Summary
The prokaryotic community composition of activated sludge from a seawater‐processing wastewater treatment plant (Almeria, Spain) was investigated by using the rRNA approach, combining different molecular techniques such as denaturing gradient gel electrophoresis (DGGE), clone libraries and in situ hybridization (FISH and CARD‐FISH). Most of the sequences retrieved in the DGGE and the clone libraries were similar to uncultured members of different phyla. The most abundant sequence recovered from Bacteria in the clone library corresponded to a bacterium from the Deinococcus–Thermus cluster (almost 77% of the clones), and the library included members from other groups such as the Alpha, Gamma and Delta subclasses of Proteobacteria, the Bacteroidetes and Firmicutes. Concerning the archaeal clone library, we basically found sequences related to different orders of methanogenic Archaea, in correspondence with the recovered DGGE bands. Enumeration of DAPI (4′,6‐diamidino‐2‐phenylindole) stained cells from two different activated sludge samples after a mechanical flocculation disruption revealed a mean cell count of 1.6 × 109 ml−1. Around 94% of DAPI counts (mean value from both samples) hybridized with a Bacteria specific probe. Alphaproteobacteria were the dominant bacterial group (36% of DAPI counts), while Beta‐, Delta‐ and Gammaproteobacteria, Bacteroidetes, Actinobacteria and Firmicutes contributed to lower proportions (between 0.5–5.7% of DAPI counts). Archaea accounted only for 6% of DAPI counts. In addition, specific primers for amplification of the amoA (ammonia monooxygenase) gene were used to detect the presence of Beta, Gamma and archaeal nitrifiers, yielding positive amplifications only for Betaproteobacteria. This, together with negative in situ hybridizations with probes for well‐known nitrifiying bacteria, suggests that nitrification is performed by still undetected microorganisms. In summary, the combination of the three approaches provided different and complementary pictures of the real assemblage composition and allowed to get closer to the main microorganisms involved in key processes of seawater‐processing activated sludge.
Introduction
Activated sludge systems are one of the most important biotechnological processes in wastewater treatment plants (wwtps). They consist of a complex mixture of microorganisms able to remove organic substances and nutrient contaminants from municipal or industrial sewage, being thus a crucial tool in environmental protection. For years, researchers have investigated the microbial communities of activated sludge in order to understand their specific biological processes (Amann et al., 1998; Wagner et al., 2002). Studies of diversity can provide insight on the correlation between microbial composition and ecosystem function, as well as knowledge about temporal and spatial variations in microbial communities. However, the vast majority of bacteria present in activated sludge cannot be isolated by conventional culture‐dependent techniques; the percentage of culturable bacteria in comparison with total cell counts is estimated to range between 1% and 15% with optimized media (Wagner et al., 1993).
The current use of molecular methods, which do not require the isolation and cultivation of microorganisms, has allowed a more comprehensive analysis of microbial diversity in wastewater research. Sequence analysis of 16S rRNA gene clone libraries (Snaidr et al., 1997), fingerprinting techniques such as denaturing gradient gel electrophoresis (Boon et al., 2002), thermal gradient gel electrophoresis (Eichner et al., 1999) and terminal restriction fragment length polymorphism (T‐RFLP) (Saikaly et al., 2005), as well as the design of group‐specific rRNA‐targeted oligonucleotide probes for fluorescence in situ hybridization (FISH) (Wagner et al., 1993; 1994), have greatly expanded our understanding of wastewater microbiology. The cultivation‐independent rRNA approach allows to determine the composition and dynamics of microbial communities in these systems and to identify the microbial key players for the different processes.
Considerable microbial diversity has been detected in wwtps, including bacteria involved in biological phosphorus removal (Bond et al., 1999; Jeon et al., 2003; Seviour et al., 2003), nitrifiers (Juretschko et al., 1998; Coskuner and Curtis, 2002; Otawa et al., 2006) and methanogens (Zheng, 2000). Sequences from Proteobacteria, Bacteroidetes, Chloroflexi, Actinobacteria and the Planctomycetes were retrieved in significant numbers in different clone libraries (Wagner et al., 2002). Nevertheless, all studies of microbial diversity in wwtps refer to freshwater treatment plants, either domestic or industrial. As far as we know, no studies have been done on wwtps that utilize seawater for their operation.
On the other hand, only a few works have applied the full‐cycle rRNA approach for the study of microbial communities in activated sludges, which includes the establishment of a 16S rRNA gene clone library, the design of a set of clone‐specific oligonucleotide probes, and the determination of the abundance of the respective bacterial populations by quantitative FISH (Snaidr et al., 1997; Juretschko et al., 1998). In this paper, the prokaryotic diversity of a wwtp from a pharmaceutical industry located in the south of Spain, which has the particularity to utilize seawater, has been characterized using a polyphasic approach with three molecular tools such as DGGE, clone libraries and FISH. This wastewater treatment plant is in operation since 1998 and today, very few plants of this type are running in the world; its main influent corresponds to intermediate products from amoxicillin synthesis. The use of seawater instead of freshwater responds to the deficiency in hydric resources prevailing in this location, one of the driest areas in Spain. To our knowledge, this is the first study that analyses an activated sludge with these characteristics.
Results
DGGE fingerprinting from seawater‐processing activated sludge
The DGGE analysis from the two samples of activated sludge, corresponding to years 2007 and 2008, yielded a total of 20 and 17 different band positions for Bacteria and Archaea respectively (Fig. 1). Both samples showed virtually the same pattern for both set of primers, although differences in band intensity could be observed in some bands. This finding suggested that the system was rather stable along time. Bands were excised from both gels in order to determine their phylogenetic affiliation, and informative sequences were obtained from 12 (Bacteria) and 5 bands (Archaea) (Table 1). These bands accounted for 58% (Bacteria) and 32% (Archaea) of the total mean band intensity and most of them showed similarities with sequences from uncultured clones by blast search.
Figure 1.
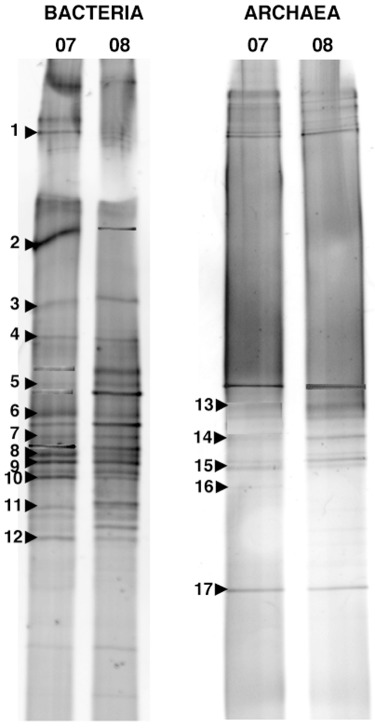
Negative images of DGGE gels with PCR products amplified with bacterial and archaeal primer sets from samples of activated sludge corresponding to years 2007 and 2008. Bands excised and sequenced are numbered and their affiliations are shown in Table 1.
Table 1.
Phylogenetic affiliation of sequences obtained from DGGE bands, with closest uncultured and cultured matches, and relative intensity of the bands.
| Band | Closest match | % similarity (No. of bases)a | Taxonomic group | Accession No. (GenBank) | Cultured closest match (% similarity) | Relative intensity (%) |
|---|---|---|---|---|---|---|
| DER_1 | Uncultured Bacteroidetes/Chlorobi group clone 3B02‐03 | 92.1 (498) | Bacteroidetes | DQ431894 | Marinicola seohaensis (84.9) | 5.6 |
| DER_2 | Uncultured Bacteroidetes clone ML617.5J‐33 | 93.3 (502) | Bacteroidetes | AF507866 | Owenweeksia hongkongensis (86.5) | 24.9 |
| DER_3 | Uncultured Bacteroidetes clone 02D2Z22 | 92.8 (482) | Bacteroidetes | DQ330313 | Owenweeksia hongkongensis (88.8) | 4.0 |
| DER_4 | Vitellibacter sp. | 77.7 (383) | Bacteroidetes | EU642844 | The same | 0.6 |
| DER_5 | Clone nsmp VI41 | 97.2 (529) | γ‐Proteobacteria | AB212895 | Luteibactor rhizovicina (85.8) | 5.6 |
| DER_6 | Clone nsmp VI41 | 82.0 (437) | γ‐Proteobacteria | AB212895 | Aquimonas sp. (75.7) | 2.1 |
| DER_7 | Clone Strom2G11 | 87.2 (449) | α‐Proteobacteria | EU918039 | Parvibaculum sp. | 7.9 |
| DER_8 | Nitratireductor sp. | 94.4 (487) | α‐Proteobacteria | EU564843 | The same | 7.9 |
| DER_9 | Clone 101‐91 | 99.8 (539) | δ‐Proteobacteria | EF157196 | Desulfonatronum cooperativum (84.1) | 2.0 |
| DER_10 | Clone nsmo VI20 | 99.6 (541) | γ‐Proteobacteria | AB212894 | Frateuria aurantia (94.1) | 2.0 |
| DER_11 | Clone OTU_23 | 85.1 (430) | Deinococcus–Thermus | EU083501 | Truepera radiovictrix (80.0) | 1.5 |
| DER_12 | Clone OTU_23 | 96.1 (493) | Deinococcus–Thermus | EU083501 | Truepera radiovictrix (89.3) | 1.4 |
| DER_13 | Clone Hua6‐s78 | 83.7 (385) | Euryarchaea | EU481593 | Methanobacterium aarhusense (78.8) | 11.8 |
| DER_14 | Clone ss037b | 93.8 (379) | Euryarchaea | AJ969783 | Aciduliprofundum boonei (84.2) | 7.8 |
| DER_15 | Clone ss037b | 94.4 (476) | Euryarchaea | AJ969783 | Methanobrevibacter sp. (79.3) | 1.3 |
| DER_16 | Clone ss037b | 95.3 (487) | Euryarchaea | AJ969783 | Aciduliprofundum boonei (79.1) | 2.8 |
| DER_17 | Clone ss037b | 96.3 (494) | Euryarchaea | AJ969783 | Methanobrevibacter sp. (79.7) | 26.7 |
Number of bases used to calculate the levels of sequence similarity.
