Abstract
Neuraminidases (sialidases) catalyze the removal of terminal sialic acid from glycoconjugates. Bacterial pathogens often utilize neuraminidases to scavenge host sialic acid, which can be utilized either as a nutrient or as a decorating molecule to disguise themselves from host immune attacks. Herein, a putative neuraminidase (TDE0471) was identified in Treponema denticola, an oral spirochete associated with human periodontitis. TDE0471 is a cell surface-exposed exo-neuraminidase that removes sialic acid from human serum proteins; it is required for T. denticola to grow in a medium that mimics gingival crevice fluid, suggesting that the spirochete may use sialic acid as a nutrient in vivo. TDE0471 protects T. denticola from serum killing by preventing the deposition of membrane attack complexes on the bacterial cell surface. Animal studies revealed that a TDE0471-deficient mutant is less virulent than its parental wild-type strain in BALB/C mice. However, it causes a level of tissue damage similar to the wild type in complement-deficient B6.129S4-C3tm1Crr/J mice albeit the damage caused by both bacterial strains is more severe in these transgenic mice. Based on these results, we propose that T. denticola has evolved a strategy to scavenge host sialic acid using its neuraminidase, which allows the spirochete to acquire nutrients and evade complement killing.
Keywords: Periodontitis, Treponema denticola, Neuraminidase, Sialic acid, Complement system
INTRODUCTION
The complement system plays a central role in host innate immune defense against microbial infections and also functions as a mediator between the innate and adaptive immune responses [for recent reviews, see (Lambris et al., 2008;Ricklin et al., 2010;Zipfel and Skerka, 2009)]. The complement cascade is activated primarily via classical, lectin, and alternative pathways. Activation of the complement system ultimately leads to the formation of the membrane attack complex (MAC), which directly disrupts targeted pathogens via cell lysis. In addition, numerous complement factors that are generated during cascade activation are involved in triggering and orchestrating microbial opsonization, phagocytosis, stimulation of the adaptive immune system, and activation of inflammatory cells. Due to its ability to rapidly recognize and eliminate microorganisms, the complement system is considered as a first line of defense against microbial intruders.
Human periodontitis is a prevalent chronic inflammatory disease that damages the supporting connective tissues around teeth and ultimately leads to tooth loss (Darveau, 2010;Pihlstrom et al., 2005). The disease is a polymicrobial infection caused by a diverse group of periodontal pathogens (Hajishengallis et al., 2012a;Pihlstrom et al., 2005). Accumulating evidence suggests that the interactions between the complement system and those periodontal pathogens play a pivotal role in periodontal inflammation and the progression of the disease (Hajishengallis, 2010;Wang et al., 2010;Hajishengallis et al., 2012b). Among those periodontal pathogens, Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola (often referred to as ‘red-complex’ bacteria) have been considered as primary etiological agents of periodontitis (Socransky and Haffajee, 2005;Holt and Ebersole, 2005). This group of bacteria predominantly inhabits periodontal pockets containing gingival crevice fluid (GCF) (Lamont and Jenkinson, 2000;Holt and Ebersole, 2005;Ellen and Galimanas, 2005). The main component (up to 70%) of GCF is plasma, which contains the complement system (Schenkein and Genco, 1977b;Schenkein and Genco, 1977a;Hajishengallis, 2010). Thus, the ‘red-complex’ bacteria presumably are exposed to continuous immune surveillance mediated by the complement system. However, this group of bacteria has evolved different mechanisms to subvert the killing. P. gingivalis suppresses complement killing by producing capsules and degrading key complement factors (i.e., IgG and C3) via cysteine proteases (Popadiak et al., 2007;Schenkein et al., 1995). The oral spirochete T. denticola is resistant to the complement killing. Recent studies showed that FhbB, a surface-exposed lipoprotein that binds factor H (FH) and FH-like proteins, contributes to the complement resistance of T. denticola (McDowell et al., 2005;McDowell et al., 2009).
