Summary
In this study, we describe the functional characterization of the Bifidobacterium breve UCC2003 gal locus, which is dedicated to the utilization of galactan, a plant‐derived polysaccharide. Using a combination of molecular approaches we conclude that the galA gene of B. breve UCC2003 encodes a β‐1,4‐endogalactanase producing galacto‐oligosaccharides, which are specifically internalized by an ABC transport system, encoded by galBCDE, and which are then hydrolysed to galactose moieties by a dedicated intracellular β‐galactosidase, specified by galG. The generated galactose molecules are presumed to be fed into the fructose‐6‐phosphate phosphoketolase pathway via the Leloir pathway, thereby allowing B. breve UCC2003 to use galactan as its sole carbon and energy source. In addition to these findings we demonstrate that GalR is a LacI‐type DNA‐binding protein, which not only appears to control transcription of the galCDEGR operon, but also that of the galA gene.
Introduction
It is now well established that the complex microbial communities that inhabit the gastrointestinal tract (GIT) play a vital role in maintaining gut health and homeostasis although the precise molecular mechanisms involved are as yet poorly understood (reviewed by Zoetendal et al., 2006; Marchesi and Shanahan, 2007). Bifidobacteria, which belong to the phylum Actinobacteria, constitute a significant bacterial group in the human GIT and have attracted a lot of attention as a result of their perceived positive contribution to the functionality of the GIT. The health‐promoting effects attributed to (certain strains of) bifidobacteria include enhancement of immune function, improvement of colonic integrity, reduction of incidence and duration of intestinal infections, downregulation of allergic responses and stimulation of digestion and elimination (reviewed by Turroni et al., 2009). These positive attributes have led to their inclusion in functional foods (Stanton et al., 2005).
Prebiotics are defined as ‘selectively fermented food ingredients that allow specific changes, both in the composition and/or activity in the gastrointestinal microbiota that confer benefits upon host well being and health’ (Macfarlane et al., 2008). A prebiotic may also be included in a probiotic‐containing functional food, and in such cases the synergistic combinations of pro‐ and prebiotics are termed synbiotics (Rastall et al., 2005). Carbohydrates that have been shown to exert prebiotic effects include those from whole grain wheat, fructo‐oligosaccharides, galacto‐oligosaccharides and type II arabinogalactans (reviewed by Macfarlane et al., 2008; Steed et al., 2008). Together probiotics and prebiotics share a unique role in human nutrition, largely focusing on the manipulation of populations and/or activities of the bacteria that colonize the GIT. The development of functional foods containing prebiotics and/or probiotics, which can change the composition and/or activity of the microbiota, in a predictable manner, represents a major scientific challenge for both the pharma and food industries. The recent explosion in the availability of genome sequences of gastrointestinal microbes should allow the selection of novel, perhaps more selective prebiotics and will also be pivotal in attaining a fundamental understanding of the probiotic effect (Ventura et al., 2009a)
(Arabino)galactans are an abundant, plant‐derived carbohydrate source derived from pectin. Despite daily consumption of galactan through ingestion of fruit, vegetables and cereals, plant cell wall polysaccharides have not been extensively exploited as a potential source of prebiotics. Pectin consists of ‘smooth’ regions of α‐1,4‐galacturonic acid (homogalacturonan) and ‘hairy’ regions of rhamnogalacturonan. Two types of arabinogalactan side‐chains are present in rhamnogalacturonan; type I consists of a chain of β‐1,4‐linked d‐galactopyranose linkages, while type II contains a backbone of β‐1,3‐linked d‐galactopyranose residues that can be substituted with β‐1,6‐linked d‐galactopyranose residues. Both types can furthermore be substituted with β‐1,3‐linked arabinofuranose chains (de Vries and Visser, 2001). Type I (arabino)galactan is degraded by bacteria using a combination of β‐1,4‐endogalactanase and β‐galactosidase activities, where the former enzyme cleaves within the galactan moiety of its substrate releasing d‐galacto‐oligosaccharides. Bacterial β‐1,4‐endogalactanases are reported to release mainly galactotriose and galactotetraose, while some may also release galactobiose (De Vries et al., 2002). Genes encoding β‐1,4‐galactanase activity have been characterized from Bacillus subtilis (Nakano et al., 1990), Peudomonas fluorescens (Braithwaite et al., 1997), Erwinia chrysanthemi (Delangle et al., 2007), Termotoga maritime (Yang et al., 2006) and Bifidobacterium longum (Hinz et al., 2005). Degnan and Macfarlane (1995) observed that B. longum was incapable of growth on type II larch wood arabinogalactan; however, crossfeeding of B. longum was observed when grown in co‐culture with Bacteroides thetaiotaomicron. More recently, Hinz and colleagues (2005) extensively characterized the galA gene, encoding a β‐1,4‐endogalactanase, from B. longum NCC490. Here we report on the endogalactanase‐encoding gene locus from Bifidobacterium breve UCC2003, which encodes a complete functional system for galactan utilization by this strain. Our work shows that galactan utilization by B. breve UCC2003 requires an extracellular endogalactanase, encoded by galA, to degrade galactan mainly into galacto‐trisaccharide, which is then internalized by a dedicated ABC transport system and hydrolysed to galactose by a specific β‐galactosidase encoded by galG. Furthermore, we present data that implicate the LacI‐type regulator, GalR, in the regulation of promoters upstream of the endogalactanase‐encoding gene, galA, and the first gene, galC, of the ABC transport system in a galactotriose/biose‐dependent manner.