Four bands of bacterial origin affiliated to Bacteroidetes, with a contribution of 17% to total mean band intensity. The remaining bacterial bands belonged to different subclasses (Alpha, Gamma and Delta) of the phylum Proteobacteria except two bands that affiliated to the Deinococcus–Thermus group.
Excision of bands from the archaeal DGGE gel yielded sequences related to methanogenic Archaea, although identities were relatively low.
Identification of taxonomic groups by clone libraries
We analysed 278 and 117 clones in bacterial and archaeal clone libraries constructed with a sample from the marine‐activated sludge (year 2007). Sequences were grouped in OTUs using a similarity criteria clustering of 98.5% (Stackebrandt and Goebel, 1994). One representative sequence of each OTU is shown in Tables 2 and 3, together with the closest relatives and the frequency of the OTU. Coverage of the libraries was 95.7% and 91.5% respectively, indicating that in both cases this particular approach provided most of the measurable diversity.
Table 2.
Phylogenetic affiliation of clones from the bacterial clone library to the closest match and to the closest cultured strain in GenBank. (Bold text represents OTU with the highest percentage of clones.)
| Representative clone | Closest match (accession number) | % similarity | Cultured closest match (accession number) | % similarity | No. of clones (%) |
|---|---|---|---|---|---|
| α‐Proteobacteria | |||||
| BACDER07_1C3 | Clone DR938CH110701SACH95 (DQ230971) | 94.0 | Nitratireductor sp. (EU564843) | 93.4 | 1 (0.4) |
| BACDER07_1C12 | Clone SC71 (EU735614) | 97.8 | Brucella sp. (DQ167235) | 90.2 | 1 (0.4) |
| BACDER07_1F8 | Clone 81 T12d‐oil (FM242433) | 95.7 | Subaequorebacter tamlense (AM293856) | 90.1 | 1 (0.4) |
| BACDER07_1H9 | Nitratireductor sp. (EU564843) | 94.2 | The same | 94.2 | 4 (1.4) |
| BACDER07_1D12 | Clone B1‐43 (AM229476) | 98.2 | Sneathiella chinensis (DQ219355) | 95.6 | 1 (0.4) |
| BACDER07_2B7 | Clone CI75cm.2.18 (EF208711) | 98.6 | Methylocystis sp. (AJ868421) | 94.7 | 2 (0.7) |
| BACDER07_2F8 | Clone Strom2G11 (EU918039) | 96.3 | Maricaulis sp. (AJ301666) | 90 | 2 (0.7) |
| BACDER07_2G10 | Clone 256ds10 (AY212705) | 98.0 | Sphingomonas sp. (D16149) | 97.1 | 1 (0.4) |
| δ‐Proteobacteria | |||||
| BACDER07_1D11 | Clone 101‐91 (EF157196) | 98.9 | Desulfonatronum cooperativum (AY725424) | 80.7 | 7 (2.5) |
| γ‐Proteobacteria | |||||
| BACDER07_1B8 | Clone nsmpVI41 (AB212895) | 98.9 | Kangiella koreensis (AY520560) | 85.4 | 16 (5.8) |
| BACDER07_1D2 | Clone nsmpVI20 (AB212894) | 99.6 | Rhodanobacter lindaniclasticus (DQ507211) | 91.6 | 21 (7.6) |
| BACDER07_3B12 | Clone nsmpVI20 (AB212894) | 98.1 | Frateuria aurantia (AB091195) | 90.2 | 1 (0.4) |
| Deinococcus–Thermus | |||||
| BACDER07_1B5 | Clone GZKB22 (AJ853517) | 95.8 | Truepera radiovictrix (DQ022077) | 89.3 | 212 (76.3) |
| BACDER07_1G6 | Clone GZKB22 (AJ853517) | 94.5 | Truepera radiovictrix (DQ022077) | 89.8 | 2 (0.7) |
| BACDER07_1F5 | Clone 6 (EU017377) | 94.5 | Truepera radiovictrix (DQ022077) | 89.5 | 1 (0.4) |
| CFB Group | |||||
| BACDER07_1F4 | Clone 6 (DQ015772) | 96.3 | Lewinella marina (AB301495) | 84.7 | 1 (0.4) |
| BACDER07_2D6 | Clone Er‐LLAYS‐51 (EU542514) | 97.5 | Owenweeksia hongkongensis (AB125062) | 87.3 | 1 (0.4) |
| BACDER07_2H3 | Clone HF500_26D14 (EU361310) | 95.6 | Sphingobacterium sp. (AM411964) | 90.0 | 1 (0.4) |
| Firmicutes | |||||
| BACDER07_2F2 | Clone p816_b_3.45 (AB305600) | 81.7 | Bacillus sp. (EF422410) | 79.0 | 1 (0.4) |
| Unclassified bacteria | |||||
| BACDER07_3D4 | Denitromonas indolicum (AY972852) | 95.2 | The same | 95.2 | 1 (0.4) |
Table 3.
Phylogenetic affiliation of clones from the archaeal clone library to the closest match and to the closest cultured strain in GenBank. (Bold text represents OTU with the highest percentage of clones.)