Sialic acid is a group of structurally related nine-carbon sugar acids that feature prominently at terminal positions of many eukaryotic surface-exposed glycoconjugates, where they are involved in a wide range of biological processes, including cell-cell interactions and small molecule-cell recognition (Vimr et al., 2004;Severi et al., 2007;Varki, 2007). Several bacterial pathogens have evolved to utilize sialic acid as decorating molecules to modify their surface-exposed macromolecules, such as lipopolysaccharides (LPS) and polysialic acid (PSA) capsules. Such modifications allow bacterial pathogens to disguise themselves and thus circumvent and/or counteract the host’s immune responses, including complement killing [for reviews, see (Severi et al., 2007;Vimr et al., 2004;Comstock and Kasper, 2006)]. Neuraminidase (sialidase) is a family of enzymes that catalyzes the removal of terminal sialic acid from glycoconjugates (Achyuthan and Achyuthan, 2001;Powell and Varki, 2001;Roggentin et al., 1993;Vimr, 1994). Various mucosal pathogens, ranging from Streptococcus pneumoniae in the airway to Vibrio cholerae in the gut, utilize neuraminidases to scavenge host sialic acid (Vimr, 1994;Severi et al., 2007;Galen et al., 1992). In these bacteria, neuraminidases are often associated with their virulence. For instance, the two neuraminidases of S. pneumoniae contribute to the progression of infection by promoting pneumococcal brain endothelial cell invasion (Soong et al., 2006;Uchiyama et al., 2009); and the neuraminidase of Pseudomonas aeruginosa plays a key role in the initial stages of pulmonary infection (Leprat and Michel-Briand, 1980;Cacalano et al., 1992).
Similar to other mucosal pathogens, the ‘red-complex’ bacteria also appear to have neuraminidase activity. P. gingivalis encodes at least one homologue (PG0352) of neuraminidase. Recent studies showed that PG0352 is involved in biofilm formation, capsule synthesis, serum resistance, and the pathogenicity of P. gingivalis (Aruni et al., 2011;Li et al., 2012). T. forsythia encodes two neuraminidases (NanH and SiaH). NanH (TF0035) is a major neuraminidase (Thompson et al., 2009); its deletion mutant fails to attach to and invade human gingival epithelial cells (Honma et al., 2011). T. denticola shows neuraminidase activity (Wyss et al., 2004). However, the enzyme(s) conferring this activity and its role in the pathogenicity of T. denticola have not been studied. T. denticola encodes a putative neuraminidase (TDE0471). In this report, we show experimental evidence that TDE0471 is a neuraminidase that removes sialic acid from human serum glycoconjugates and that it affects nutrient acquisition, complement activation, deposition of MAC, and the virulence of T. denticola.
RESULTS
TDE0471 is a neuraminidase
T. denticola appears to have neuraminidase activity (Wyss et al., 2004). However, the enzyme(s) responsible for this activity has not yet been identified. A Blast search of the T. denticola ATCC 35405 (hereafter referred to as Td35405) genome (Seshadri et al., 2004) using neuraminidase genes from other bacteria suggested that TDE0471 encodes a putative neuraminidase that consists of 543 amino acids (aa) and has a predicted molecular weight (MW) of 59.8 kDa. The TDE0471 gene is monocistronic. Transcriptional analyses indicated that it is regulated by a sigma70 transcription factor (Fig.S1). The C-terminus (aa 124 to 543) of TDE0471 contains a conserved neuraminidase catalytic domain. Sequence alignment analysis revealed that it contains two conserved functional motifs associated with bacterial neuraminidases (Copley et al., 2001;Crennell et al., 1993): one is the RIP motif and the other is a motif consisting of three Asp-boxes (S-X-D-X-G-X-T-W/F) (Fig.S2), suggesting that TDE0471 is a neuraminidase (hereafter referred to as Tdneu)
Neuraminidase activity can be detected by a filter paper spot sialidase test (Moncla and Braham, 1989;Moncla et al., 1990). In this assay, neuraminidases cleave 4-MUNANA, a fluorogenic substrate, to produce 4-methylumbelliferone (4-MU), which is fluorescent under ultraviolet light. The substrate treated with recombinant Tdneu (rTdneu) produced fluorescence (spot 1, Fig.1A), indicative of neuraminidase activity. The observed activity was completely abolished when the substrate was treated with rTdMneu (spot 2), a truncated rTdneu in which the C-terminal catalytic domain was deleted. The activity was substantially inhibited when rTdneu was added together with Neu5Ac2en (spots 3 and 4), a neuraminidase-specific inhibitor (Barrett et al., 2011;Trappetti et al., 2009). Further studies demonstrated that rTdneu is active from pH 4.0 to 8.0, with an optimum pH of 5.0 to 6.0 (Fig.1B). It has an affinity toward 4-MUNANA with a Km of 18.72 ± 4.24 nM and a rapid reaction rate with Vmax of 0.6 ± 0.03 nmol/nM/min (Fig.1C) under the tested conditions. These results indicate that TDE0471 is a neuraminidase with properties similar to its counterparts in other bacteria.
Figure 1. TDE0471 (Tdneu) enzymatic assay.