Results
Growth of bifidobacterial strains on arabinogalactan, pectic galactan, galactan and glucose
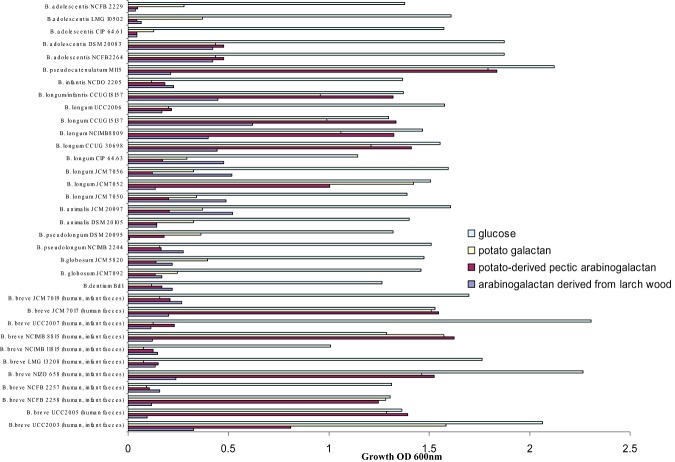
In order to establish if bifidobacteria are capable of (arabino)galactan metabolism, growth in Modified Rogosa medium supplemented with arabinogalactan derived from larch wood (backbone of β‐1,3‐linked d‐galactopyranose residues that can be substituted with β‐1,6‐linked d‐galactopyranose residues and can be further substituted with β‐1,3‐linked arabinofuranose chains), potato‐derived pectic arabinogalactan (β‐1,4‐linked d‐galactopyranose linkages substituted with β‐1,3‐linked arabinofuranose chains), potato galactan (β‐1,4‐linked d‐galactopyranose chains) or glucose was assessed for 34 bifidobacterial strains most of which are human‐derived and which represent nine bifidobacterial species. Growth assessment by measuring optical density following 24 h of anaerobic growth at 37°C revealed that all bifidobacterial strains grew well on glucose, reaching OD600 values in excess of 1.0. In contrast, none of the bifidobacterial strains tested grew well on larch arabinogalactan, while just 11 of the 34 strains tested were able to reach an OD600 higher than 1.0 when grown on potato‐derived pectic arabinogalactan or galactan as the sole carbohydrate source (Fig. 1). Interestingly, of these latter 11 strains, six belong to the B. breve species, including B. breve UCC2003 (Fig. 1). These data indicate that only certain bifidobacteria can metabolize (arabino)galactan derived from potato, and that this sugar may thus represent a selective growth substrate for such strains. Furthermore, our data show that (arabino)galactan derived from larch wood did not support growth of the bifidobacteria tested in this study, although it cannot be ruled out that in vivo intestinal communities may contain bifidobacteria that may be capable of fermenting this carbohydrate.
Figure 1.
Final optical density (OD600) values obtained following 24 h growth of various bifidobacterial strains in modified MRS containing 0.5% glucose, potato galactan, potato‐derived pectic arabinogalactan or arabinogalactan derived from larch wood as the sole carbon source. The results are mean values obtained from three separate experiments.
Genome response of B. breve UCC2003 to growth on Galactan
In order to investigate which genes may be involved in galactan metabolism in B. breve UCC2003, global gene expression was determined by microarray analysis during growth of this bifidobacterial strain on potato galactan and compared with its expression pattern when grown on ribose. Total RNA was isolated from B. breve UCC2003 cultures grown on potato galactan or ribose as sole carbohydrate source. Analysis of the DNA microarray data revealed that the expression of five adjacent genes was significantly upregulated (fold change > 15.0, P < 0.001; Table 2). These genes constitute the galactan metabolism cluster (see below) and were designated galC, galD, galE, galG, galR and galA (Fig. 2), of which the latter had previously been shown to be involved in galactan metabolism (O'Connell Motherway et al., 2009). To confirm the microarray results, quantitative RT‐PCR (qRT‐PCR) analysis was performed using primer pairs representing individual genes of the gal gene cluster (Table S1). cDNA templates were derived from RNA isolated from B. breve UCC2003 following growth on galactan or ribose. As expected, the galCDEG and galA genes were shown to be upregulated, consistent with the microarray results (Table 2).
Table 2.
Effect of potato galactan on the transcriptome of B. breve UCC2003.
| Locus tag_gene | Putative function | Galactana | QRT‐PCRb |
|---|---|---|---|
| bbr_0417_galC | Solute binding protein | 37.6 | 26.62 |
| bbr_0418_galD | Sugar permease protein | 20.0 | 18.0 |
| bbr_0419_galE | Sugar permease protein | 19.3 | 15.6 |
| bbr_0420_galG | β‐galactosidase GH 42 family | 17.8 | 15.0 |
| bbr_0421_galR | Transcriptional regulator, LacI family | 6.6 | 3.0 |
| bbr_0422_galA | Endogalactanase | 28.9 | 25.3 |
Expression ratios presented in bold have a Bayesian P‐value < 0.001 according to the Cyber‐T‐test (Long et al., 2001).
Expression ratios of selected genes quantified by QRT‐PCR. cDNA templates were derived from RNA samples of B. breve UCC2003 culture grown on ribose as a comparator.
Figure 2.
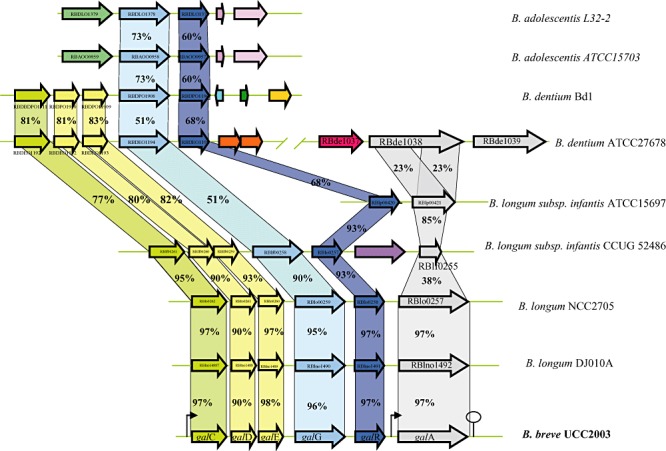
Comparison of the gal locus of B. breve UCC2003 with corresponding putative galacto‐oligosaccharides/galactan utilization loci from other bifidobacteria. Each solid arrow indicates an open reading frame. The lengths of the arrows are proportional to the length of the predicted open reading frame and the gene locus name, which is indicative of its putative function, is indicated within the arrow. Orthologs are marked with the same colour while the amino acid identity of each predicted protein is indicated as a percentage relative to its equivalent protein encoded by B. breve UCC2003. The bent arrows indicate the galc and galA promoters; the lollipop sign designates putative rho‐independent terminator region.