| Representative clone | Closest match (accession number) | % similarity | Cultured closest match (accession number) | % similarity | No. of clones (%) |
|---|---|---|---|---|---|
| Methanosarcinales | |||||
| ARCHDER07_1A12 | Methanococcoides sp. (Y16946) | 99.5 | The same | 99.5 | 5 (4.3) |
| ARCHDER07_1C3 | Clone Z3‐Arc‐1 (EU999009) | 98.6 | Methanolobus profundi (AB370245) | 97.8 | 2 (1.7) |
| ARCHDER07_1D4 | Clone TFC20L31Ar (EU362350) | 97.0 | Methanosaeta harundinacea (AY970347) | 96.2 | 2 (1.7) |
| Methanomicrobiales | |||||
| ARCHDER07_2C9 | Clone WIP (EF420166) | 98.5 | Methanoculleus marisnigri (CP000562) | 97.5 | 2 (1.7) |
| ARCHDER07_1B2 | Clone GoM‐GC234‐015R (AY211693) | 96.1 | Methanoculleus sp. (AJ133793) | 93.8 | 6 (5.1) |
| ARCHDER07_2D6 | Clone PMMV‐Arc14 (AJ937680) | 92.2 | Methanoculleus sp. (AJ133793) | 89.8 | 1 (0.9) |
| Methanobacteriales | |||||
| ARCHDER07_1A2 | Clone 4B09 (AY835426) | 94.7 | Methanothermus fervidus (M32222) | 81.8 | 8 (6.8) |
| ARCHDER07_1B11 | Clone ALAS95 (EU616776) | 99.2 | Methanobacterium aarhusense (DQ649334) | 84.4 | 1 (0.9) |
| DER_1 | |||||
| ARCHDER07_1D11 | Clone ss037b (AJ969783) | 97.1 | Methanomethylovorans sp. (EU544305) | 78.0 | 5 (4.3) |
| ARCHDER07_1G10 | Clone HARR41 (AJ699117) | 99.5 | Aciduliprofundum boonei (DQ451875) | 81.2 | 2 (1.7) |
| DER_2 | |||||
| ARCHDER07_2C8 | Clone GNA03E09 (EU731492) | 94.3 | Methanobrevibacter gottschalkii (U55239) | 70.8 | 1 (0.9) |
| ARCHDER07_1C11 | Clone MOB4‐5 (DQ841225) | 91.7 | Methanobrevibacter gottschalkii (U55239) | 70.1 | 1 (0.9) |
| ARCHDER07_1E6 | Clone GNA03E09 (EU731492) | 96.0 | Methanobacterium sp. (DQ517520) | 71.0 | 3 (2.6) |
| ARCHDER07_1A5 | Clone GNA03E09 (EU731492) | 98.2 | Methanococcus infernus (AF025822) | 72.8 | 47 (40.2) |
| ARCHDER07_1A10 | Clone GNA03G10 (EU731491) | 94.9 | Methanococcus vulcanus (AF051404) | 71.9 | 1 (0.9) |
| ARCHDER07_2D4 | Clone CaR3b.h02 (EU244267) | 90.1 | Methanobrevibacter sp. (AJ550156) | 70.8 | 2 (1.7) |
| ARCHDER07_2A4 | Clone CaS1s.h02 (EF014578) | 88.9 | Methanococcus infernus (AF025822) | 72.5 | 1 (0.9) |
| ARCHDER07_1A4 | Clone A21 (EU328111) | 88.6 | Methanococcus aeolicus (CP000743) | 72.8 | 12 (10.3) |
| ARCHDER07_1D1 | Clone KAB187‐14 (AB366595) | 89.8 | Methanobacterium sp. (EU366499) | 73.8 | 1 (0.9) |
| DER_3 | |||||
| ARCHDER07_1B4 | Clone 1ACC‐29 (AB175599) | 97.2 | Methanothermococcus sp. (AB175514) | 78.0 | 9 (1.7) |
| ARCHDER07_1G6 | Clone GNA03F04 (EU731409) | 95.1 | Methanocaldococcus jannaschii (L77117) | 78.8 | 1 (0.9) |
| ARCHDER07_1D10 | Clone GNA02E03 (EU731293) | 97.4 | Methanococcus aeolicus (CP000743) | 78.0 | 2 (1.7) |
| ARCHDER07_2B5 | Clone GNA01D07 (EU731138) | 91.6 | Methanothermococcus sp. (AB260046) | 77.3 | 1 (0.9) |
| ARCHDER07_1A1 | Clone ML23_ANME 9 (AY245465) | 83.2 | Methanocaldoccus indiensis (AF547621) | 76.1 | 1 (0.9) |
A significant number of clones from both libraries showed similarity to uncultured sequences deposited in GenBank. The most abundant sequence recovered from Bacteria corresponded to a bacterium from the Deinococcus–Thermus cluster (almost 77% of the clones) with the same sequence as band DER_12. The library included also members from other groups such as the Alpha, Gamma and Delta subclasses of Proteobacteria, the Bacteroidetes and Firmicutes. Within the Proteobacteria, which represented 21% of the clones, members of the Gammaproteobacteria predominated (14% of total clones). One clone was affiliated to the unclassified bacterium Denitromonas indolicum. Similarities based on sequence comparison of these clones varied between 81.7% and 99.6%. The 16S rRNA similarities were approximately at the species level (≥ 97%, Stackebrandt and Goebel, 1994) for 19% of the clones, while 79% were similar at the genus level (95–97%). In general, there was agreement between the different sequences retrieved by DGGE and the clone library. Inclusion of all the sequences in a phylogenetic tree indicated that most of the DGGE band sequences corresponded to several of the most abundant clones recovered from the library (tree not shown).
Concerning the archaeal clone library, we basically found sequences related to different orders of methanogenic Archaea, in correspondence with the recovered DGGE bands. In this case, similarities ranged between 83.2% and 99.5%. We paid especial attention to these archaeal sequences, since most of them were only moderately related to known archaea. Phylogenetic analyses were performed by several methods and summarized in a maximum‐likelihood tree with Bayesian posterior probabilities and neighbour‐joining bootstrap values in the relevant nodes (Fig. 2). Based on the tree structure and bootstrap values, a high percentage of archaeal sequences (77%) were grouped into three separate clusters, named DER_1, 2 and 3, which formed three independent branches composed exclusively by environmental clones. Other sequences clustered within three major phylogenetic groups of methanogens: Methanosarcinales, Methanomicrobiales and Methanobacteriales, while sequences belonging to Methanococcales and Methanopyrales were not retrieved.
Figure 2.
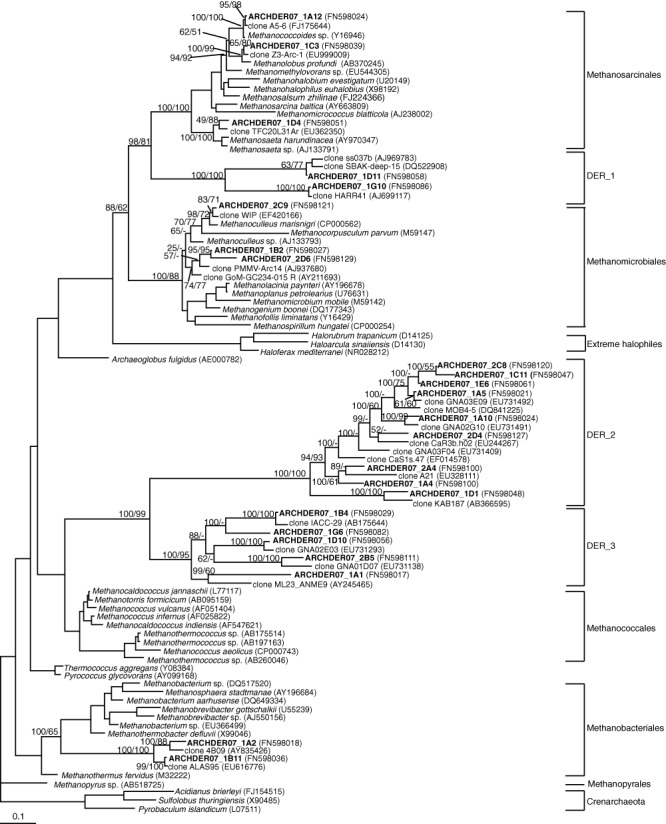
Maximum‐likelihood phylogenetic tree with partial 16S rDNA archaeal sequences (695 informative positions). Clones in bold are from this study. Posterior probability values and neighbour‐joining bootstrap values (1000 replicates) are shown in the relevant nodes. The scale bar indicates 0.1 substitutions per position.
Detection of amoA genes
Using specific primers for the amplification of the gene amoA from the Beta, Gamma and archaeal nitrifiers, we were able to confirm the presence of ammonia‐oxidizers from the Beta subclass of Proteobacteria (data not shown). The gene amoA encodes the catalytic α‐subunit of ammonia monooxygenase, the enzyme responsible for catalysing the rate‐limiting step in ammonia oxidation, and it has been used extensively as a molecular marker for cultivation‐independent studies of ammonia‐oxidizing communities. However, we could not find amplification for Gamma and Archaea nitrifiers in our samples.
Quantitative analysis of marine‐activated sludge composition by in situ hybridization
The activated sludge samples from years 2007 and 2008 were also analysed by DAPI staining and fluorescent in situ hybridization. A sound quantification of activated sludge samples was complicated by the heterogeneous cell distribution caused by the flocculation. Several treatments were tested for cell dispersal, e.g. sonication and vortexing at different times (5, 15 or 30 min, data not shown). No differences in hybridization signals were found in treatments at 5 and 15 min, while the fraction of hybridized cells decreased after sonicating or vortexing samples during 30 min. In any case, microscopic examination clearly showed that there were still aggregates. In order to increase accuracy we counted separately the hybridized free cells and the hybridized cells in aggregates for each probe, evaluating several thousand cells in the case of aggregates. Enumeration of DAPI‐stained preparations revealed total cell counts of 1.89 × 109 ml−1 (year 2007) and 1.28 × 109 ml−1 (year 2008), being 65% (mean value from both years) in the form of aggregates and the rest as free cells. The mean number of cells per aggregate was 444. However, one has to keep in mind that microscopic enumeration of aggregated cell clusters is likely resulting in underestimations, but since this effect applies equally to DAPI and FISH counts, the conclusions drawn from the different fractions of probe positive‐DAPI stained cells remain valid.
The results obtained from CARD‐FISH analyses for both years (Fig. 3) indicated that the microbial assemblage had virtually the same composition, and that the percentage of the hybridized groups remained very similar, confirming that the system was stable along time. Hybridization with the universal set of probes EUB (Eub+) detected a mean value from both years of 86% of DAPI‐stained free cells and 97% of cells in aggregates (Eub+), while Archaea reached only 4% and 3% of mean DAPI counts for free and aggregated cells, respectively, indicating that the majority of fixed cells were Bacteria. A considerable amount of total Eub+ free cells (54%) were identified with probes for broad phylogenetic groups (Alpha‐, Beta‐, Gamma‐ and Deltaproteobacteria, Bacteroidetes, Firmicutes and Actinobacteria). This value was similar for aggregates (62%). Alphaproteobacteria were the dominant bacterial group [27% (free cells) and 44% (aggregates) of mean DAPI counts], while Gammaproteobacteria and Bacteroidetes contributed to lower and similar proportions in the free‐cells fraction (around 8% of mean DAPI counts). These values were lower in the case of aggregated cells (around 4% of mean DAPI counts). Other groups, such as Beta and Deltaproteobacteria, Firmicutes and Actinobacteria, were present even at lower numbers (1–4% of mean DAPI counts in free cells, and 0–1% in aggregates). A specific probe (DT01) corresponding to a sequence that accounted for 77% of the clones in the bacterial clone library was designed in this work. This sequence had approximately 96% similarity with an uncultured bacterium from the Deinococcus–Thermus phylum (Table 2). However, CARD‐FISH results showed that this microorganism was not particularly abundant (3% and 7% of mean DAPI counts for free and aggregated cells respectively), denoting a large positive bias for this bacterium in the clone library.