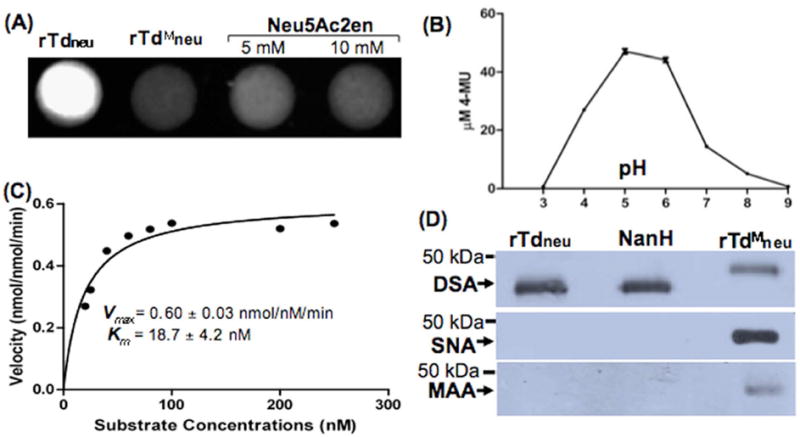
(A) Filter paper spot sialidase test of rTdneu. The substrate 4-MUNANA was treated with: rTdneu (spot 1, from the left), truncated rTdneu (rTdMneu, spot 2), and Neu5Ac2en (spots 3 and 4). The image was processed using the ChemiDoc XRS system (Bio-Rad) with an excitation wavelength of 302 nm and an emission wavelength of 548 nm. (B) rTdneu is active in a wide pH range. For this assay, the spot test was carried out from pH 3.0 to pH 9.0. The release of 4-MU (the product 4-MUNANA) was quantified based on a standard curve (fluorescence intensity vs. defined amounts of 4-MU). The data shown are the means from at least three samples. Error bars represent ± the standard error of the mean (SEM). The amounts of 4-MU (the product of 4-MUNANA) were determined using a standard curve. (C) Kinetic analysis of rTdneu. This assay was carried out according to a standard method for enzyme kinetics studies. The saturation curves were fitted to Michaelis-Menten kinetics using GraphPad Prism (GraphPad Software), and Km and Vmax were calculated. (D) Lectin blot analysis of rTdneu. The assay was performed as previously described (Gut et al., 2008;Li et al., 2012). Biotin-labeled SNA, MAA, and DSA lectins were used to detect terminal α2,6- and α2,3-linked sialic acid and galactose linked to GlcNAc of human α-1 acid glycoprotein (AGP), respectively. The C. perfringens neuraminidase NanH was used as a positive control, and rTdMneu was used as a negative control. Arrows point to the products detected by the three lectins.
Tdneu is an exo-neuraminidase
Based on substrate specificity, bacterial neuraminidases can be divided into two major classes (Taylor, 1996;Vimr et al., 2004): exo-neuraminidases that cleave α2,3-, α2,6-, and α2,8-glycosidic linkages of terminal sialic acids; and endo-neuraminidases that hydrolyze α2,8-sialosyl linkages in oligo- or poly-sialic acids. The substrate specificity of neuraminidases can be detected by lectin blot assays with human α-1acid glycoprotein (AGP) and three plant lectins: DSA specifically binds to galactose-linked GlcNAc, MAA to α2,3-linked sialic acid, and SNA to α2,6-linked sialic acid (Gut et al., 2008). For this assay, the recombinant NanH protein of Clostridium perfringens, a well-studied exo-neuraminidase (Cassidy et al., 1965;Chien et al., 1996), was used as a positive control. As shown in Fig.1D, rTdneu and NanH completely abolished the binding activity of SNA/MAA to AGP (lanes 1 and 2), but had no impact on that of DSA to AGP (top panel). Furthermore, rTdMneu did not affect the binding activity of SNA/MAA to AGP (lane 3). These results indicate that Tdneu is a NanH-like exo-neuraminidase that is able to cleave both α2,3- and α2,6-linked sialic acid in AGP.
Isolation and characterizations of a TDE0471 isogenic mutant and its complemented strain
To investigate the function of Tdneu, TDE0471 was inactivated by allelic exchange mutagenesis (Fig.S3A). Erythromycin-resistance colonies that appeared on the plates were first screened by PCR for the presence of the antibiotic resistance gene. One positive clone (Tde471mut) was selected for further characterization by PCR with different pairs of primers. the PCR results showed that the TDE0471 gene was inactivated as expected (Fig.S4A). Tde471mut was then complemented by inserting the full length TDE0471 into the erythromycin resistance cassette (ermR) on the chromosome of the mutant, as illustrated in Fig.S3B. The resultant complemented colonies became erythromycin sensitive and gentamicin resistant. Tde471com, a complemented clone, was selected for further characterization. Immunoblotting analysis using a specific antibody against Tdneu (αTdneu) showed that Tdneu was absent in Tde471mut and restored to the wild-type level in Tde471com (Fig.2A). The paper spot test using whole cell lysates of T. denticola strains showed that neuraminidase activity was detected in Td35405 and Tde471com, but not in Tde471mut (Fig.2B). These results indicated that the TDE0471 gene product and enzymatic activity were abrogated in Tde471mut and restored in Tde471com. In addition, as no residual neuraminidase activity was detected in Tde471mut (lane 2, Fig.2B), it is most likely that Tdneu is the only neuraminidase of T. denticola.