Genetic organization of the putative galactan utilization cluster
Our presumption, based on microarray results, that the genes upstream of galA were also involved in galactan metabolism was substantiated by their conservation among sequenced bifidobacterial genomes. The gal gene cluster (Fig. 2) contains galA, which is a clear homologue of the B. longum NCC490 galA gene, which encodes a characterized endogalactanase (Hinz et al., 2005). The B. breve UCC2003 galA gene is located downstream of a gene, galR, predicted to encode a LacI‐type transcriptional regulator and representing the presumed regulator of the galCDEGR operon and galA of B. breve UCC2003 (see below). The galC, galD and galE genes, which together are believed to specify an ABC‐type uptake system for galacto‐oligosaccharides, encode a galacto‐oligosaccharide‐binding and two permease proteins respectively. Interestingly, a gene encoding a putative ATP‐binding protein is present upstream of galC, although the array data indicate that this gene is not under galactan‐inducible control (data not shown), and it may therefore be that this gene specifies a general ATP‐binding protein involved in providing energy to multiple ABC‐type sugar uptake systems (Quentin et al., 1999; Webb et al., 2008). The galG gene is predicted to encode a putative β‐galactosidase, which belongs to the glycosyl hydrolase family GH42 (Cantarel et al., 2009), and is predicted to be responsible for hydrolysis of internalized galacto‐oligosaccharides to galactose monomers. Comparative genome analysis showed that the B. breve UCC2003 gal gene cluster is most similar to the similarly organized putative endogalactanase gene clusters of B. longum strains DJ010A and NCC2705 (Fig. 2). In B. longum ssp. infantis strains CCUG52486 and ATTCC15697 the galA gene would appear to have undergone an internal deletion with the loss of the GH53 domain, as the putative proteins encoded by blon_0440 and rblf0255 are much shorter than GalA and show similarity only to the C‐terminal putative calcium‐binding extension of GalA. The absence of a functional GalA homologue in strains CCUG52486 and ATTCC15697 is consistent with their inability to grow on galactan as sole carbohydrate source (data not shown). Interestingly and consistent with the observation that Bifidobacterium dentium Bd1 is incapable of growth on galactan (Fig. 1), analysis of the Bd1 genome sequence (Ventura et al., 2009b) established that this strain lacks a galA homologue. In contrast, B. dentium ATCC27678 can metabolize potato galactan (data not show) and as expected encodes an endogalactanase, although with a different domain organization compared with its counterparts encoded by B. breve or B. longum, as it contains two GH53 domains, where in each domain the two catalytic glutamate residues characteristic of GH53 enzymes can be identified. A similar domain organization is present in an endogalactanase encoded by Bacillus coagulans 36D1 (Copeland et al., 2010). Interestingly, the B. dentium ATCC27678 galA homologue (encoded by bde_1038) is at a distinct location in the chromosome and not located adjacent to other homologues of the B. breve UCC2003 gal locus in this strain. No obvious galA homologue or homologues encoding the presumed galCDE ABC transporter components are present in the Bifidobacterium adolescentis strains DSM20083 or L32‐2, although both strains harbour clear homologues of the β‐galactosidase‐encoding gene galG and the associated LacI‐type regulator encoding gene, galR (Fig. 2). The presence of the galA gene in B. breve UCC2003 had previously been shown to be essential for the ability of this strain to metabolize galactan (O'Connell Motherway et al., 2009). The link between the presence of a galA homologue and the ability to metabolize galactan was confirmed by comparative genome hybridization using B. breve UCC2003‐based microarrays, demonstrating that tested B. breve strains, which are either capable or incapable of growth on galactan (Fig. 1), contain or lack DNA sequences with significant identity to galA, respectively, although all these strains contain sequences that are homologous to the galCDEGR genes (A. Zomer, M. Ventura, B. Kearney, F. Turroni, M. O'Connell Motherway and D. Van Sinderen, unpubl. data).
The C‐terminal domain of GalA is not required for growth on galactan
The C‐terminal extension specified by the available galA homologues in bifidobacteria is not observed in GH53 enzymes from other bacterial strains. Hinz and colleagues (2005) have speculated that this C‐terminal extension may be a calcium‐binding domain, which may have a role in cell envelope anchoring of GalA, consistent with the presence of a possible cell wall anchoring motif (LSNTG) at the C‐terminus of GalA. To establish the role, if any, of this C‐terminal extension in galactan metabolism a galA insertion mutant was constructed in such a way that the insertion caused the separation of the GH53‐encoding domain of galA from the 3′‐end of the galA gene. The resulting mutant strain, which was designated UCC2003‐galA‐967 and which was expected to express a truncated GalA (amino acids 1 to 443) lacking the C‐terminal domain, was still capable of growth on galactan as its sole carbohydrate source thereby indicating that the C‐terminal extended structure of GalA is not necessary for the enzyme's activity, a notion which is further substantiated below.
Substrate specificity of recombinant GalA and GalG from B. breve UCC2003
In order to verify that the GH53 domain of GalA is sufficient for galactan metabolism and establish a role for the putative β‐galactosidase‐encoding gene, galG, we individually cloned the complete galA gene, a truncated version of galA, encoding just the GH53 domain‐encoding section (generating the same truncated galA as was created for the UCC2003‐galA‐967 mutant described above; for details see Experimental procedures), and galG in the nisin‐inducible expression vector pNZ8150 to generate pNZ‐galA, pNZ‐galAT and pNZ‐galG respectively (See Experimental procedures). The His10‐tagged endogalactanase GalA, truncated endogalactanase (designated as GalAT) and β‐galactosidase GalG proteins were each overexpressed and purified from the soluble cell extract fraction of Lactococcus lactis NZ9000 harbouring the recombinant plasmids pNZ‐galA, pNZ‐galAT or pNZ‐galG by means of metal chelate affinity chromatography. SDS‐PAGE analysis of GalA, GalAT and GalG revealed for each protein a single band at an apparent molecular mass of approximately 93 kDa, 44.6 kDa and 79 kDa, respectively, which is in agreement with their expected size as calculated from the recombinant galA and galG gene sequences (data not shown). The end products formed by the hydrolysis of galactan following incubation with the purified endogalactanase or truncated endogalactanase were analysed by HP‐TLC (Fig. 3). Consistent with the observations of Hinz and colleagues (2005) the results clearly demonstrate that both the endogalactanase and the C‐terminally truncated endogalactanase can liberate galacto‐oligosaccharides, predominantly galactotriose from galactan (Fig. 3, lanes 3 and 5). Upon addition of GalG to the reaction mix the galactotriose is further hydrolysed to the monosaccharide galactose (Fig. 3, lanes 4 and 6). Under the conditions tested GalG was incapable of hydrolysing lactose to any significant degree, but instead showed a preference for galactotriose/biose (Fig. 3, lanes 8 and 9). Collectively, these results demonstrate that the endogalactanase gene cluster encodes an endogalactanase for the extracellular metabolism of galactan, and that galG specifies a β‐galactosidase that cleaves β1‐4 linkages in galactotriose/biose.
Figure 3.
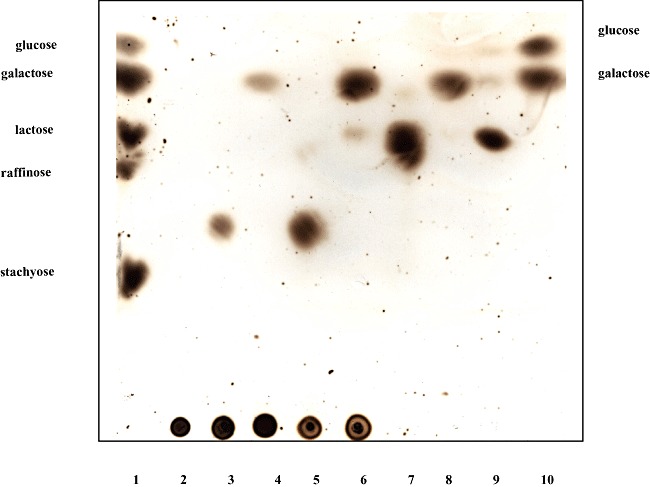
Substrate specificity of GalA, GalAT or GalG as determined by HPTLC. Lane 3–6 contain hydrolysis products of potato galactan following incubation with GalA (lane 3), GalA plus GalG (lane 4), GalAT (lane 5), GalAT plus GalG (lane 6). The hydrolysis products of galactobiose and lactose following incubation with GalG are in lanes 8 and 9 respectively. Carbohydrate standards (lanes 1 and 10) are listed to the left and right of the panel, while lanes 2 and 7 contain potato galactan and galactobiose respectively.