Figure 3.
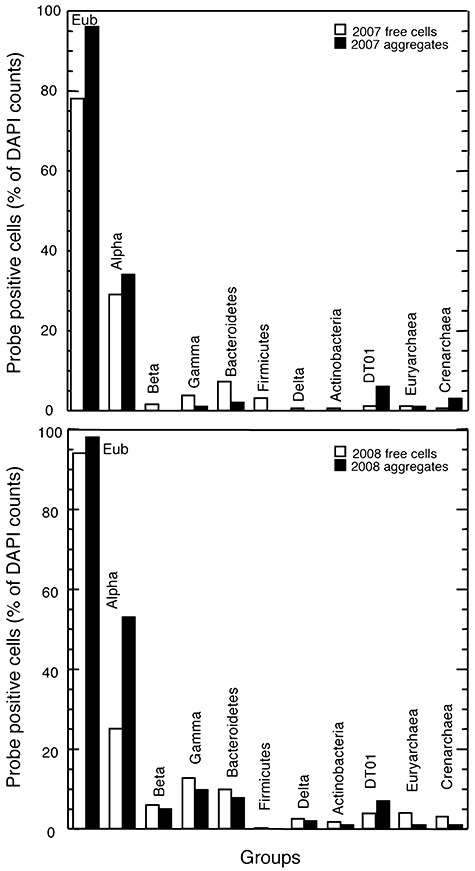
Proportions of bacterial groups detected by CARD‐FISH with HRP probes in free cells and in aggregates from samples of activated sludge corresponding to years 2007 and 2008.
In situ hybridization with a set of well‐known hierarchical 16S rRNA‐targeted probes for ammonia‐oxidizing bacteria (including the genera Nitrobacter and most of the betaproteobacterial ammonia‐oxidizers, such as members of the genera Nitrosomonas, Nitrosococcus mobilis and Nitrosospira), usually used for activated sludge (Juretschko et al., 1998), showed no signal in our samples, in accordance with the results observed in the DGGE and the bacterial clone library for these particular microorganisms.
Quantitative comparison between DGGE, CARD‐FISH and clone libraries
In the case of Bacteria, we quantitatively compared the results obtained by the three different molecular methods in order to test the strong and weak points of each approach, and how they affect the overall picture of activated sludge diversity (Fig. 4). For FISH representation, we took into account the contribution of free and aggregated cells for every probe. The most remarkable trend in this figure is the over‐representation of a sequence corresponding to the Deinococcus–Thermus group in the clone library as compared with CARD‐FISH and DGGE. Alphaproteobacteria and Bacteroidetes, by contrast, seemed to be underrepresented in the clone library, while Alphaproteobacteria were overrepresented with CARD‐FISH. On the other hand, the detection of Gamma‐ and Deltaproteobacteria was more proportionate by the three methods. Other groups could only be detected by CARD‐FISH, such as Betaproteobacteria, and Actinobacteria, although at very low relative abundance.
Figure 4.
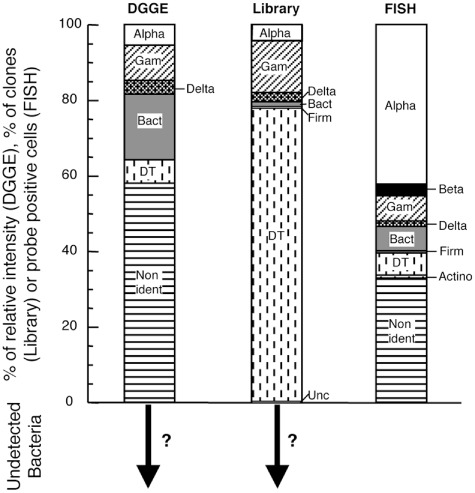
Percentage of relative intensity of DGGE bands, proportions of clones (Library) and probe positive cells scaled to Eub probes (FISH) affiliated to different phylogenetic groups [Alphaproteobacteria (Alpha), Betaproteobacteria (Beta), Gammaproteobacteria (Gam), Deltaproteobacteria (Delta), Bacteroidetes (Bact), Firmicutes (Firm), Actinobacteria and Deinococcus–Thermus clone (DT)]. FISH data correspond to the mean of samples from years 2007 and 2008. The arrows with question marks indicate that an unknown proportion of phylogenetic groups cannot be retrieved by the set of primers used in both DGGE and clone libraries.
Discussion
The advent of molecular techniques in the past two decades has provided many insights into the diversity and functions of predominantly uncultured wastewater microorganisms. However, relatively few works have studied activated sludge microorganisms by a full‐cycle rRNA approach, and never before the diversity of a marine‐activated sludge has been detailed. In our study, the objective was to obtain a comprehensive picture of the diversity of the prokaryotic assemblage in a seawater‐processing wwtp, combining and comparing different molecular approaches (DGGE, clone library and FISH).
In this particular wwtp, we predict that the influent composition becomes crucial in order to understand the composition of the microbial community. The presence of amoxicillin, together with a high salt concentration, will surely affect microbial diversity. Actually, concern is growing over environmental contamination with pharmaceuticals because of their widespread use and incomplete removal during wastewater treatment. Thus, Kraigher and colleagues (2008) investigated the influence of pharmaceutical residues on the structure of activated sludge bacterial communities in wastewater treatment bioreactors and observed a minor but consistent shift in the community structure in bioreactors supplied with pharmaceuticals, as well as a reduction in diversity.
Diversity in seawater‐utilizing activated sludge
Different authors have addressed the study of activated sludge diversity either in wwtp or laboratory‐scale reactors by means of molecular techniques (Bond et al., 1995; Kämpfer et al., 1996; Snaidr et al., 1997; Christensson et al., 1998; Dabert et al., 2001; Daims et al., 2001; Liu and Seviour, 2001; Juretschko et al., 2002; Eschenhagen et al., 2003; Sanapareddy et al., 2009). These studies indicated considerable microbial diversity in wwtps and the dominance of the Beta subclass of the class Proteobacteria. Apart from the Proteobacteria, other groups, such as the Bacteroidetes, the Chloroflexi, the Actinobacteria and the Planctomycetes, could be detected either in clone libraries or using FISH with group‐specific probes. The composition of the bacterial community of the seawater‐activated sludge described here differed strongly from those previously reported, since Betaproteobacteria did not seem to be the predominant group. It was even not detected in the clone library and represented only a 3% of the total hybridized bacteria. This is most likely due to the fact that our wwtp is fed with seawater, which is known to contain very few Betaproteobacteria (Rappé and Giovannoni, 2003). In contrast, other subclasses of Proteobacteria, such as Alpha‐, Gamma‐ or Deltaproteobacteria, were detected with the three methodologies, although in the case of Alphaproteobacteria, at different proportions. On the other hand, the Bacteroidetes group seemed also to be represented by the three methods.
The most remarkable feature of clone libraries is the severe over‐representation of one sequence belonging to the Deinococcus–Thermus group (around 77% of total bacterial clones). Its closest match in GenBank was an uncultured bacterium, and its closest cultured match was Truepera radiovictrix, although at low similarities (less than 90%, Table 1). The phylum Deinococcus–Thermus includes extremely radiation resistant bacteria, as well as slightly thermophilic or thermophilic members, and it also comprises a number of environmental 16S rRNA gene sequences, several of which are not closely related to any cultured strains and form distinct lineages. Nevertheless, when comparing with DGGE and CARD‐FISH data, this sequence turned out to be less abundant and discrepancies with the clone library become clear.
Most of the sequences retrieved in the bacterial clone library were similar to uncultured members of different phyla, but in some cases, cultured closest matches were related at the genus or even at the species level (Table 1). For example, one clone of Alphaproteobacteria (BACDER07_1D12) was similar at the genus level to Sneathiella chinensis, a marine chemoheterotrophic bacterium, and another clone (BACDER07_2B7) to Methylocystis sp., a methanotrophic bacterium. On the other hand, one clone (BACDER07_2G10) was similar at the species level to Sphingomonas sp., a genus recognized by its capability to degrade a wide variety of refractory environmental pollutants and to carry out diverse other biotechnologically useful activities, such as the biosynthesis of valuable biopolymers (Laskin and White, 1999); sphingomonads have been identified in situ by FISH in activated sludge samples and turned out to be rather abundant, accounting for about 5–10% of the total cells (Neef et al., 1999). Since exopolysaccharides are a significant part of the polymeric extracellular matrix material of flocs, and members of the genus Sphingomonas are known to be able to produce slimes and/or capsules, the authors suggested that they could be involved in the formation process of sludge flocs. Finally, another clone was similar at the genus level to Denitromonas indolicum, an unclassified bacterium. This genus was found to be able to grow with perchlorate as the sole electron acceptor (Zuo et al., 2009).
