) and potential auto‐induction related issues connected with Nisin Q production in a derivative of a producer of another Nisin variant (Yoneyama et al., 2008). For this reason we focused on MIC assays with equimolar concentrations of purified peptide. Nisin F proved to be most impressive when assayed in this way, consistently being the most, or second most, active peptide. While the activity of Nisin F was, like the other variants, greatly reduced against the VISA isolates, its activity against other clinical S. aureus (i.e. hVISA and MRSA) was impressive. Previous Nisin F studies established its in vitro efficacy against two clinical sinusitis‐associated S. aureus strains (de Kwaadsteniet et al., 2008) and in vivo activity against an MRSA strain using a Wistar rat pneumonia model (de Kwaadsteniet et al., 2009). However, here, as a consequence of the direct comparison of the activities of Nisin A, F, Q and Z against a range of S. aureus targets, it is apparent that, of these four peptides, Nisin F is most deserving of further investigation as an anti‐S. aureus chemotherapeutic option.
Experimental procedures
Bacterial strains and culture conditions
Two MRSA strains, ST 299 and ST 525, and two VRE strains, EC 676 and EC 725, were obtained from the Antibiotic Resistance Monitoring and Reference Laboratory, Health Protection Agency (HPA), Colindale, UK. Two VISA strains, VISA 22781 and VISA 22900, and two hVISA strains, hVISA 35197 and hVISA 32683, were obtained from the Bristol Centre of Antimicrobial Research and Evaluation (BCARE), UK. Two methicillin‐sensitive S. aureus (MSSA) strains were also assayed; i.e. S. aureus NCTC 8325‐4 and S. aureus Newman. Staphylococcus aureus strains were cultured in Mueller Hinton broth at 37°C with aeration. VRE strains were cultured in cation‐adjusted Mueller Hinton broth in accordance with NCCLS microbroth dilution guidelines (National Committee for Clinical Laboratory Standards, 2003) at 37°C without aeration. Lactococcus lactis HP and NZ9800 were grown at 30°C in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) without aeration.
Molecular biology techniques
pDF05 (pCI372‐nisA) (Field et al., 2008) was used as a template for PCR‐based bioengineering of nisA. Oligonucleotides (primers listed in Table 2) were designed to introduce the individual amino acid changes to generate Nisin F‐ and Nisin Z‐encoding genes (MWG Biotech, Germany) and PCR amplification using the high‐fidelity KOD polymerase (Novagen). Amplified products were treated with DpnI (Stratagene) at 37°C for 60 min in order to digest the template DNA and then purified using QIAquick PCR purification kit (Qiagen). Following transformation into Escherichia coli Top 10 cells, plasmid DNA from candidates was isolated and successful alterations were detected using a ‘check’ oligonucleotide (i.e. an oligonucleotide containing a 3′ sequence designed to amplify the bioengineered gene only) together with the 5′ Nisin A to Z for primer in the case of Nisin Z and the 5′ Nisin A to F for primer in the case of Nisin F. The integrity of the bioengineered genes was confirmed by DNA sequencing (MWG Biotech, Germany). Nisin Q‐, Nisin U‐ and Nisin U2‐encoding genes (GenBank Accession Nos GU384319, GU384320 and GU384321 respectively) were generated by gene synthesis (GENEART, Regensburg, Germany) and were cloned into pCI372 to generate pCI372nisQ, pCI372nisU and pCI372nisU2. The various pCI372 constructs were then introduced into L. lactis strain NZ9800 as described previously (Field et al., 2008) and the purification of each of the individual variants was carried out as described (Field et al., 2010).
Table 2.
PCR primers used in this study.