Disruption of the GalC and GalG encoding genes
In order to establish if disruption of particular genes from the galCDEGR gene cluster in B. breve UCC2003 would result in loss of this strain's ability to metabolize galactan, insertion mutants in the galC and galG genes were generated, resulting in strains B. breve UCC2003‐galC‐701 and UCC2003‐galG‐420 respectively (Table 1). To verify the expected galactan‐negative phenotype of these insertion mutants, strains UCC2003 (wild type), UCC2003‐galC‐701 and UCC2003‐galG‐420 were analysed for their ability to grow in mMRS supplemented with galactan or glucose (positive control) as the sole carbon source. As expected, and in contrast to the wild type, the B. breve UCC2003‐galC and UCC2003‐galG insertion mutants were shown to be incapable of growth on galactan as the sole carbon source (Fig. 4). We predict that the galC disruption in B. breve UCC2003‐galC is likely to have a polar effect on the transcription of the downstream genes of the galCDEGR operon. To demonstrate that the protein products of this ABC transporter gene cluster are uniquely necessary for the transport of the galacto‐oligosaccharides generated through hydrolysis of galactan by GalA in B. breve UCC2003, complementation experiments were performed. The β‐galactosidase‐encoding gene, galG, was expressed under the control of the p44 promoter on pCIB‐p44 in B. breve UCC2003‐galC and UCC2003‐galG (see Experimental procedures). Expression of GalG in UCC2003‐galG restored the ability of this mutant strain to grow on galactan as a sole carbohydrate source, while expression of GalG in B. breve UCC2003‐galC did not restore the ability of this strain to grow on galactan (Fig. 4). This complementation experiment provides supporting evidence that the ABC transport system, encoded by gaICDE, is the sole transporter of galacto‐oligosaccharides derived from galactan in B. breve UCC2003 and that the β‐galactosidase specified by galG is essential for the intracellular metabolism of the galacto‐trisaccharides derived from GalA activity.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains and plasmids | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| E. coli EC101 | Cloning host, repA+ kmr | Law et al. (1995) |
| E. coli EC101‐pNZ‐M.BbrII + M.BbrIII | EC101 harbouring pNZ8048 derivative containing bbrIIM and bbrIIIM. | O'Connell Motherway et al. (2009) |
| Lactococcus lactis strains | ||
| L. lactis NZ9000 | MG1363, pepN::nisRK, nisin‐inducible overexpression host | de Ruyter et al. (1996) |
| L. lactis NZ9000‐ pNZ‐galA | NZ9000 containing pNZ‐galA | This study |
| L. lactis NZ9000‐ pNZ‐galAT | NZ9000 containing pNZ‐galAT | This study |
| L. lactis NZ9000‐ pNZ‐galG | NZ9000 containing pNZ‐galG | This study |
| L. lactis NZ9000‐ pNZ‐galR | NZ9000 containing pNZ‐galR | This study |
| Bifidobacterium sp. strains | ||
| B. breve UCC 2003 | Isolate from nursling stool | Mazéet al. (2007) |
| B. breve UCC2003‐galA‐967 | pORI19‐tet‐galA‐967 insertion mutant of UCC2003 | This study |
| B. breve UCC2003‐galG‐410 | pORI19‐tet‐galG‐410 insertion mutant of UCC2003 | This study |
| B. breve UCC2003‐galC‐701 | pORI19‐tet‐galC‐701 insertion mutant of UCC2003 | This study |
| B. breve UCC 2004 | Isolate from human faeces | UCC |
| B. breve UCC 2005 | Isolate from human faeces | UCC |
| B. breve UCC 2007 | Isolate from human faeces | UCC |
| B. breve JCM 7017 | Isolate from human faeces | JCM |
| B. breve JCM 7019 | Isolate from infant faeces | JCM |
| B. breve NCFB 2257 | Isolate from infant intestine | NCFB |
| B. breve NCFB 2258 | Isolate from infant intestine | NCFB |
| B. breve NCTC 11815 | Isolate from infant intestine | NCTC |
| B. breve CCUG 43878 | Isolate from human faeces | CCUG |
| B. adolescentis CIP 64.61 | Isolate from human intestine | CIP |
| B. adolescentis DSM 20083 | Isolate from human intestine | DSM |
| B. adolescentis NCFB 2229 | Isolate from human intestine | NCFB |
| B. adolescentis NCFB 2204 | Isolate from human intestine | NCFB |
| B. adolescentis LMG 10502 | Isolate from human intestine | LMG |
| B. animalis JCM 20097 | Isolate from calf faeces | JCM |
| B. animalis DSM 20105 | Isolate from chicken faeces | DSM |
| B. bifidum NCIMB 8810 | Isolate from human intestine | NCIMB |
| B. bifidum LMG 11041 | Isolate from animal intestine | LMG |
| B. dentium Bd1 | Isolate from human dental caries | Ventura et al. (2009b) |
| B. dentium ATCC 27678 | Isolate from human dental caries | ATCC |
| B. longum JCM 7050 | Isolate from human faeces | JCM |
| B. longum JCM 7052 | Isolate from human faeces | JCM |
| B. longum JCM 7053 | Isolate from infant faeces | JCM |
| B. longum JCM 7056 | Isolate from infant faeces | JCM |
| B. longum CIP 64.63 | Isolate from infant intestine | CIP |
| B. longum CCUG 30698 | Isolate from human abdomen | CCUG |
| B. longum NCIMB 8809 | Isolate from human faeces | NCIMB |
| B. longum CCUG 15137 | Isolate from human | CCUG |