Concerning the diversity of Archaea, less attention has been paid to their role in wastewater treatment processes, since it seems clear that Bacteria are responsible for the majority of carbon removal in the activated sludge process (Gray et al., 2002). In our samples, Archaea represented only a small fraction of total mean DAPI counts from the two samples (4% in free cells and 3% in aggregates). Virtually all sequences retrieved in our archaeal clone library were related to methanogenic bacteria (Fig. 2), consistent with previous reports of the existence of anoxic microenvironments in the flocs, in which methanogens might be active (Schramm et al., 1999). All methanogens are strictly anaerobic Archaea pertaining to the Euryarchaeota. Although they are very diverse phylogenetically, they can only utilize a restricted number of substrates of three major types: CO2, methyl‐group containing compounds and acetate. Their common habitats include marine and freshwater sediments, flooded soils, human and animal gastrointestinal tracts, termites, anaerobic digestors, landfill, geothermal systems and heartwood of trees. Nevertheless, it has been demonstrated the presence of methanogenic bacteria in aerated activated sludge. Thus, Gray and colleagues (2002) retrieved archaeal 16S rRNA gene sequences related to Methanosarcinales, Methanomicrobiales and Methanobacteriales. However, the relatively low rates of methanogenesis measured by these authors indicated that, although active, the methanogens played a minor role in carbon turnover in activated sludge.
In our study, the recovered sequences affiliated also within the orders Methanosarcinales, Methanomicrobiales and Methanobacteriales with the same proportion in the clone library (8% each). However, most of the sequences clustered into novel branches (DER_1, 2 and 3), which were closely related to environmental clones. One of these sequences (representative clone: ARCHDER07_1A5), belonging to DER_2, accounted for 40% of the total clones and was related to a sequence from Guerrero Negro hypersaline microbial mats (Robertson et al., 2009).
On the other hand, Crenarchaeota were also detected by CARD‐FISH (2% of total DAPI counts from free and aggregated cells), but this group was not represented in the archaeal clone library or in the retrieved sequences from the DGGE. Polymerase chain reaction (PCR) bias against Crenarchaeota could explain this disagreement.
Linking diversity and function
The functional assignment of detected microorganisms is complicated by the fact that 16S rRNA sequence‐based identification does not generally allow to infer their functional properties. Phylogenetically closely related microorganisms may possess different metabolic traits, while on the other hand several physiological features like the ability to denitrify are dispersed in different phylogenetic lineages. Therefore, the full‐cycle rRNA‐approach needs to be supplemented with other techniques that allow a functional assignment of the detected microorganisms. In this particular sense, we included in our study the amplification of the functional gene coding for the active‐site polypeptide of ammonia monooxygenase (amoA) as a physiological marker, as well as the use of specific rRNA‐targeted probes for the detection of ammonia oxidizers in order to link diversity with function, in particular those aspects referred to nitrogen removal.
In engineered systems such as wwtps, the coupled nitrification and denitrification processes are considered the major mechanisms of nitrogen removal. The nitrifiers encompass two groups of microorganisms, the ammonia and the nitrite‐oxidizing bacteria, which catalyse the oxidation of ammonia to nitrite and of nitrite to nitrate respectively. Many wwtps harbour diverse Beta‐ and Gammaproteobacteria ammonia‐oxidizers, such as the Nitrosomonas europaea/Nitrosomonas eutropha lineage (Beta), the Nitrosococcus mobilis lineage (Beta), the Nitrosomonas marina cluster (Beta) and the Nitrosococcus group (Gamma). Also, heterotrophic microorganisms have been reported to oxidize nitrogen compounds under very specific conditions (Kim et al., 2005). In addition, Nitrobacter and Nitrospira have been recognized as nitrite‐oxidizers (Wagner et al., 2002). Recently, ammonia‐oxidizing organisms belonging to the archaeal domain have also been described (You et al., 2009). The denitrification process, i.e. the removal of nitrate to the atmosphere, seems to be mainly done by members of the genera Alcaligenes, Pseudomonas, Methylobacterium, Bacillus, Paracoccus, Hyphomicrobium, as well as by many members of the betaproteobacterial order Rhodocyclales (Wagner et al., 2002; Hosselhoe et al., 2009).
In our study, the seawater‐processing wwtp was known to have a nitrogen sludge load of 150–170 kg h−1, a nitrification fraction of 98% and a total nitrogen removal over 80% (M.I. Maldonado, pers. comm.). Thus, nitrification and denitrification are important processes in this system. Although amplification from beta ammonia‐oxidizers was detected in the two samples, we have not been able to find sequences corresponding to recognized microorganisms known to catalyse the oxidation of ammonia to nitrite or of nitrite to nitrate in freshwater wwtps. Sequences from Betaproteobacteria (to which a diversity of nitrifiers from freshwater wwtps belong) have not been recovered from the DGGE gels and the clone library, although CARD‐FISH analyses showed a certain amount of bacteria belonging to this group (3% of total DAPI‐stained cells). It is possible that the DNA extraction technique applied was not sufficient rigorous to lyse the cells of ammonia–oxidizers in the activated sludge, or that PCR or cloning biases occurred. However, hybridization with specific probes for beta ammonia‐oxidizers showed no signal, suggesting that the microorganisms carrying out this function in our samples could not be assigned to any of the well‐known lineages from freshwater activated sludges. Thus, nitrifiers in this specific seawater‐processing wwtp correspond to different genera. In fact, it has been shown that several heterotrophic Bacillus strains can carry out aerobic nitrification, as well as denitrification (Kim et al., 2005), and we have found sequences of Bacillus in the clone library (although with a low similarity) and also by culture‐dependent techniques (data not shown). Actually, Bacillus strains are able to remove nitrogen and phosphorus as well as organic matter. On the other hand, it is also possible that, although being crucial for nitrogen removal, the well‐known nitrifiers could not be detected due to their low abundance. Concerning the Gammaproteobacteria, no amplification of the amoA gene was found and most of the sequences recovered in this study corresponded to unidentified clones.
In contrast, sequences of Nitratireductor sp., able to reduce nitrate to nitrite, have been retrieved from the DGGE and the bacterial clone library. However, most candidates for denitrifying bacteria in this work have been found by culture‐dependent approaches (data not shown). Thus, members of the genera Alcaligenes, Pseudomonas, Bacillus, Paracoccus, Halomonas and Marinobacter have been isolated in rich media, although we do not know whether these genera are representative for the in situ active denitrifiers of this system.
Recently, it has been shown that autotrophic oxidation of ammonia is not restricted to the domain Bacteria. Könneke and colleagues (2005) isolated an ammonia‐oxidizing crenarchaeon named Nitrosopumilus maritimus able to oxidize ammonia to nitrite under mesophilic conditions, and Park and colleagues (2006) reported molecular evidence that ammonia‐oxidizing archaea occur in activated sludge bioreactors used to remove ammonia from wastewater. However, amplification of the archaeal amoA was not found in our samples. Nevertheless, it is important to note that significant diversity exists in each of these functional groups of organisms and that a detailed knowledge of their biology needs to be gained.
Correspondence between DGGE, clone library and FISH
The comparison between clone library, DGGE and FISH results is not straightforward because of the different levels of phylogenetic resolution of each technique. There is general agreement regarding the limitations of each methodology (Amann et al., 1995; Wintzingerode et al., 1997), but few studies have compared these techniques in activated sludge systems (Snaidr et al., 1997; Juretschko et al., 2002; Eschenhagen et al., 2003), and none has compared them in seawater‐processing activated sludge.
DGGE allowed an assessment of the composition of the prokaryotic assemblage of the activated sludge sample with sufficient resolution. However, a failure to obtain sequences from faint bands prevents the use of DGGE for describing bacterial diversity accurately (Sánchez et al., 2009). Because not all bands were sequenced, it cannot be discarded that differences between community composition shown by DGGE and by other techniques are due to insufficient sequencing. On the other hand, the clone library provided the highest phylogenetic resolution and a detailed picture of the species within each phylogenetic group. However, PCR bias and the varying copy number of the rRNA operon in different organisms produced severe overestimations (Deinococcus–Thermus) or underestimations (Alphaproteobacteria) of specific groups compared with the direct quantification obtained by CARD‐FISH. On the other hand, the picture of the bacterial assemblage composition provided by CARD‐FISH was limited by the number and phylogenetic resolution of the probes. A substantial proportion of the Eub338‐II‐III positive cells remained unidentified by the general probes used, while no cell remained undetectable.
