Abstract
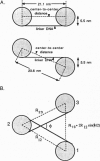
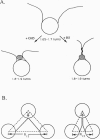
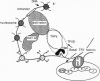
In this review, we attempt to summarize, in a critical manner, what is currently known about the processes of condensation and decondensation of chromatin fibers. We begin with a critical analysis of the possible mechanisms for condensation, considering both old and new evidence as to whether the linker DNA between nucleosomes bends or remains straight in the condensed structure. Concluding that the preponderance of evidence is for straight linkers, we ask what other fundamental process might allow condensation, and argue that there is evidence for linker histone-induced contraction of the internucleosome angle, as salt concentration is raised toward physiological levels. We also ask how certain specific regions of chromatin can become decondensed, even at physiological salt concentration, to allow transcription. We consider linker histone depletion and acetylation of the core histone tails, as possible mechanisms. On the basis of recent evidence, we suggest a unified model linking targeted acetylation of specific genomic regions to linker histone depletion, with unfolding of the condensed fiber as a consequence.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavykin S. G., Usachenko S. I., Zalensky A. O., Mirzabekov A. D. Structure of nucleosomes and organization of internucleosomal DNA in chromatin. J Mol Biol. 1990 Apr 5;212(3):495–511. doi: 10.1016/0022-2836(90)90328-J. [DOI] [PubMed] [Google Scholar]
- Bednar J., Horowitz R. A., Dubochet J., Woodcock C. L. Chromatin conformation and salt-induced compaction: three-dimensional structural information from cryoelectron microscopy. J Cell Biol. 1995 Dec;131(6 Pt 1):1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield V., Dalton W. O., Van Holde K. E. Frictional coefficients of multisubunit structures. I. Theory. Biopolymers. 1967 Feb;5(2):135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. I. Synchrotron radiation X-ray scattering results. Eur Biophys J. 1986;13(3):157–173. doi: 10.1007/BF00542560. [DOI] [PubMed] [Google Scholar]
- Bordas J., Perez-Grau L., Koch M. H., Vega M. C., Nave C. The superstructure of chromatin and its condensation mechanism. II. Theoretical analysis of the X-ray scattering patterns and model calculations. Eur Biophys J. 1986;13(3):175–185. doi: 10.1007/BF00542561. [DOI] [PubMed] [Google Scholar]
- Brownell J. E., Zhou J., Ranalli T., Kobayashi R., Edmondson D. G., Roth S. Y., Allis C. D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996 Mar 22;84(6):843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Butler P. J. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. EMBO J. 1984 Nov;3(11):2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Buttinelli M., Leoni L., Sampaolese B., Savino M. Influence of DNA topology and histone tails in nucleosome organization on pBR322 DNA. Nucleic Acids Res. 1991 Aug 25;19(16):4543–4549. doi: 10.1093/nar/19.16.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A., Kimura T., Gould H., Allan J. Perturbation of chromatin structure in the region of the adult beta-globin gene in chicken erythrocyte chromatin. J Mol Biol. 1987 Jan 5;193(1):57–70. doi: 10.1016/0022-2836(87)90626-7. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Walker I. O. The modification of deoxyribonucleohistone by trypsin and chymotrypsin. Eur J Biochem. 1973 May 2;34(3):519–526. doi: 10.1111/j.1432-1033.1973.tb02789.x. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990 Feb 20;211(4):883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Noll M. Nucleosome arcs and helices. Science. 1978 Oct 20;202(4365):280–286. doi: 10.1126/science.694532. [DOI] [PubMed] [Google Scholar]
- Ebralidse K. K., Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Nucleosomal structure at hyperacetylated loci probed in nuclei by DNA-histone crosslinking. Nucleic Acids Res. 1993 Oct 11;21(20):4734–4738. doi: 10.1093/nar/21.20.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E. A., Felsenfeld G. Comparison of the folding of beta-globin and ovalbumin gene containing chromatin isolated from chicken oviduct and erythrocytes. Biochemistry. 1986 Dec 2;25(24):8010–8016. doi: 10.1021/bi00372a033. [DOI] [PubMed] [Google Scholar]
- Forte P., Leoni L., Sampaolese B., Savino M. Cooperativity in nucleosomes assembly on supercoiled pBR322 DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8683–8694. doi: 10.1093/nar/17.21.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer P., Bednar J., Dubochet J., Hamiche A., Prunell A. DNA at the entry-exit of the nucleosome observed by cryoelectron microscopy. J Struct Biol. 1995 May-Jun;114(3):177–183. doi: 10.1006/jsbi.1995.1017. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Dong F., Ausio J. Role of the histone "tails" in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992 Sep 25;267(27):19587–19595. [PubMed] [Google Scholar]
- Gill G., Tjian R. Eukaryotic coactivators associated with the TATA box binding protein. Curr Opin Genet Dev. 1992 Apr;2(2):236–242. doi: 10.1016/s0959-437x(05)80279-5. [DOI] [PubMed] [Google Scholar]