| B. longum/infantis CCUG 18157 | Isolate from human faeces | CCUG |
| B. longum subsp. infantis CCUG 52486 | Isolate from human faeces | CCUG |
| B. longum subsp. infantis ATCC 15697 | Isolate from human faeces | ATCC |
| B. infantis NCDO 2205 | Isolate from infant intestine | NCDO |
| B. pseudocatenulatum LMG 10505 | Isolate from infant faeces | LMG |
| B. pseudocatenulatum NCIMB 8811 | Isolate from infant faeces | NCIMB |
| B. pseudolongum NCIMB 2244 | Isolate from swine faeces | NCIMB |
| B. pseudolongum DSM 20095 | Isolate from chicken faeces | DSM |
| B. glodosum JCM 5820 | Isolate from animal rumen | JCM |
| B. glodosum JCM 7092 | Isolate from bovine rumen | JCM |
| B. thermophilum JCM 7027 | Isolate from swine faeces | JCM |
| Plasmids | ||
| pNZ8150 | Cmr, nisin‐inducible translational fusion vector | Mierau and Kleerebezem (2005) |
| pNZ‐galA | Cmr, pNZ8150 derivative containing translational fusion of gala‐encoding DNA fragment without signal sequence to nisin‐inducible promoter | This study |
| pNZ‐galAT | Cmr, pNZ8150 derivative containing translational fusion of truncated galA‐encoding DNA fragment (from bases 90 to 1330)to nisin‐inducible promoter | This study |
| pNZ‐galG | Cmr, pNZ8150 derivative containing translational fusion of galG‐encoding DNA fragment without signal sequence to nisin‐inducible promoter | This study |
| pNZ‐galR | Cmr, pNZ8150 derivative containing translational fusion of galR‐encoding DNA fragment without signal sequence to nisin‐inducible promoter | This study |
| pNZ44 | pNZ8048 containing constitutive p44 promoter from Lactococcal chromosome | McGrath et al. (2001) |
| pSKEM | E. coli bifidobacterial shuttle vectoe harbouring pCIBA089 rep | Cronin et al. (2007) |
| pCIB‐p44 | Complementation vector; pNZ44 where repA has been replaced with pCIBA089 rep | This study |
| pCIB‐p44‐galG | pCIB‐p44 derivative with galG transcriptionally fused to p44 promoter | This study |
| pAM5 | pBC1‐puC19‐Tcr | Alvarez‐Martín et al. (2007) |
| pORI19 | Emr, repA‐, ori+, cloning vector | Law et al. (1995) |
| pORI19‐tet‐galA | Internal 967 bp fragment of galA and tetW cloned in pORI19 | This study |
| pORI19‐tet‐galG | Internal 410 bp fragment of galG and tetW cloned in pORI19 | This study |
| pORI19‐tet‐galC | Internal 701 bp fragment of galA and tetW cloned in pORI19 | This study |
ATCC, American type culture collection; CCUG, Culture Collection of the University of Goteborg; CIP, Collection de l'Institut Pasteur; DSM, German Collection of Microorganisms and Cell Cultures; JCM, Japan Collection of Microorganisms; LMG, Belgian Co‐ordinated Collection of Microorganisms; NCDO, National Collection of Dairy Organisms; NCFB, National Collection of Food Bacteria; NCIMB, National Collection of Industrial and Marine Bacteria; NCTC, National Collection of Type Cultures; UCC, University College Cork Culture Collection.
Figure 4.
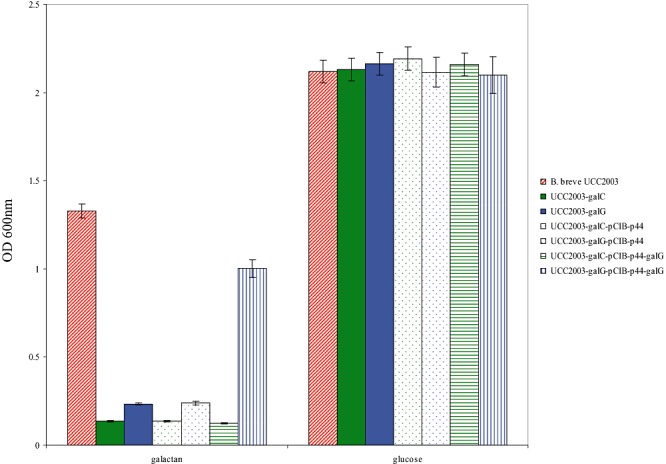
Growth profile analysis of B. breve UCC2003, UCC2003‐galC, UCC2003‐galG and mutant strains harbouring pCIB‐p44 (control) or pCIB‐p44‐galG in modified rogosa broth supplemented with potato galactan or glucose.
Identification of the transcription start site of galA and galC
From the genetic organization (Fig. 2) and the observed expression pattern (Table 2) of the gal locus it was deduced that this locus contained at least two galactan‐inducible promoters: one in front of the galC gene and one in front of the galA gene. In order to determine the transcription start site of these presumed galA and galC promoters, primer extension analysis was performed using RNA extracted from B. breve UCC2003 grown in Modified Rogosa medium containing 0.5% galactan as the sole carbohydrate source. Two extension products were identified 92 and 93 nucleotides 5′ to the predicted translational start site for the galA gene, while for the galC gene two transcription initiation sites were observed 234 and 235 bp upstream of its predicted translational start site (Fig. 5). Analysis of the galC promoter regions revealed potential promoter recognition sequences resembling consensus −10 and −35 hexamers, while for the galA promoter a clear −10 sequence could be identified, with no obvious −35 sequence present within the expected range of this −10 sequence although a potential −35 sequence is present further upstream (Fig. 5).
Figure 5.
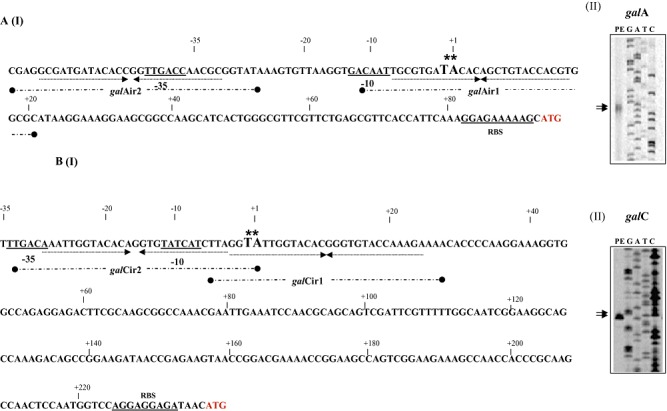
Schematic representation of the B. breve UCC2003 galA (panel A I) and galC (panel B I) promoter regions. Boldface type and underlining indicate the −10 and −35 hexamers as deduced from the primer extension results (Panel A II‐galA and B II‐galC) and ribosomal binding site (RBS); the transcriptional start sites (TSS) are indicated by asterisks; arrows under sequence in bold indicate the inverted repeat sequence that represents the GalR binding sequence. The inverted repeat sequences galAIR1 and IR2 and galCIR1 and IR2 are indicated as broken thick black line underneath the respective sequences.