Although DGGE is also subject to PCR bias, in our study the group proportion with this technique was more similar to what was found for CARD‐FISH than to clone libraries. This discrepancy was also shown by Massana and colleagues (2006), who observed that clone libraries obtained with a primer set amplifying one‐third of the 18S rRNA gene from eukaryotes (the set that is regularly used in DGGE studies) provided very good correlation between clonal representation and cell abundance determined by FISH. In contrast, the primer set amplifying the complete 18S rRNA gene gave a very biased view of the phylogenetic groups under study when compared with FISH abundance, with some phylotypes being severely overestimated and others underestimated.
Snaidr and colleagues (1997), however, despite using a primer set which amplified almost‐full‐length 16S rRNA gene fragments, found a general agreement between clone library and FISH when analysing the bacterial community structure of activated sludge from a municipal wwtp, although discrepancies became clear when using more specific probes. In their study, almost 20% of DAPI cells remained undetected.
In summary, the combination of the three techniques was very useful for assessing a comprehensive appraisal of prokaryotic diversity, and thus a polyphasic approach is essential to have a complete picture of the prokaryotic assemblage. These methods also showed that this particular activated sludge can contain significant hidden diversity of unknown and uncultured marine‐related microorganisms that can contribute to its functioning. Therefore, further attempts to isolate the key microorganisms involved will be essential in order to understand their specific biological processes.
Experimental procedures
Sampling
Samples of aerated mixed activated sludge from a seawater processing wastewater treatment plant located in Almeria (southeast Spain) were collected in December 2007 and November 2008 in a 1 l sterile bottle and stored at 4°C until processing. The plant treats wastewater from a pharmaceutical industry and the performance of the reactor is constant in function of time, with a continuous entrance of intermediate products from amoxicillin synthesis (between 0–250 mg l−1). The mean influent flow of the plant is 300 m3 h−1 and has a treatment volume of 32 000 m3. Nitrogen and chemical oxygen demand (COD) sludge loads were about 150–170 kg h−1 and 900–1000 kg h−1 respectively. Ionic concentrations in the influent were as follows: NH4+: 0.6–1.1 g l−1; K+: 121.1 mg l−1; Mg2+: 97.3 mg l−1; Ca2+: 386.6 mg l−1; NO3‐: 0 g l−1; NO2‐: 0 mg l−1; SO42−: 4–6.5 g l−1 and PO43−: 3.5–5.4 g l−1. In the efluent, the ionic concentrations of nitrogen compounds were: NH4+: 0–40 mg l−1; NO3‐: 200–600 mg l−1; NO2‐: 0–20 mg l−1; total nitrogen: 100–250 mg l−1; being the total nitrogen and COD removals above 80% and 90% respectively.
DNA extraction and PCR amplification
Fifty‐millilitre samples were centrifuged and the pellets were stored at −20°C until use. Upon thawing, community DNA was extracted using the DNA Power Soil kit from MOBIO (12888‐50).
Fragments of the bacterial 16S rRNA gene suitable for DGGE analysis were obtained by using the specific primer 358F with a 40 bp GC clamp, and the universal primer 907RM (Sánchez et al., 2007). PCR was carried out with a Biometra thermocycler using the following program: initial denaturation at 94°C for 5 min; 10 touchdown cycles of denaturation (at 94°C for 1 min), annealing (at 63.5°C to 53.5°C for 1 min, decreasing 1°C each cycle), and extension (at 72°C for 3 min); 20 standard cycles (annealing at 53.5°C, 1 min) and a final extension at 72°C for 5 min.
Primers 344f‐GC and 915R were used for archaeal 16S rRNA amplification (Casamayor et al., 2002). The PCR protocol included an initial denaturation step at 94°C for 5 min, followed by 20 touchdown cycles of denaturation (at 94°C for 1 min), annealing (at 71°C to 61°C for 1 min, decreasing 1°C each cycle), and extension (at 72°C for 3 min); 20 standard cycles (annealing at 55°C, 1 min) and a final extension at 72°C for 5 min.
PCR mixtures for 16S rRNA amplification contained 1–10 ng of template DNA, each deoxynucleoside triphosphate at a concentration of 200 µM, 1.5 mM MgCl2, each primer at a concentration of 0.3 µM, 2.5 U Taq DNA polymerase (Invitrogen) and PCR buffer supplied by the manufacturer. BSA (bovine serum albumin) at a final concentration of 600 µg ml−1 was added to minimize the inhibitory effect of humic substances (Kreader, 1996). The volume of reactions was 50 µl. PCR products were verified and quantified by agarose gel electrophoresis with a low DNA mass ladder standard (Invitrogen).
Primers amoA‐1F and amoA‐2R were used for amplification of ammonia oxidizers of the Beta subclass of Proteobacteria (Rotthauwe et al., 1997). The PCR protocol included an initial denaturation step at 94°C for 5 min, followed by 42 cycles of denaturation (at 94°C for 60 s), annealing (at 60°C for 90 s) and extension (at 72°C for 90 s), and a final step consisting of 90 s at 60°C and 10 min at 72°C.
For detection of ammonia oxidizers of the gamma‐subclass of Proteobacteria, primers amoA‐3F and amoB‐4R were utilized (Purkhold et al., 2000). Thermal cycling was carried out by an initial denaturation step at 94°C for 30 s, followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 48°C for 20 s, and elongation at 72°C for 40 s. Cycling was completed by a final elongation step at 72°C for 10 min.
The presence of archaeal amoA fragments was checked by using the primers Arch‐amoAF and Arch‐amoAR (Francis et al., 2005) with the following protocol: 95°C for 4 min, 30 cycles consisting of 94°C for 30 s, 56°C for 30 s and 72°C for 60 s, and a final step of 72°C for 10 min.
PCR mixtures for amplification of the amoA gene contained 1 µl of template DNA, each deoxynucleoside triphosphate at a concentration of 200 µM, 1.5 mM MgCl2, each primer at a concentration of 0.3 µM, 1.25 U Taq DNA polymerase (Promega) and PCR buffer supplied by the manufacturer. For archaeal amoA amplification, BSA at a final concentration of 150 µg ml−1 was added. The volume of reactions was 25 µl. PCR products were verified and quantified by agarose gel electrophoresis with a low DNA mass ladder standard (Invitrogen).
DGGE fingerprinting
DGGEs were run in a DCode system (Bio‐Rad) as described by Muyzer and colleagues (1998). A 6% polyacrylamide gel with a gradient of 30–70% (Bacteria) or 40–80% (Archaea) DNA‐denaturant agent was cast by mixing solutions of 0% and 80% denaturant agent (100% denaturant agent is 7 M urea and 40% deionized formamide). Seven hundred nanograms of PCR product was loaded for each sample, and the gels were run at 100 V for 18 h at 60°C in 1× TAE buffer [40 mM Tris (pH 7.4), 20 mM sodium acetate, 1 mM EDTA]. The gel was stained with SybrGold (Molecular Probes) for 45 min, rinsed with 1× TAE buffer, removed from the glass plate to a UV‐transparent gel scoop, and visualized with UV in a Gel Doc EQ (Bio‐Rad). Prominent bands were excised from the gels, resuspended in milli‐q water overnight and re‐amplified for its sequencing.
Clone libraries
Bacterial 16S rRNA was amplified using universal primers 27F and 1492R (Lane, 1991). Reactions were carried out in an automated thermocycler (Biometra) with the following cycle: an initial denaturation step at 94°C for 5 min, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C and 2 min at 72°C, and a final extension step of 10 min at 72°C.
Primers 109F and 915R were used for Archaeal amplification (Großkopf et al., 1998). The cycle was as follows: 5 min at 94°C, 38 cycles consisting of primer annealing at 52°C for 1 min, DNA elongation at 72°C for 90 s and denaturation at 94°C for 1 min, and a final cycle of 52°C for 1 min and 72°C for 6 min.
PCR mixtures contained 1–10 ng of template DNA, each deoxynucleoside triphosphate at a concentration of 200 µM, 1.5 mM MgCl2, each primer at a concentration of 0.3 µM, 2.5 U Taq DNA polymerase (Invitrogen) and PCR buffer supplied by the manufacturer.
PCR products were cloned with the TOPO TA cloning kit (Invitrogen) according to the manufacturer's instructions. Putative colonies were picked, transferred to a multi‐well plate containing Luria–Bertani medium and 7% glicerol, and stored at −80°C for further amplification and sequencing.
rRNA sequencing
Purification of PCR products and sequencing reactions from DGGE bands and clones were performed by Macrogen (South Korea) with primers 907rM (DGGE), 27F (bacterial clone library) and 109F (archaeal clone library). Macrogen utilized the Big Dye Terminator version 3.1 sequencing kit and reactions were run in an automatic ABI 3730XL Analyzer‐96 capillary type. Gene sequences were deposited in GenBank under accession numbers FN597722‐FN597999 and FN598017‐FN598150.