| Primer name | Primer sequence |
|---|---|
| Nis A to Z for | 5′‐P‐CAGCAACTTGTAACTGTAGTATTCACGTAAGCAAATAACCA‐3′ |
| Nis A to Z rev | 5′‐TGAATACTACAGTTACAAGTTGCTGTTTTCATGTTACAACC‐3′ |
| Nis A to Z check | 5′‐GAAAACAGCAACTTGTAAC‐3′ |
| Nis A to F for | 5′‐P‐CAG CAA CTT GTA ACT GTA GTG TTC ACG TAA GCA AAT AA‐3′ |
| Nis A to F rev | 5′‐GCT TAC GTG AAC ACT ACA GTT ACA AGT TGC TGT TTT CAT‐3′ |
| Nis A to F check | 5′‐AAC TTG TAA CTG TAG TGT T‐3′ |
| pCI372for | 5′‐ACCTCTCGGTTATGAGTTAG‐3′ |
| pCI372rev | 5′‐CGGGAAGCTAGAGTAAGTAG‐3′ |
| Nisin_for (EcoRI) | 5′‐AGGAATTCTAGTCTTATAAC‐3′ |
| NisinQ_rev (XbaI) | 5′‐GCTCTAGATTATTTGCTTAC‐3′ |
| NisinU/U2_rev (XbaI) | 5′‐GATCTAGATTATCCAAAATG‐3′ |
Antimicrobial activity assays
Deferred antagonism assays were performed by spotting 3 µl from an overnight culture of each variant‐producing L. lactis onto the surface of GM17 agar and growing overnight at 30°C. The resultant spots were subsequently overlaid with GM17 agar (0.75% w/v agar) seeded with the indicator strain L. lactis HP. Plates were then incubated at 30°C overnight after which time zones of clearing were measured and compared. Minimum inhibitory concentration studies were carried out in triplicate in 96‐well microtitre plates (Sarstedt) as described previously (Piper et al., 2009b). In brief, a 10 ml overnight culture of the producer strain in question was subcultured into fresh media, 10 ml of GM17 broth, and allowed to grow to log phase, i.e. OD600 0.5. A Nisin stock solution of 2.5 µM (or 10 µM in the case of more resistant strains) was prepared by weighing pure lyophilized powder and suspending in the correct volume of GM17 broth to obtain the desired concentration. Twofold serial dilutions of the peptide were added to the target strain in the microtitre plate. Assays were carried out in triplicate and biological replicates were also employed. The MIC concentration was the lowest concentration which prevented growth of the target after 16 h incubation at 37°C.
Investigations to detect the presence of peptides which are unmodified and/or uncleaved
Overnight cultures of the bioengineered strains were subject to mass spectrometric analysis following centrifugation of 1 ml volumes of the culture and lysis of the resulting pellet with glass beads and vigorous vortexing. Dilutions were made prior to mass spectrometric analysis.
To address the possibility that induction by Nisin A may be required, further investigations were carried out. Overnight cultures of the relevant strains were re‐inoculated into fresh media and allowed to reach log phase (OD600 0.5), at which point varying levels of exogenous Nisin A (supernatant obtained from an overnight of Nisin A) were added to these log‐phase cultures. After incubation for a further hour at 30°C, cell lysis was again carried out as described above before proceeding for mass spectrometric analysis. Yet again however, no peptides of relevant mass were detected.
Acknowledgments
We are grateful to Paula O' Connor and Kenneth Collins for assistance. This research was funded by a Health Research Board (HRB) Ireland Research Project Grant and by the Irish Government under the National Development Plan, through a Science Foundation Ireland Investigator award to C.H., R.P.R. and P.D.C. (06/IN.1/B98).
References
- Breukink E., Wiedemann I., van Kraaij C., Kuipers O.P., Sahl H., de Kruijff B. Use of the cell wall precursor lipid II by a pore‐forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- Brotz H., Josten M., Wiedemann I., Schneider U., Gotz F., Bierbaum G., Sahl H.G. Role of lipid‐bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- Brumfitt W., Salton M.R., Hamilton‐Miller J.M. Nisin, alone and combined with peptidoglycan‐modulating antibiotics: activity against methicillin‐resistant Staphylococcus aureus and vancomycin‐resistant enterococci. J Antimicrob Chemother. 2002;50:731–734. doi: 10.1093/jac/dkf190. [DOI] [PubMed] [Google Scholar]