Figure 2. Characterizations of the TDE0471 mutant (Tde471mut) and its complemented strain (Tde471com).
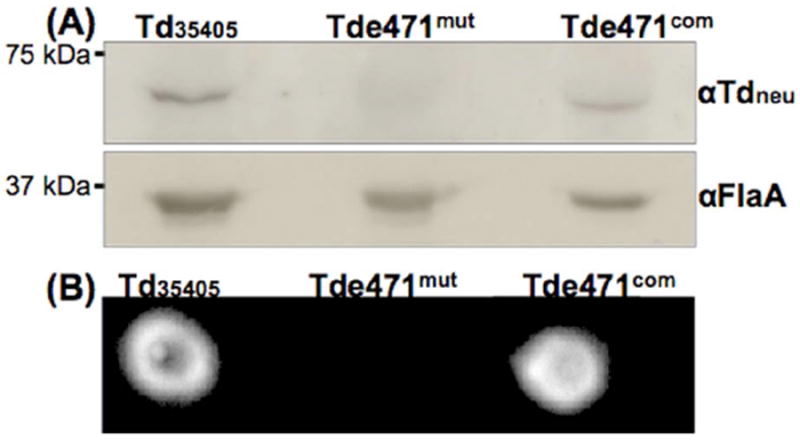
(A) Immunoblotting analysis of Td35405, Tde471mut, and Tde471com strains. Equivalent amounts of Td35405, Tde471mut, and Tde471com whole cell lysates were analyzed by SDS-PAGE and then probed with αTdneu, a specific antibody against Tdneu. The flagellin protein, FlaA, was used as a sample loading control and detected by αFlaA, a specific antibody against FlaA. (B) Filter paper spot test of Td35405, Tde471mut, and Tde471com strains. The assay was carried out as described in Figure 1A using the whole cell lysates of the three strains.
Tdneu is a cell surface-exposed protein
Signal peptide prediction using the SignalP 4.0 server suggested that the N-terminal 24 aa of Tdneu is a signal peptide (Fig.S5), implying that it is likely secreted. To test this prediction, the wild-type cells were first treated with proteinase K (PK) and the resultant cell pellets were analyzed by immunoblots. After the treatment, no trace of Tdneu was detected in the cell pellets (Fig.3A), suggesting that Tdneu resides on the cell surface and can be readily degraded by PK. This proposition was further confirmed by IFA studies using αTdneu, in which strong fluorescence signals were observed in the wild type cells, but not in the mutant cells (Fig.3B).
Figure 3. Localization of Tdneu.
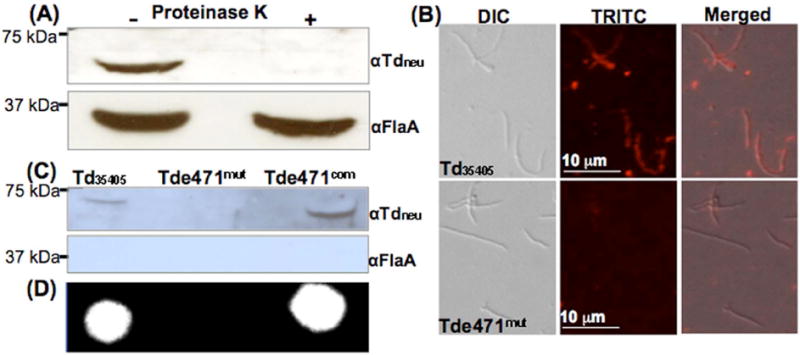
(A) Immunoblotting analysis of proteinase K-treated Td35405 whole-cell lysates. The spirochetes were either incubated with proteinase K (240 μg/ml) or PBS at 37°C for 1 h. The resultant samples were separated on SDS-PAGE and then probed with αTdneu and αFlaA. (B) IFA analysis. Td35405 and Tde471mut cells were fixed with methanol, stained with αTdneu, and counterstained with a goat-anti-rat Texas red antibody as previously described. The micrographs were taken under DIC light microcopy or fluorescence microscopy with a tetramethylrhodamine isothiocyanate (TRITC) emission filter, and the resultant images were merged. (C) Detection of Tdneu in the supernatants prepared from the cultures of Td35405, Tde471mut, and Tde471com strains by immunoblotting analysis. FlaA was used as a sample preparation control. (D) Detecting neuraminidase activity in the supernatants by the filter paper spot test as described in Figure 1A.