Sequences were subjected to a blast search (Altschul et al., 1997) to obtain an indication of the phylogenetic affiliation, and to the Bellerophon program (Huber et al., 2004) to determine potential chimeric artefacts. Sequences sharing similarities over 98.5% were considered similar phylogenetic entities (OTU). The coverage of the clone libraries was calculated according to the following equation: C = 1 − (n/N), where n is the number of unique clones and N is the total number of clones examined. Seventeen sequences (accession numbers: FN598000–FN598016), corresponding to three OTU from the Deinoccoccus–Thermus group (representative clones: BACDER07_1B5, BACDER07_1G6 and BACDER07_1F5), were fully sequenced using primers 27F and 1492R for probe design purposes.
Phylogenetic analyses
Partial 16S rDNA sequences from each OTU of the archaeal clone library were aligned by using MAFFT version 6 (http://align.bmr.kyushu‐u.ac.jp/mafft/online/server/) with a selection of euryarchaeal sequences from databases (including the closest matches obtained by blast search) and three crenarchaeal sequences as an outgroup. Very variable regions of the alignment were automatically removed with Gblocks (Castresana, 2000), using parameters optimized for rDNA alignments (minimum length of a block of 5; allowing gaps in half positions), leaving 695 informative positions. Maximum‐likelihood analysis was carried out with PAUP 4.0b10 (Swofford, 2002), with the general time‐reversible model assuming a discrete gamma distribution with six rate categories and a proportion of invariable sites. Parameters were estimated from an initial neighbour‐joining tree. Bayesian analysis was carried out with MrBayes v3.0B (Huelsenbeck and Ronquist, 2001), using the same model described above but with four rate categories in the gamma distribution. Bayesian posterior probabilities were computed by running 2 000 000 generations by using the program default priors on model parameters. Trees were sampled every 100 generations. A total of 3000 trees were discarded as ‘burn‐in’ upon examination of the log likelihood curve of the sampled trees, so only the stationary phase was considered in the final tree. Neighbour‐joining bootstrap values from 1000 replicates were calculated with PAUP following the same model used for the maximum‐likelihood analysis.
In situ hybridization
For separation of the sludge flocs, the original sample was vortexed during 5 min and subsequently diluted, fixed with formaldehyde and filtered on a 0.2 µm pore‐size polycarbonate filter. CARD‐FISH of prokayotic populations was carried out following the protocol described by Pernthaler and colleagues (2004). Several horseradish peroxidase probes were used to characterize the composition of the prokaryotic assemblage in activated sludge: CREN554 (Massana et al., 1997), EURY806 (Teira et al., 2004), EUB 338‐II‐III (Amann et al., 1990; Daims et al., 1999), ALF968 (Neef, 1997), GAM42a (Manz et al., 1992), CF319 (Manz et al., 1996), BET42a (Manz et al., 1992), DELTA495a (Loy et al., 2002), LGC354B (Meier et al., 1999) and HGC69a (Roller et al., 1994). The EUB antisense probe NON338 (Wallner et al., 1993) was used as a negative control. The probe DT01 (5′‐ACCAAGCGCATCACACCG‐3′) targeting the clones BACDER07_1B5 and BACDER07_1G6 from the Deinococcus–Thermus phylum, was newly designed in this study by using the PROBE_DESIGN tool of the ARB software package (http://www.arb‐home.de), and optimized following the protocol described in Pernthaler and colleagues (2001). This probe does not target the clone BACDER07_1F5.
Fluorescence in situ hybridization of nitrifying bacteria was carried out following the protocol detailed by Pernthaler and colleagues (2001). The 16S rRNA‐targeted oligonucleotide probes used were: NEU, complementary to a signature region of most halophilic and halotolerant ammonia oxidizers, Nso190 and Nso1225, specific for ammonia oxidizers in the Beta subclass of Proteobacteria, NIT3, complementary to a region of Nitrobacter species, and Nsv443, specific for the Nitrosospira cluster (Juretschko et al., 1998)
All probes were purchased from Thermo Fisher Scientific (Ulm, Germany). Filters were permeabilized with lysozyme (10 mg ml−1, 37°C, 1 h) and achromopeptidase (60 U ml−1, 37°C, 0.5 h) before hybridization. Hybridizations were carried out at 35°C overnight and specific hybridization conditions were established by addition of formamide to the hybridization buffers (20% formamide for NON338 and EURY806 probes, 30% for Nsv443, 35% for Nso1225, 40% for NEU and NIT3, 45% for ALF968 and LGC354B, 50% for Delta495a and HGC69a, and 55% for the other probes). The optimal hybridization conditions (30% formamide) of the newly designed probe DT01 were experimentally determined. Counterstaining of CARD‐FISH preparations was done with DAPI (1 µg ml−1). Free cells and aggregates were counted separately in each field. Also, several transects were inspected and mean numbers of aggregates were calculated. Between 500 and 1000 free DAPI‐positive cells were counted manually in a minimum of 10 fields, while several thousands of cells (between 4000 and 10 000) were counted in aggregates.
Acknowledgments
This work was supported by the Spanish projects Consolider TRAGUA (CSD2006‐00044), MICRORESP (PET2008‐0165‐02) and GEMMA (CTM2007‐63753‐C02‐01/MAR). We thank P. López for her lab support.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R., Stahl D.A. Combination of 16S rRNA‐targeted oligonucleotice probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R.I., Ludwig W., Schleifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Lemmer H., Wagner J. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol Ecol. 1998;25:205–215. [Google Scholar]
- Bond P.L., Hugenholtz P., Keller J., Blackhall L.L. Bacterial community structures of phosphate‐removing and non‐phosphate.removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond P.L., Erhart R., Wagner M., Keller J., Blackall L.L. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol. 1999;65:4077–4084. doi: 10.1128/aem.65.9.4077-4084.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon N., De Windt W., Verstraete W., Top E.M. Evaluation of nested PCR‐DGGE (denaturing gradient gel electrophoresis) with group‐specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol Ecol. 2002;39:101–112. doi: 10.1111/j.1574-6941.2002.tb00911.x. [DOI] [PubMed] [Google Scholar]
- Casamayor E.O., Massana R., Benlloch S., Øvreás L., Díez B., Goddard V.J. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods ina multipond solar saltern. Environ Microbiol. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. et al. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Christensson M., Blackall L.L., Welander T. Metabolic transformation and characterisation of the sludge community in an enhanced biological phosphorus removal system. Appl Microbiol Biotechnol. 1998;49:226–234. [Google Scholar]
- Coskuner G., Curtis T.P. In situ characterization of nitrifiers in an activated sludge plant: detection of Nitrobacter Spp. J Appl Microbiol. 2002;93:431–437. doi: 10.1046/j.1365-2672.2002.01715.x. [DOI] [PubMed] [Google Scholar]
- Dabert P., Sialve B., Delgenes J.P., Moletta R., Godon J.J. Characterisation of the microbial 16S rDNA diversity of an aerobic phosphorus‐removal ecosystem and monitoring of its transition to nitrate respiration. Appl Microbiol Biotechnol. 2001;55:500–509. doi: 10.1007/s002530000529. [DOI] [PubMed] [Google Scholar]
- Daims H., Bruhl A., Amann R., Schleifer K.H., Wagner M. The domain‐specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Daims H., Nielsen J.L., Nielsen P.H., Schleifer K.H., Wagner M. In situ characterization of Nitrospira‐like nitrite‐oxidation bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner C.A., Erb R.W., Timmis K.N., Wagner‐Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65:102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenhagen M., Schuppler M., Röske I. Molecular characterization of the microbial community structure in two activated sludge systems for the advanced treatment of domestic eflluents. Water Res. 2003;37:3224–3232. doi: 10.1016/S0043-1354(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Francis C.A., Roberts K.J., Beman M., Santoro A.E., Oakley B.B. Ubiquity and diversity of ammonia‐oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;41:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.D., Miskin E.P., Kornilova O., Curtis T.P., Head I.M. Occurrence and activity of Archaea in aerated activated sludge wastewater treatment plants. Environ Microbiol. 2002;4:158–168. doi: 10.1046/j.1462-2920.2002.00280.x. [DOI] [PubMed] [Google Scholar]
- Großkopf R., Janssen P.H., Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosselhoe M., Füreder S., Schloter M., Brodossy L., Iversen N., Roslev P. Isotope array analysis of Rhodocyclales uncovers functional redundancy and versatility in an activated sludge. ISME J. 2009;3:1349–1364. doi: 10.1038/ismej.2009.78. et al. [DOI] [PubMed] [Google Scholar]
- Huber T., Faulkner G., Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jeon C.O., Lee D.S., Park J.M. Microbial communities in activated sludge performing enhanced biological phosphorus removal in a sequencing batch reactor. Water Res. 2003;37:2195–2205. doi: 10.1016/S0043-1354(02)00587-0. [DOI] [PubMed] [Google Scholar]