- Bryan E.M., Bae T., Kleerebezem M., Dunny G.M. Improved vectors for nisin‐controlled expression in gram‐positive bacteria. Plasmid. 2000;44:183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- Chatterjee C., Paul M., Xie L., van der Donk W.A. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Hill C., Ross R.P. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005a;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Hill C., Ross R.P. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci. 2005b;6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- Eichenbaum Z., Federle M.J., Marra D., de Vos W.M., Kuipers O.P., Kleerebezem M., Scott J.R. Use of the lactococcal nisA promoter to regulate gene expression in gram‐positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke G., Gutowski‐Eckel Z., Kiesau P., Siegers K., Hammelmann M., Entian K.D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D., Connor P.M., Cotter P.D., Hill C., Ross R.P. The generation of nisin variants with enhanced activity against specific gram‐positive pathogens. Mol Microbiol. 2008;69:218–230. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- Field D., Quigley L., O'Connor P.M., Rea M.C., Daly K., Cotter P.D. Studies with bioengineered Nisin peptides highlight the broad‐spectrum potency of Nisin V. Microb Biotechnol. 2010;3:473–486. doi: 10.1111/j.1751-7915.2010.00184.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao M., Obita T., Yoneyama F., Kohda D., Zendo T., Nakayama J., Sonomoto K. Complete covalent structure of nisin Q, new natural nisin variant, containing post‐translationally modified amino acids. Biosci Biotechnol Biochem. 2008;72:1750–1755. doi: 10.1271/bbb.80066. [DOI] [PubMed] [Google Scholar]
- Giacometti A., Cirioni O., Barchiesi F., Scalise G. In‐vitro activity and killing effect of polycationic peptides on methicillin‐resistant Staphylococcus aureus and interactions with clinically used antibiotics. Diagn Microbiol Infect Dis. 2000;38:115–118. doi: 10.1016/s0732-8893(00)00175-9. [DOI] [PubMed] [Google Scholar]
- Guder A., Wiedemann I., Sahl H.G. Posttranslationally modified bacteriocins – the lantibiotics. Biopolymers. 2000;55:62–73. doi: 10.1002/1097-0282(2000)55:1<62::AID-BIP60>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Hasper H.E., Kramer N.E., Smith J.L., Hillman J.D., Zachariah C., Kuipers O.P. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. et al. [DOI] [PubMed] [Google Scholar]
- Immonen T., Ye S., Ra R., Qiao M., Paulin L., Saris P.E. The codon usage of the nisZ operon in Lactococcus lactis N8 suggests a non‐lactococcal origin of the conjugative nisin‐sucrose transposon. DNA Seq. 1995;5:203–218. doi: 10.3109/10425179509030968. [DOI] [PubMed] [Google Scholar]
- Jansen E.F., Hirschmann D.J. Subtilin, an antibacterial product of Bacillus subtilis: culturing conditions and properties. Arch Biochem. 1944;4:297–304. [Google Scholar]
- Kaletta C., Entian K.D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989;171:1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner R., Jung G., Horner T., Zahner H., Schnell N., Entian K.D., Gotz F. Gallidermin: a new lanthionine‐containing polypeptide antibiotic. Eur J Biochem. 1988;177:53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- Kluskens L.D., Nelemans S.A., Rink R., de Vries L., Meter‐Arkema A., Wang Y. Angiotensin‐(1‐7) with thioether bridge: an angiotensin‐converting enzyme‐resistant, potent angiotensin‐(1‐7) analog. J Pharmacol Exp Ther. 2009;328:849–854. doi: 10.1124/jpet.108.146431. et al. [DOI] [PubMed] [Google Scholar]
- Kuipers O.P., Beerthuyzen M.M., de Ruyter P.G., Luesink E.J., de Vos W.M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- Kuipers A., Wierenga J., Rink R., Kluskens L.D., Driessen A.J., Kuipers O.P., Moll G.N. Sec‐mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis. Appl Environ Microbiol. 2006;72:7626–7633. doi: 10.1128/AEM.01802-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers A., Rink R., Moll G.N. Translocation of a thioether‐bridged azurin peptide fragment via the sec pathway in Lactococcus lactis. Appl Environ Microbiol. 2009;75:3800–3802. doi: 10.1128/AEM.00341-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano K., Tanaka N., Shimizu T., Nagatoshi K., Nou S., Sonomoto K. Dual antibacterial mechanisms of nisin Z against Gram‐positive and Gram‐negative bacteria. Int J Antimicrob Agents. 2005;26:396–402. doi: 10.1016/j.ijantimicag.2005.08.010. [DOI] [PubMed] [Google Scholar]
- de Kwaadsteniet M., Doeschate K.T., Dicks L.M. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus. Appl Environ Microbiol. 2008;74:547–549. doi: 10.1128/AEM.01862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kwaadsteniet M., Doeschate K.T., Dicks L.M. Nisin F in the treatment of respiratory tract infections caused by Staphylococcus aureus. Lett Appl Microbiol. 2009;48:65–70. doi: 10.1111/j.1472-765X.2008.02488.x. [DOI] [PubMed] [Google Scholar]