T. denticola surface-exposed proteins (e.g., the major surface protein, Msp) are often dissociated from cell surfaces and released to the culture media (Fenno et al., 1998;Bian et al., 2005). To determine if this is the case for Tdneu, supernatants prepared from the T. denticola cultures were analyzed by immunoblots to detect Tdneu and by the filter paper spot assay to test for the enzymatic activity. Consistent with the hypothesis, the protein and enzymatic activity were detected in the supernatants of Td35405 and Tde471com, but not in that of the mutant (Fig.3C, D). To rule out the possibility that the detected protein and enzymatic activity are due to cell lysis during the growth and sample preparation, FlaA, a flagellar sheath protein in the periplasmic space of T. denticola (Ruby et al., 1997), was used as a sample control; no trace of FlaA was detected in the supernatants (Fig.3C). Collectively, these results indicate that Tdneu is a surface-exposed protein that is readily dissociated from the cells and released into its growth milieus.
Tdneu cleaves sialic acid from human serum proteins
Total sialic acid in the plasma is abundant (approximately 2 mM). Under normal physiological conditions, almost all sialic acids (99.9%) are bound to a diverse range of glycoproteins and glycolipids (often named sialoglycoconjugates) (Ponnio et al., 1999;Sillanaukee et al., 1999). To determine if Tdneu is able to desialylate those sialoglycoconjugates, human serum was incubated with rTdneu and then probed with SNA and MAA. SNA and MAA bound to a variety of serum sialoglycoconjugates (Fig.4). The observed binding activities showed a dose-dependent reduction when the serum was treated with different amounts of rTdneu for 1 hour (Fig.4A), and the binding activities were almost completely abolished when the serum was co-incubated with 0.2 μg rTdneu for 3 hours (Fig.4B, C). Of note, there was no desialylation activity observed in the samples treated with rTdMneu (Fig.4B, C). In addition, the treatment of the serum with rTdneu had no impact on the binding activity of Concanavalin A (ConA) (Fig.4D), suggesting that rTdneu has no proteolytic activity against the serum proteins. These results indicate that the corresponding α2,3- and α2,6-linked terminal sialic acid residues on the serum sialoglycoconjugates were cleaved by Tdneu. This activity is consistent with the catalytic feature of Tdneu as an exo-neuraminidase, as shown in Fig.2.
Figure 4. Lectin blot analysis of rTdneu-treated human serum.
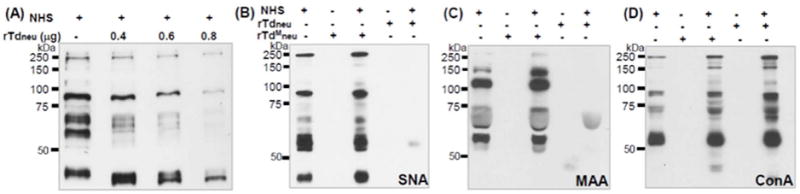
(A) Normal human serum (NHS) was treated with various amounts of rTdneu at 37°C for 1 h. The resultant samples were separated on SDS-PAGE gels followed by Coomassie blue staining; (B-D) The NHS was treated with 0.2 μg rTdneu or the same amount of rTdMneu at 37°C for 3 hours. The resultant samples were separated on SDS-PAGE gels, transferred to PVDF membranes, and probed with biotin-labeled SNA (B, 0.2 μg/ml), MAA (C, 2 μg/ml), or ConA (D, 0.5 μg/ml). The final concentrations of NHS in the reactions were 0.15% for SNA and ConA, and 1.5% for MAA.
The Tde471mut fails to grow in the serum growth medium
Several bacteria are able to utilize sialic acid as a nutrient when other carbohydrates are limited (Vimr et al., 2004;Severi et al., 2007). In the TYGVS medium, Td35405, Tde471mut and Tde471com strains exhibited similar growth patterns (Fig.5A), suggesting that the lack of Tdneu has no significant impact on T. denticola growth in a nutrient-enriched medium. As mentioned earlier, T. denticola primarily lives in the GCF, which contains 50~70% serum (Schenkein and Genco, 1977b;Schenkein and Genco, 1977a). To mimic this growth condition, the T. denticola strains were cultivated in serum growth medium that contains 50% heat-inactivated rabbit serum. In this medium, both Td35405 and Tde471com were able to grow and reached approximately 108 cells/ml at the stationary phase. However, Tde471mut completely failed to grow (Fig.5B). Interestingly, the mutant recovered its growth ability and reached the same cell density as Td35405 and Tde471com at the stationary phase when the medium was supplemented with 100 or 500 μM of exogenous N-acetyl-neuraminic acid (Neu5Ac, a well-studied sialic acid and the most abundant one; Fig.5B) (Vimr et al., 2004; Comb and Roseman, 1960). These results indicate that T. denticola may require Tdneu to release sialic acid from the serum, which can be further metabolized to provide essential nutrients for its growth in vivo.