- Hacques M. F., Muller S., De Murcia G., Van Regenmortel M. H., Marion C. Accessibility and structural role of histone domains in chromatin. biophysical and immunochemical studies of progressive digestion with immobilized proteases. J Biomol Struct Dyn. 1990 Dec;8(3):619–641. doi: 10.1080/07391102.1990.10507832. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Schultz P., Ramakrishnan V., Oudet P., Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996 Mar 22;257(1):30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994 Apr 15;13(8):1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Clayton A. L., Crane-Robinson C. Histone acetylation and globin gene switching. Nucleic Acids Res. 1992 Mar 11;20(5):1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Thomas J. O. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur J Biochem. 1990 Jan 12;187(1):145–153. doi: 10.1111/j.1432-1033.1990.tb15288.x. [DOI] [PubMed] [Google Scholar]
- Hong L., Schroth G. P., Matthews H. R., Yau P., Bradbury E. M. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem. 1993 Jan 5;268(1):305–314. [PubMed] [Google Scholar]
- Horowitz R. A., Agard D. A., Sedat J. W., Woodcock C. L. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994 Apr;125(1):1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan L. J., Utley R. T., Adams C. C., Vettese-Dadey M., Workman J. L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994 Dec 15;13(24):6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Leuba S. H., Yang G., Robert C., Samori B., van Holde K., Zlatanova J., Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994 Dec;103(7):441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- Makarov V. L., Dimitrov S. I., Tsaneva I. R., Pashev I. G. The role of histone H1 and non-structured domains of core histones in maintaining the orientation of nucleosomes within the chromatin fiber. Biochem Biophys Res Commun. 1984 Aug 16;122(3):1021–1027. doi: 10.1016/0006-291x(84)91193-8. [DOI] [PubMed] [Google Scholar]
- Marion C., Bezot P., Hesse-Bezot C., Roux B., Bernengo J. C. Conformation of chromatin oligomers. A new argument for a change with the hexanucleosome. Eur J Biochem. 1981 Nov;120(1):169–176. doi: 10.1111/j.1432-1033.1981.tb05685.x. [DOI] [PubMed] [Google Scholar]
- Marion C., Roux B., Coulet P. R. Role of histones H1 and H3 in the maintenance of chromatin in a compact conformation. Study with an immobilized enzyme. FEBS Lett. 1983 Jul 4;157(2):317–321. doi: 10.1016/0014-5793(83)80568-7. [DOI] [PubMed] [Google Scholar]
- Marion C., Roux B., Pallotta L., Coulet P. R. Study of chromatin organization with trypsin immobilized on collagen membranes. Biochem Biophys Res Commun. 1983 Aug 12;114(3):1169–1175. doi: 10.1016/0006-291x(83)90685-x. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Noll M., Zimmer S., Engel A., Dubochet J. Self-assembly of single and closely spaced nucleosome core particles. Nucleic Acids Res. 1980 Jan 11;8(1):21–42. doi: 10.1093/nar/8.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R. Probing the conformation of nucleosome linker DNA in situ with pyrimidine dimer formation. J Biol Chem. 1995 Sep 22;270(38):22440–22444. [PubMed] [Google Scholar]
- Pehrson J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9149–9153. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Ridsdale J. A., Hendzel M. J., Delcuve G. P., Davie J. R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990 Mar 25;265(9):5150–5156. [PubMed] [Google Scholar]
- Saccone G. T., Skinner J. D., Burgoyne L. A. Resistance of chromatin superstructure to tryptic digestion modulated by conjugated polyacrylamide. FEBS Lett. 1983 Jun 27;157(1):111–114. doi: 10.1016/0014-5793(83)81126-0. [DOI] [PubMed] [Google Scholar]
- Strätling W. H. Role of histone H1 in the conformation of oligonucleosomes as a function of ionic strength. Biochemistry. 1979 Feb 20;18(4):596–603. doi: 10.1021/bi00571a008. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. P., Athey B. D., Muglia L. J., Schappe R. S., Gough A. H., Langmore J. P. Chromatin fibers are left-handed double helices with diameter and mass per unit length that depend on linker length. Biophys J. 1986 Jan;49(1):233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig B., Wittig S. Nucleosome mono, di, tri-, and tetramers from chicken embryo chromatin. Nucleic Acids Res. 1977 Nov;4(11):3901–3917. doi: 10.1093/nar/4.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9021–9025. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Horowitz R. A. Chromatin organization re-viewed. Trends Cell Biol. 1995 Jul;5(7):272–277. doi: 10.1016/s0962-8924(00)89038-8. [DOI] [PubMed] [Google Scholar]
- Yang G., Leuba S. H., Bustamante C., Zlatanova J., van Holde K. Role of linker histones in extended chromatin fibre structure. Nat Struct Biol. 1994 Nov;1(11):761–763. doi: 10.1038/nsb1194-761. [DOI] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Direct detection of linker DNA bending in defined-length oligomers of chromatin. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Linker DNA bending induced by the core histones of chromatin. Biochemistry. 1991 Aug 27;30(34):8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]
- Yao J., Lowary P. T., Widom J. Twist constraints on linker DNA in the 30-nm chromatin fiber: implications for nucleosome phasing. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9364–9368. doi: 10.1073/pnas.90.20.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990 Jul;15(7):273–276. doi: 10.1016/0968-0004(90)90053-e. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992 Dec;103(Pt 4):889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., van Holde K. The linker histones and chromatin structure: new twists. Prog Nucleic Acid Res Mol Biol. 1996;52:217–259. doi: 10.1016/s0079-6603(08)60968-x. [DOI] [PubMed] [Google Scholar]
- van Holde K., Zlatanova J. Chromatin higher order structure: chasing a mirage? J Biol Chem. 1995 Apr 14;270(15):8373–8376. doi: 10.1074/jbc.270.15.8373. [DOI] [PubMed] [Google Scholar]