- Majchrzykiewicz J.A., Lubelski J., Moll G.N., Kuipers A., Bijlsma J.J., Kuipers O.P., Rink R. Production of a class II two‐component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother. 2010;54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll G.N., Kuipers A., Rink R. Microbial engineering of dehydro‐amino acids and lanthionines in non‐lantibiotic peptides. Antonie Van Leeuwenhoek. 2010;97:319–333. doi: 10.1007/s10482-010-9418-4. [DOI] [PubMed] [Google Scholar]
- Morency H., Mota‐Meira M., LaPointe G., Lacroix C., Lavoie M.C. Comparison of the activity spectra against pathogens of bacterial strains producing a mutacin or a lantibiotic. Can J Microbiol. 2001;47:322–331. [PubMed] [Google Scholar]
- Mulders J.W., Boerrigter I.J., Rollema H.S., Siezen R.J., de Vos W.M. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur J Biochem. 1991;201:581–584. doi: 10.1111/j.1432-1033.1991.tb16317.x. [DOI] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards. National Committee for Clinical Laboratory Standards; 2003. [Google Scholar]
- Piper C., Cotter P.D., Ross R.P., Hill C. Discovery of medically significant lantibiotics. Curr Drug Discov Technol. 2009a;6:1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- Piper C., Draper L.A., Cotter P.D., Ross R.P., Hill C. A comparison of the activities of lacticin 3147 and nisin against drug‐resistant Staphylococcus aureus and Enterococcus species. J Antimicrob Chemo. 2009b;64:546–551. doi: 10.1093/jac/dkp221. [DOI] [PubMed] [Google Scholar]
- Rink R., Kuipers A., de Boef E., Leenhouts K.J., Driessen A.J., Moll G.N., Kuipers O.P. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry. 2005;44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- Rink R., Kluskens L.D., Kuipers A., Driessen A.J., Kuipers O.P., Moll G.N. NisC, the cyclase of the lantibiotic nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry. 2007;46:13179–13189. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- Rogers L.A., Whittier E.O. Limiting factors in the lactic fermentation. J Bacteriol. 1928;16:211–229. doi: 10.1128/jb.16.4.211-229.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollema H.S., Metzger J.W., Both P., Kuipers O.P., Siezen R.J. Structure and biological activity of chemically modified nisin A species. Eur J Biochem. 1996;241:716–722. doi: 10.1111/j.1432-1033.1996.00716.x. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K.‐D., Schneider U., Gotz F., Zahner H., Kellner R., Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide‐rings. Nature (London) 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- Severina E., Severin A., Tomasz A. Antibacterial efficacy of nisin against multidrug‐resistant Gram‐positive pathogens. J Antimicrob Chemother. 1998;41:341–347. doi: 10.1093/jac/41.3.341. [DOI] [PubMed] [Google Scholar]
- Stein T., Borchert S., Conrad B., Feesche J., Hofemeister B., Hofemeister J., Entian K.D. Two different lantibiotic‐like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J Bacteriol. 2002;184:1703–1711. doi: 10.1128/JB.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos W.M., Mulders J.W., Siezen R.J., Hugenholtz J., Kuipers O.P. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl Environ Microbiol. 1993;59:213–218. doi: 10.1128/aem.59.1.213-218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey J.M., van der Donk W.A. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Wirawan R.E., Klesse N.A., Jack R.W., Tagg J.R. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl Environ Microbiol. 2006;72:1148–1156. doi: 10.1128/AEM.72.2.1148-1156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama F., Fukao M., Zendo T., Nakayama J., Sonomoto K. Biosynthetic characterization and biochemical features of the third natural nisin variant, nisin Q, produced by Lactococcus lactis 61‐14. J Appl Microbiol. 2008;105:1982–1990. doi: 10.1111/j.1365-2672.2008.03958.x. [DOI] [PubMed] [Google Scholar]
- Zendo T., Fukao M., Ueda K., Higuchi T., Nakayama J., Sonomoto K. Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61‐14 isolated from a river in Japan. Biosci Biotechnol Biochem. 2003;67:1616–1619. doi: 10.1271/bbb.67.1616. [DOI] [PubMed] [Google Scholar]