Figure 5. Growth curves of Td35405, Tde471mut, and Tde471com strains in TYGVS medium (A) and the serum growth medium (B).
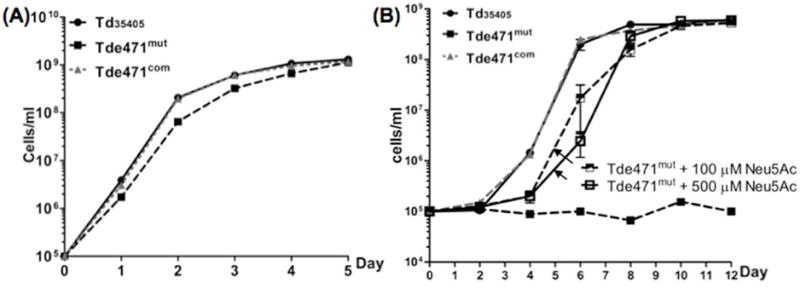
The serum growth medium contains 50% heat-inactivated rabbit serum supplemented with 6.0 μg/ml TPP. Cell counting was repeated in triplicate with at least three independent samples. The results are expressed as the mean ± SEM. The data were analyzed by one-way ANOVA followed by Tukey’s multiple comparison at p < 0.01.
Tdneu affects serum bactericidal activity against T. denticola
Several complement factors (e.g., C1q, IgG, and FH) are glycosylated and contain sialic acid. In some cases, sialylation is essential for their biological activities, such as the complement activation and formation of MAC (Ritchie et al., 2002;Varki, 2007). To determine if Tdneu affects serum bactericidal activity against T. denticola, serum killing assays were conducted. Similar to a recent report (McDowell et al., 2011), Td35405 was resistant to the serum killing and its average survival rate was 87% following 1-hour incubation in 25% normal human serum (NHS). Compared to Td35405, Tde471mut was more vulnerable to the killing – its survival rate decreased to 39% (Fig.6). The diminished resistance was fully restored when the mutant was complemented (Fig.6). These results indicate that Tdneu acts to attenuate the serum bactericidal activity against T. denticola.
Figure 6. Survival rates of Td35405, Tde471mut, and Tde471com strains in serum.
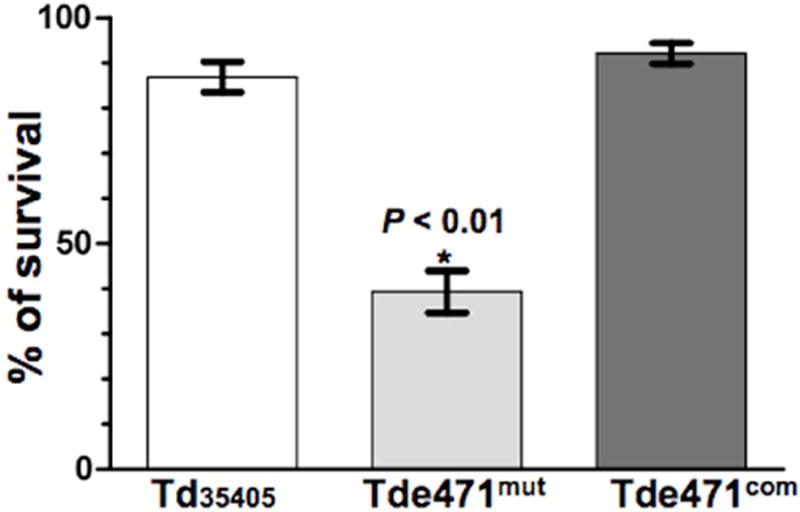
The strains were co-incubated with either 25% normal human serum (NHS) or heat-inactivated human serum (HIS) for 1 h at 37°C. The number of surviving bacteria was counted using a Petroff Hausser counting chamber. The survival rates were calculated as follows: the total number of living cells in NHS divided by the total number of living cells in HIS. Cell counting was repeated in triplicate with at least three independent samples, and the results are expressed as the mean ± SEM. The data were analyzed by one-way ANOVA followed by Tukey’s multiple comparison at p < 0.01.