- Juretschko S., Timmermann G., Schmid M., Schleifer K.‐H., Pommerening‐Roser A., Koops H.‐P., Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira‐like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretschko S., Loy A., Lehner A., Wagner M. The microbial community composition of a nitrifying‐denitrifying activated sludge from an industrial sewage treatment plant by the full‐cycle rRNA approach. Syst Appl Microbiol. 2002;25:84–99. doi: 10.1078/0723-2020-00093. [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Erhart R., Beimfohr C., Böhringer J., Wagner M., Amann R. Characterization of bacterial communities from activated sludge: culture‐dependent numerical identification versus in situ identification using group‐ and genus‐specific rRNA‐targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- Kim J.K., Park K.P., Cho K.S., Nam S.W., Park T.J., Bajpai R. Aerobic nitrification‐denitrification by heterotrophic Bacillus strains. Bioresource Technol. 2005;96:1897–1906. doi: 10.1016/j.biortech.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Könneke M., Bernhard A.E., de la Torre J.R., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia‐oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Kraigher B., Kosjek T., Heath E., Kompare B., Mandic‐Mulec I. Influence of pharmaceutical residues on the structure of activated sludge bacterial communities in wastewater treatment bioreactors. Water Res. 2008;42:4578–4588. doi: 10.1016/j.watres.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Kreader C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Laskin A.I., White D.C. Preface to special issue on Sphingomonas. J Ind Microbiol Biotechnol. 1999;23:231. doi: 10.1038/sj.jim.2900748. [DOI] [PubMed] [Google Scholar]
- Liu Y., Seviour R.J. Design and application of oligonucleotide probes for fluorescent in situ indentification of the filamentous bacterial morphotype Nostocoida limicola in activated sludge. Environ Microbiol. 2001;3:551–560. doi: 10.1046/j.1462-2920.2001.00229.x. [DOI] [PubMed] [Google Scholar]
- Loy A., Lehner A., Lee N., Adamczyk J., Meier H., Ernst J. Oligonucleotide microarray for 16S rRNA gene‐based detection of all recognized lineages of sulfate‐reducing prokaryotes in the environment. Appl Environ Microbiol. 2002;68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W., Amann R., Ludwig W., Wagner M., Schleifer H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- Manz W., Amann R., Ludwig W., Vancanneyt M., Schleifer H. Application of a suite of 16S rRNA‐specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga‐Flavobacter‐Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- Massana R., Murray A.E., Preston C.M., DeLong E.F. Vertical distribution and phylogenetic characterization of marine planktonic Archae in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R., Guillou L., Terrado R., Forn I., Pedrós‐Alió C. Growth of uncultured heterotrophic flagellates in unamended seawater incubations. Aquat Microb Ecol. 2006;45:171–180. [Google Scholar]
- Meier H., Amann R., Ludwig W., Schleifer K.‐H. Specific oligonucleotide probes for in situ detection of a major group of gram‐positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- Muyzer G., Brinkhoff T., Nübel U., Santegoeds C., Schäfer H., Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A.D.L., van Elsas J.D., Bruijn F.J., editors. Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- Neef A. 1997. ) Anwendung der in situ‐Einzelzell‐Identifizierung von Bakterien zur Populationsanlayse in komplexen mikrobiellen biozönosen. PhD Thesis. Munich, Germany: Technische Universität München.
- Neef A., Witzenberger R., Kämpfer P. Detection of sphingomonads and in situ identification in activated sludge using 16S rRNA‐targeted oligonucleotide probes. J Ind Microbiol Biotechnol. 1999;23:261–267. doi: 10.1038/sj.jim.2900768. [DOI] [PubMed] [Google Scholar]
- Otawa K., Asano R., Ohba Y., Sasaki T., Kawamura E., Koyama F. Molecular analysis of ammonia‐oxidizing bacteria community in intermittent aeration sequencing batch reactors used for animal wastewater treatment. Environ Microbiol. 2006;8:1985–1996. doi: 10.1111/j.1462-2920.2006.01078.x. et al. [DOI] [PubMed] [Google Scholar]
- Park H.‐D., Wells G.F., Bae H., Criddle C.S., Francis C.A. Occurrence of ammonia‐oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J., Glöckner F.O., Schönhuber W., Amann R. Fluorescence in situ hybridization (FISH) with rRNA‐targeted oligonucleotide probes. In: Paul J.H., editor. Academic Press; 2001. pp. 207–226. [Google Scholar]
- Pernthaler A., Pernthaler J., Amann R. Sensitive multi‐color fluorescence in situ hybridization for the identification of environmental microorganisms. In: Akkermans A.D.L., de Bruijn F.J., van Elsas J.D., editors. 2nd. Kluwer Academic Publishers; 2004. pp. 711–726. [Google Scholar]
- Purkhold U., Pommerening‐Röser A., Juretschko S., Schmid M.C., Koops H.P., Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé M.S., Giovannoni S.J. The uncultured microbial diversity. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Robertson C.E., Spear J.R., Harris J.K., Pace N.R. Diversity and stratification of Archaea in a hypersaline microbial mat. Appl Environ Microbiol. 2009;75:1801–1810. doi: 10.1128/AEM.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller C., Wagner M., Amann R., Ludwig W., Schleifer K.‐H. In situ probing of Gram‐positive bacteria with a high DNA G+C content. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- Rotthauwe J.H., Witzel K.P., Liesack W. The ammonia monooxigenase structural gene amoA as a functional marker: molecular fine‐scale analysis of natural ammonia‐oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikaly P.E., Stroot P.G., Oerther D.B. Use of 16S rRNA gene terminal restriction fragment analysis to assess the impact of solids retention time on the bacterial diversity of activated sludge. Appl Environ Microbiol. 2005;71:5814–5822. doi: 10.1128/AEM.71.10.5814-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanapareddy N., Hamp T.J., Gonzalez L.C., Hilger H.A., Fodor A.A., Clinton S.M. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol. 2009;75:1688–1696. doi: 10.1128/AEM.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez O., Gasol J.M., Massana R., Mas J., Pedrós‐Alió C. Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities. Appl Environ Microbiol. 2007;73:5962–5967. doi: 10.1128/AEM.00817-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez O., Gasol J.M., Balagué V., Massana R., Mas J., Pedrós‐Alió C. Influence of primer mismatch and microdiversity on DGGE results: a case study with SAR. Aquat Microb Ecol. 2009;54:211–216. [Google Scholar]
- Schramm A., Santegoeds C.M., Nielsen H.K., Ploug H., Wagner M., Pribyl M. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl Env Microbiol. 1999;65:4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seviour R.J., Mino T., Onuki M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol Ecol. 2003;27:99–127. doi: 10.1016/S0168-6445(03)00021-4. [DOI] [PubMed] [Google Scholar]
- Snaidr J., Amann R., Huber I., Ludwig W., Schleifer K.‐H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goebel B.M. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present definition of bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Swofford D.L. Sinauer Associates; 2002. [Google Scholar]
- Teira E., Reinthaler T., Pernthaler A., Pernthaler J., Herndl G. Combining catalyzed reported deposition‐fluorescence in situ hybridization and microautoradiography to detect substrate utilization by Bacteria and Archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Amann R., Lemmer H., Schleifer K.‐H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture‐dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Aβmuss B., Hartmann A., Hutzler P., Amann R. In situ analysis of microbial consortia in activated sludge using fluorescently labelled, rRNA‐targeted oligonucleotide probes and confocal laser scanning microscopy. J Microsc. 1994;176:181–187. doi: 10.1111/j.1365-2818.1994.tb03513.x. [DOI] [PubMed] [Google Scholar]
- Wagner M., Loy A., Nogueira R., Purkhold U., Lee N., Daims H. Microbial community composition and function in wastewater treatment plants. Antonie van Leeuwenhoek. 2002;81:665–680. doi: 10.1023/a:1020586312170. [DOI] [PubMed] [Google Scholar]
- Wallner G., Amann R., Beisker W. Optimizing fluorescent in situ hybridization with rRNA‐targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- Wintzingerode F., Göbel U.B., Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR‐based rRNA analysis. FEMS Microb Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- You J., Das A., Dolan E.M., Hu Z. Ammonia‐oxidizing archaea involved in nitrogen removal. Water Res. 2009;43:1801–1809. doi: 10.1016/j.watres.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Zheng D. Quantification of Methanosaeta species in anaerobic bioreactors using genus‐and species‐specific hybridization probes. Microbial Ecol. 2000;39:246–262. doi: 10.1007/s002480000003. [DOI] [PubMed] [Google Scholar]
- Zuo G., Roberts D.J., Lehman S.G., Jackson G.W., Fox G.E., Willson R.C. Molecular assessment of salt‐tolerant, perchlorate‐ and nitrate‐ reducing microbial cultures. Water Sci Technol. 2009;60:1745–1756. doi: 10.2166/wst.2009.635. [DOI] [PubMed] [Google Scholar]