GalR binds to the galC and galA promoter regions
The presence of galR, encoding a putative LacI‐type regulator within the endogalactanase gene cluster suggests that this gene is involved in the transcriptional regulation of the gal gene cluster as obvious from the microarray data (Table 2). In order to establish if GalR is capable of direct interaction with specific operators within the promoter region(s) of the gal gene cluster, we first cloned the galR gene in the nisin‐inducible vector pNZ8150 with the introduction of a His‐tag‐encoding sequence to facilitate subsequent protein purification. The purified GalR protein was then used to perform electrophoretic mobility shift assays, which clearly demonstrate that the GalR protein can complex with IRD800‐labelled DNA fragments encompassing the galA and galC promoter regions (Fig. 6B). Further delineation of the GalR recognition sequence suggested that GalR binding required an 85 bp DNA segment present within the galA and galC promoter regions (Fig. 6A). Inspection and comparison of these two 85 bp fragments revealed the presence of two inverted repeats in each fragment, which represent putative operator sequences for the GalR protein. This notion was further validated by electrophoretic mobility shift assays using 34 bp DNA fragments that just contained the predicted operator sequences (Fig. 6C). Introduction of two point mutations in the putative GalR‐binding motif (a T‐C and a G‐A mutation at positions five and six in Fig. 6C) that are highly conserved in the motif were shown to significantly reduce binding of GalR (results not shown). To investigate whether GalR interaction with its target DNA sequence is influenced by a carbohydrate effector molecule, as is known for other LacI‐type regulators (reviewed by Wilson et al., 2007; Swint‐Kruse and Matthews, 2009), several carbohydrates were tested for their effects on GalR–DNA complex formation. The results obtained clearly demonstrate that the binding ability of GalR for the g1 fragment of the galA promoter region is completely lost in the presence of galactobiose at concentrations ranging from 20 mM to 2 mM (Fig. 6D), whereas under the same experimental conditions lactose or galactose did not affect GalR binding to its target sequence (results not shown).
Figure 6.
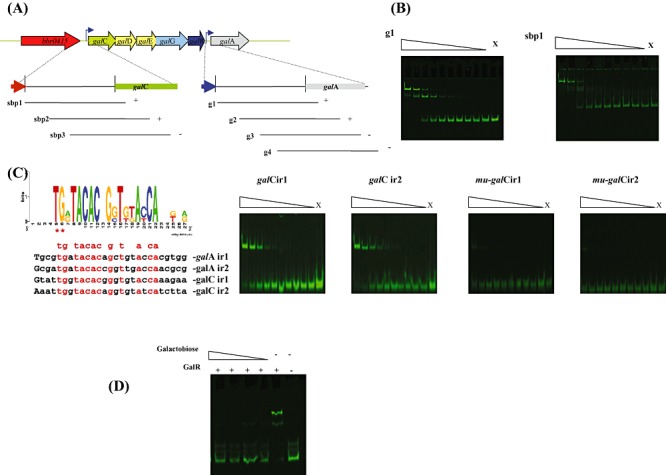
Panel A: Representation of the B. breve UCC2003 endogalactanase operon and DNA fragments used in electrophoretic mobility shift assays (EMSAs) for the galC and galA promoter resions. Plus and minus signs indicate whether or not GalR was able to bind to the particular DNA fragment respectively. Panel B: EMSA showing GalR interaction with DNA fragments encompassing fragment g1 and sbp1. Panel C: Alignment and web logo representation of predicted binding sequences of GalR together with EMSAs illustrating GalR interaction with galCIR1, galCIR2 and mutated derivatives mu‐galCIR1 and mu‐galCIR2. In each panel lane X represents a binding reaction to which no protein was added, while the remaining lanes represent binding reactions with the respective DNA probes incubated with increasing amounts of GalR (concentrations ranging from 0.04 nM 0.01 µM). Each successive lane, from right to left, corresponds to a doubling in the concentration of GalR. Panel D: EMSA showing GalR interaction with the DNA fragment g1 with the addition of galactobiose at concentrations ranging from 20–2 µM.
Discussion
Bifidobacteria rapidly colonize the intestine of infants during the first days to weeks of life. In breast‐fed infants B. breve is a frequently detected species followed by B. infantis, B. longum and B. bifidum (Sakata et al., 2005; Klaassens et al., 2009). Differential capacities for complex carbohydrate utilization have been observed for different bifidobacteria and several studies have demonstrated that bifidobacteria dedicate a significant portion of their coding capacity to the metabolism of a wide variety of carbohydrates (Schell et al., 2002; Ventura et al., 2007a,b). Over 50 different bifidobacterial carbohydrases have been described in the literature to date (reviewed by van den Broek et al., 2008). Using B. breve UCC2003 as a model to study bifidobacterial carbohydrate metabolism, we previously characterized an operon encoding a β‐fructofuranosidase (Ryan et al., 2005), an extracellular amylopullulanase that hydrolyses α‐1,4 and α‐1,6 glucosidic linkages in starch and related polysaccharides (Ryan et al., 2006; O'Connell Motherway et al., 2008), two novel α‐glucosidases exhibiting hydrolytic activities towards panose, isomaltose, isomaltotriose and trehalose (Pokusaeva et al., 2009), and a gene cluster dedicated to ribose metabolism (Pokusaeva et al., 2010). In addition, a PEP‐PTS system involved in fructose metabolism was identified and studied in this bacterium (Mazéet al., 2007).
In this study, we describe the functional characterization of a locus dedicated to the utilization of galactan by B. breve UCC2003. The galA gene of B. breve UCC2003 was previously shown to be involved in the degradation of potato galactan (O'Connell Motherway et al., 2009) and is presumed to encode an extracellular β‐1,4‐endogalactanase. The data presented here establish that galactotriose derived from this endogalactanase activity is specifically transported to the cytoplasm through an ABC transport system, which is specified by the gene products of galCDE, and that galactotriose is then degraded to galactose by a dedicated intracellular β‐galactosidase, encoded by galG. The galactose is then presumed to be fed into the fructose‐6‐phosphate phosphoketolase pathway via the Leloir pathway, thereby allowing B. breve UCC2003 to use galactan as a sole carbon and energy source.
In line with the findings described previously for B. longum NCC490 (Hinz et al., 2005), the purified GalA from B. breve UCC2003 liberates predominately galactotriose from galactan with very small amounts of galactobiose and galactose being produced. In addition, through construction of a UCC2003 galA insertion mutant that separates the GH53‐encoding domain from its C‐terminal domain, as well as purification of a truncated version of GalA (GalAT) we demonstrate that only the GH53 domain of GalA is essential for galactan metabolism to galactotriose. Hinz and colleagues (2005) hypothesized that GalA functions by means of a processive mechanism; initially, the galactan undergoes a mid chain or endo cleavage, allowing the enzyme to remain attached to one end of the cleaved galactan chain, after which it liberates galacto‐oligosaccharides in an exo‐fashion. The authors speculate that the C‐terminal extension may play a role in forming a fold over the catalytic site and maintaining galactan at the catalytic site for multiple cleavage events. This substrate entrapment strategy may provide such galactan‐metabolizing bifidobacteria a selective advantage in the highly complex and competitive environment of the gut.