Tde471mut exhibits enhanced complement deposition on the cell surface
The hallmark of complement-dependent killing is the formation of MAC (C5b-9) on bacterial cell surfaces (Lambris et al., 2008;Ricklin et al., 2010;Vimr and Lichtensteiger, 2002). Tde471mut exhibited decreased serum resistance (Fig.6). Conceivably, the loss of Tdneu may enhance complement deposition and formation of MAC on the mutant cells, which would make Tde471mut more vulnerable to the serum killing. To test this hypothesis, complement deposition on the Td35405, Tde471mut, and Tde471com strains was measured and compared. As shown in Fig.7A, SDS-PAGE analyses showed that various proteins were evident in the serum-incubated mutant cells, but not in the samples of Td35405 and Tde471com after 20 min incubation in NHS. Notably, these proteins disappeared when the mutant cells were treated with EDTA (Fig.7A), a chelating agent that blocks the complement cascade activation (Berger et al., 1985). In addition, the immunoblotting analyses using a rat serum against T. denticola (Fig.7B) and a polyclonal antibody against Msp (data not shown) demonstrated the presence of similar amounts of T. denticola proteins in the experimental samples (Fig.7B). Collectively, these results suggest that the proteins present on the mutant were derived from the complement system, not from the bacterial cells.
Figure 7. Deposition of membrane attack complex (MAC) on Td35405, Tde471mut, and Tde471com strains.
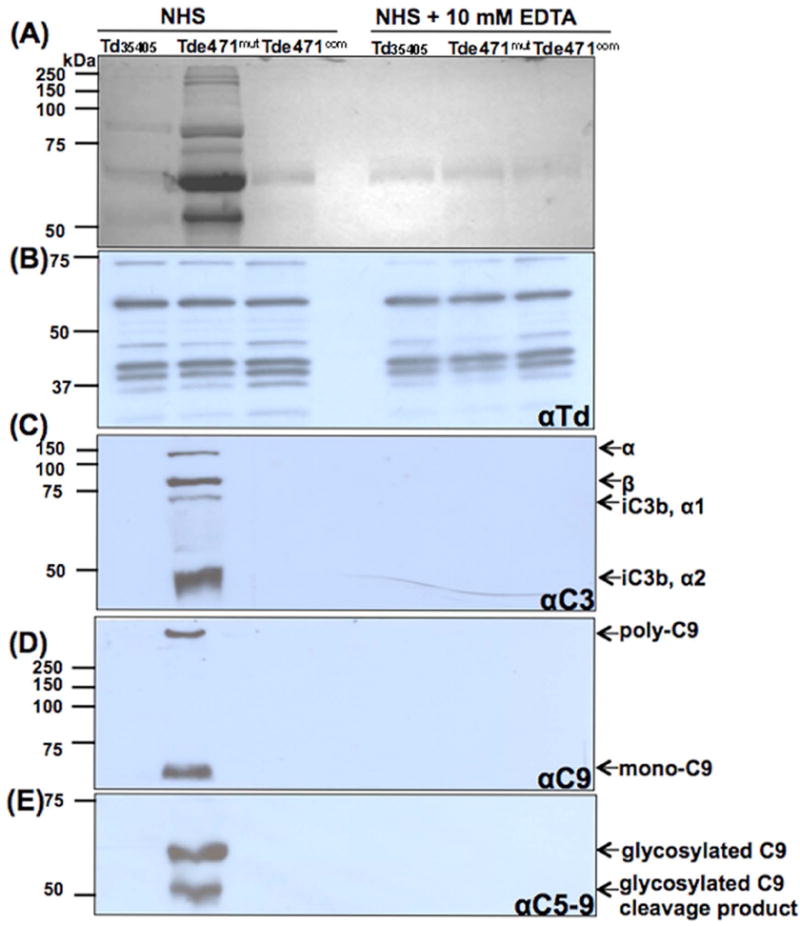
106 cells of Td35405, Tde471mut, or Tde471com were co-incubated with 25% NHS for 20 min at 37°C. As a control, 10 mM EDTA was added to the reactions to block complement activation. The resultant serum-treated cells (approximately 5 × 104) were subjected to SDS-PAGE, followed by coomassie blue staining (A), or probed with four different antibodies as labeled: (B) αTd, a rat polyclonal antibody against Td35405; (C) αC3, a polyclonal antibody against C3; (D) αC9, a monoclonal antibody against C9; and (E) αC5-9, a monoclonal antibody against C5-9 complex. Notably, complement components were detected only in the serum-treated Tde471mut, but not in the serum-treated Td35405 and Tde471com strains. The complement factors detected by the antibodies and their molecular weights are indicated.