To investigate the involvement of the ABC transporter encoded by galCDE and the β‐galactosidase specified by galG in galactan metabolism, insertion mutants were created in galC, the first gene of the ABC transporter, specifying the substrate‐binding protein and galG. In contrast to the parent strain UCC2003, the galC and galG mutant strains were no longer able to grow on galactan, thereby establishing that the ABC transporter and β‐galactosidase encoded by the gal locus are dedicated towards galactotriose transport and metabolism.
The deduced protein GalR is related to members of the LacI‐type regulatory protein family and our results obtained with the purified GalR protein are consistent with its role as a transcriptional regulator of the gal locus. Two GalR binding sites each were found to be present in 85 bp regions of the galA and galC promoter regions. The sequence required for recognition of GalR was shown to be a 9 bp inverted repeat, for each promoter region the two inverted repeat sequences overlap the −10 and −35 promoter recognition sequences. Most members of the LacI family bind carbohydrate or nucleoside effectors, which modulate their binding properties (Wilson et al., 2007; Swint‐Kruse and Matthews, 2009). Our results demonstrate that GalR–DNA interaction was lost in the presence of low concentrations of galactobiose, and was not affected by the presence of lactose or galactose. Because it is not commercially available, we did not test the effect of galactotriose, but we predict that, as galactotriose is the predominant product of endogalactanase activity, this trisaccharide also abolishes the GalR–DNA interaction. Therefore, it is presumed that galactotriose or galactobiose, and perhaps other β‐1,4 galacto‐oligosaccharides, are inducers of the gal operon, as they promote release of GalR from the operator sequences upstream of galC and galA. In the absence of galactotriose/biose, GalR is presumed to bind to its operator sites thereby blocking transcription of the gal genes. This simple control mechanism through negative regulation of transcription appears to be common in bifidobacteria (Parche et al., 2006; 2007; Pokusaeva et al., 2009; Ventura et al., 2009a) allowing these bacteria to quickly and efficiently respond to the presence of particular carbohydrates.
Fermentation of complex carbohydrates in the gut is assumed to be a result of the combined action of several bacteria (Xu et al., 2007). However, knowledge on how individual intestinal species of bacteria utilize complex poly‐ and oligosaccharides is limited, despite its importance for our understanding of the various metabolic activities that take place in the colon. The data presented here illustrate that the ability to metabolize the plant‐derived polysaccharide galactan is not ubiquitous among bifidobacteria or indeed B. breve strains. While certain components of the B. breve UCC2003 gal locus, i.e. those that specify the ABC transporter system, the β‐galactosidase and the GalR regulator, were present in all tested B. breve strains, a clear correlation was found between the presence of the endogalactanase gene and the (in)ability of such individual strains to grow on galactan as a sole carbohydrate source. Therefore, in the gastrointestinal environment we speculate that bifidobacterial strains lacking endogalactanase activity can still metabolize the galactotriose that is generated by extracellular endogalactanase activity of other bacteria, because of the retention of genes specifying the galactotriose ABC transporter and β‐galactosidase.
Interestingly, we observed that bifidobacterial strains that can metabolize galactan have a preference for β‐1,4‐linked galactans derived from potato (tubers), while none of the strains we examined in this study could grow to an appreciable level on arabinogalactan derived from larch wood that comprises β‐1,3‐linked galactose units. The ability of probiotic strains to ferment particular oligo‐ and polysaccharides has been the basis for selection as prebiotics. The observed preference for galactan containing predominantly β‐1,4 galactose units by the bifidobacterial strains tested here may have application in the development of targeted bifidogenic galacto‐oligosaccharides for specific probiotic strains using single or combinations of bifidobacterial β‐galactosidases. The incorporation of such galacto‐oligosaccharides in foods has potential for the development of novel functional foods or infant food formulas. Our previous studies on starch metabolism established that B. breve UCC2003 produces an extracellular starch‐degrading enzyme, ApuB, which has a preference for starch derived from potatoes (Ryan et al., 2005; O'Connell Motherway et al., 2008). It is particularly interesting to note that UCC2003 produces at least two extracellular enzymes that are dedicated to metabolize polysaccharides commonly found in potatoes, which in fact have only become the staple diet of Europeans since their introduction to Europe from Peru in the 16th century (Lekhnovitch, 1961).
Experimental procedures
The description of the Experimental procedures resides in Appendix S1 in Supporting information.
Acknowledgments
The Alimentary Pharmabiotic Centre is a research centre funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan. The authors and their work were supported by SFI (Grant no. 02/CE/B124 and 07/CE/B1368). The authors wish to acknowledge Breda Kearney, Aldert Zomer and John MacSharry for their technical assistance and sharing of unpublished data.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Oligonucleotide primers used in this study.
Appendix S1. Experimental procedures.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alvarez‐Martín P., O'Connell‐Motherway M., van Sinderen D., Mayo B. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol. 2007;76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
- Braithwaite K.L., Barna T., Spurway T.D., Charnock S.J., Black G.W., Hughes N. Evidence that galactanase A from Pseudomonas fluorescens subspecies cellulosa is a retaining family 53 glycosyl hydrolase in which E161 and E270 are the catalytic residues. Biochemistry. 1997;36:15489–15500. doi: 10.1021/bi9712394. et al. [DOI] [PubMed] [Google Scholar]
- van den Broek L.A., Hinz S.W., Beldman G., Vincken J.P., Voragen A.G. Bifidobacterium carbohydrases‐their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res. 2008;52:146–163. doi: 10.1002/mnfr.200700121. [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate‐Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland A., Lucas S., Lapidus A., Barry K., Glavina del Rio T., Dalin E. 2010. et al) Sequencing of the draft genome and assembly of Bacillus coagulans 36D1. NCBI database.
- Cronin M., Knobel M., O'Connell‐Motherway M., Fitzgerald G.F., van Sinderen D. Molecular dissection of a bifidobacterial replicon. Appl Environ Microbiol. 2007;73:7858–7866. doi: 10.1128/AEM.01630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries R.P., Visser J. Aspergillus enzymes involved in degradation of cell wall polysaccharides. Microbiol Mol Biol Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries R.P., Parenicová L., Hinz S.W., Kester H.C., Beldman G., Benen J.A., Visser J. The beta‐1,4‐endogalactanase A gene from Aspergillus niger is specifically induced on arabinose and galacturonic acid and plays an important role in the degradation of pectic hairy regions. Eur J Biochem. 2002;269:4985–4993. doi: 10.1046/j.1432-1033.2002.03199.x. [DOI] [PubMed] [Google Scholar]
- Degnan B.A., Macfarlane G.T. Arabinogalactan utilization in continuous cultures of Bifidobacterium longum: effect of co‐culture with Bacteroides thetaiotaomicron. Anaerobe. 1995;1:103–112. doi: 10.1006/anae.1995.1005. [DOI] [PubMed] [Google Scholar]
- Delangle A., Prouvost A.F., Cogez V., Bohin J.P., Lacroix J.M., Cotte‐Pattat N.H. Characterization of the Erwinia chrysanthemi Gan locus, involved in galactan catabolism. J Bacteriol. 2007;189:7053–7061. doi: 10.1128/JB.00845-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz S.W., Pastink M.I., van den Broek L.A., Vincken J.P., Voragen A.G. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl Environ Microbiol. 2005;71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassens E.S., Boesten R.J., Haarman M., Knol J., Schuren F.H., Vaughan E.E., de Vos W.M. Mixed‐species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast‐ and formula‐fed infants. Appl Environ Microbiol. 2009;75:2668–2676. doi: 10.1128/AEM.02492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Buist G., Haandrikman A., Kok J., Venema G., Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekhnovitch V.S. Introduction of the potato into western and central Europe. Nature. 1961;191:518–519. [Google Scholar]
- Long A.D., Mangalam H.J., Chan B.Y., Tolleri L., Hatfield G.W., Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coliK12. J Biol Chem. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. , and . [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Steed H., Macfarlane S. Bacterial metabolism and health‐related effects of galacto‐oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- McGrath S., Fitzgerald G.F., van Sinderen D. Improvement and Optimization of Two Engineered Phage Resistance Mechanisms in Lactococcus lactis. Appl Environ Microbiol. 2001;67:608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J., Shanahan F. The normal intestinal microbiota. Curr Opin Infect Dis. 2007;20:508–513. doi: 10.1097/QCO.0b013e3282a56a99. [DOI] [PubMed] [Google Scholar]
- Mazé A., O'Connell‐Motherway M., Fitzgerald G.F., Deutscher J., van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau I., Kleerebezem M. 10 years of the nisin‐controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- Nakano H., Takenishi S., Kitahata S., Kinugasa H., Watanabe Y. Purification and characterization of an exo‐1,4‐beta‐galactanase from a strain of Bacillus subtilis. Eur J Biochem. 1990;193:61–67. doi: 10.1111/j.1432-1033.1990.tb19304.x. [DOI] [PubMed] [Google Scholar]
- O'Connell Motherway M., Fitzgerald G.F., Neirynck S., Ryan S., Steidler L., van Sinderen D. Characterisation of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M., O'Driscoll J., Fitzgerald G.F., van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2:321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parche S., Beleut M., Rezzonico E., Jacobs D., Arigoni F., Titgemeyer F., Jankovic I. Lactose‐over‐glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J Bacteriol. 2006;188:1260–1265. doi: 10.1128/JB.188.4.1260-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parche S., Amon J., Jankovic I., Rezzonico E., Beleut M., Barutçu H. Sugar Transport Systems of Bifidobacterium longum NCC2705. J Mol Microbiol Biotechnol. 2007;12:9–19. doi: 10.1159/000096455. et al. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K., O'Connell‐Motherway M., Zomer A., Fitzgerald G.F., van Sinderen D. Characterization of two novel alpha‐glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2009;75:1135–1143. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokusaeva K., Neves A.R., Zomer A., O'Connell‐Motherway M., MacSharry J., Curley P. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y., Fichant G., Denizot F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- Rastall R.A., Gibson G.R., Gill H.S., Guarner F., Klaenhammer T.R., Pot B. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol Ecol. 2005;52:145–152. doi: 10.1016/j.femsec.2005.01.003. et al. [DOI] [PubMed] [Google Scholar]
- de Ruyter P.G., Kuipers O.P., de Vos W.M. Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.M., Fitzgerald G.F., van Sinderen D. Transcriptional regulation and characterization of a novel beta‐fructofuranosidase‐encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2005;71:3475–3482. doi: 10.1128/AEM.71.7.3475-3482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.M., Fitzgerald G.F., van Sinderen D. Screening for and identification of starch‐, amylopectin‐, and pullulan‐degrading activities in bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata S., Tonooka T., Iishizeki S., Takada M., Sakamoto M., Fukuyama M., Benno Y. Culture independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol Lett. 2005;243:417–423. doi: 10.1016/j.femsle.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Schell M.A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton C., Ross R.P., Fitzgerald G.F., van Sinderen D. Fermented functional foods based on probiotics and their biogenic metabolites. Curr Opin Biotechnol. 2005;16:198–203. doi: 10.1016/j.copbio.2005.02.008. , and . [DOI] [PubMed] [Google Scholar]
- Steed H., Macfarlane G.T., Macfarlane S. Prebiotics, synbiotics and inflammatory bowel disease. Mol Nutr Food Res. 2008;52:898–905. doi: 10.1002/mnfr.200700139. , and . [DOI] [PubMed] [Google Scholar]
- Swint‐Kruse L., Matthews K.S. Allostery in the LacI/GalR family: variations on a theme. Curr Opin Microbiol. 2009;12:129–137. doi: 10.1016/j.mib.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., van Sinderen D., Ventura M. Bifidobacteria: from ecology to genomics. Front Biosci. 2009;14:4673–4684. doi: 10.2741/3559. [DOI] [PubMed] [Google Scholar]
- Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G.F., Chater K.F., van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007a;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., O'Connell‐Motherway M., Leahy S., Moreno‐Munoz J.A., Fitzgerald G.F., van Sinderen D. From bacterial genome to functionality; case bifidobacteria. Int J Food Microbiol. 2007b;120:2–12. doi: 10.1016/j.ijfoodmicro.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Ventura M., O'Flaherty S., Claesson M.J., Turroni F., Klaenhammer T.R., van Sinderen D., O'Toole P.W. Genome‐scale analyses of health‐promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009a;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Ventura M., Turroni F., Zomer A., Foroni E., Giubellini V., Bottacini F. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009b;5:12 e1000785. doi: 10.1371/journal.pgen.1000785. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A.J., Homer K.A., Hosie A.H. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J Bacteriol. 2008;190:168–178. doi: 10.1128/JB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.J., Zhan H., Swint‐Kruse L., Matthews K.S. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Mahowald M.A., Ley R.E., Lozupone C.A., Hamady M., Martens E.C. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:156. doi: 10.1371/journal.pbio.0050156. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ichinose H., Yoshida M., Nakajima M., Kobayashi H., Kaneko S. Characterization of a thermostable endo‐beta‐1,4‐D‐galactanase from the hyperthermophile Thermotoga maritima. Biosci Biotechnol Biochem. 2006;70:538–534. doi: 10.1271/bbb.70.538. [DOI] [PubMed] [Google Scholar]
- Zoetendal E.G., Vaughan E.E., de Vos W.M. A microbial world within us. Mol Microbiol. 2006;59:1639–1650. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotide primers used in this study.
Appendix S1. Experimental procedures.