This proposition was further confirmed by immunoblotting analysess with specific antibodies against C3, C9, and C5b-9 complex (Fig.7C-E). For C3 antibody (αC3), four products were detected in the serum-incubated Tde471mut cells, but not in the serum-incubated Td35405 and Tde471com cells. The sizes of the detected bands are similar to those of C3-related products (Fig.7C), including C3b α chain (119 kDa) and β chain (75 kDa), and factor I-mediated C3b cleaved products (iC3b-α1, 68 kDa; α2, 43 kDa) (Barnes and Weiss, 2001;Ben and Klimpel, 2008). A similar pattern was observed when the samples were probed with antibodies against C9 (αC9) and C5b-9 (αC5b-9) – in the serum-incubated mutant cells, mono-C9 (71 kDa) and poly-C9 (>1,000 kDa) were detected by αC9 (Fig.7D); and two glycosylated C9 products were detected by αC5b-9 (Fig.7E). In contrast, none of these proteins was evident in the serum-incubated Td35405 and Tde471com cells. Taken together, these results indicate that Tdneu impairs the complement activation and deposition. This constitutes a novel protection mechanism of T. denticola from the serum killing.
Tdneu contributes to the virulence of T. denticola
The importance of Tdneu in the virulence of T. denticola was further assessed in vivo using a previously reported mouse skin infection model (Kesavalu et al., 1997). For this study, each BALB/C mouse was subcutaneously injected with 109 T. denticola cells at its dorsal site. Skin abscesses caused by the spirochetes were measured at ten days post-injection. Compared to the Td35405 and Tde471com strains, the tissue damage caused by Tde471mut was less (p < 0.01), which was reflected by smaller skin abscesses on mice injected with the mutant (Fig.8A). The average size of the abscesses induced by Td35405 was 71.6 ± 6.9 mm2 (n=5). The average size was reduced to 37.5 ± 2.0 mm2 (n=5) in the mutant and was restored to the wild-type level in Tde471com (67.5 ± 4.7 mm2, n=4).
Figure 8. Evaluating the virulence of Td35405, Tde471mut, and Tde471com strains using a mouse-skin-infection model.
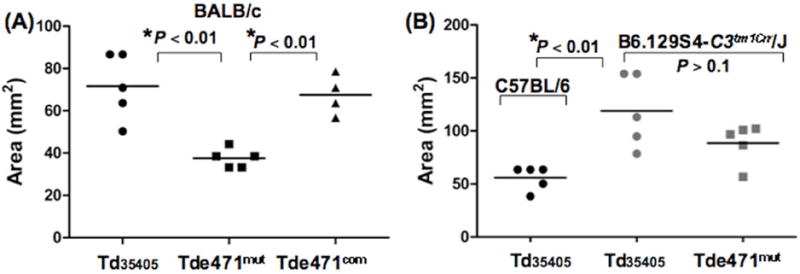
The infectious studies were carried out as previously documented (Kesavalu et al., 1997). 109 bacterial cells were subcutaneously injected into each mouse. Three different mouse strains were included: (A) BALB/C mice; (B) The complement-deficient mice (B6.129S4-C3tm1Crr/J) and its parental wild-type C57BL/6 mice. Ten days post-infection, the sizes of the abscesses were measured. The average sizes of the observed abscesses were calculated. The statistic differences between groups were analyzed by t-test, two-tailed at p < 0.01
To determine if the complement system has any impact on the pathogenicity of T. denticola, a similar experiment was repeated in B6.129S4-C3tm1Crr/J, a complement-deficient mouse strain (Wessels et al., 1995). The tissue damage caused by both Td35405 and Tde471mut was more severe in B6.129S4-C3tm1Crr/J mice than in its parental wild-type strain C57BL/6 (Fig.8B). In the mutant mice, the average sizes of skin abscesses induced by Td35405 and Tde471mut were 118.8 ± 15.3 mm2 (n=5) and 88.6 ± 8.4 mm2 (n=5), respectively, which are significantly (p < 0.01) larger than what is observed in C57BL/6 mice (55.9 ± 5.0 mm2, n=5). The average size of the abscesses induced by Tde471mut is smaller than that induced by Td35405 (Fig.8B), but it is not statistically significant (p > 0.1). These results along with the in vitro studies presented above indicate that the immune defense mediated by the complement system affects the pathogenicity of T. denticola and that Tdneu is an important virulence factor for complement evasion in vivo.
DISCUSSION
Do the neuraminidases of ‘red-complex’ bacteria share similar biochemical characteristics?
As mentioned earlier, the ‘red-complex’ bacteria primarily inhabit the gingival crevices bathed in plasma, which contains a high concentration of sialic acid that is bound to a diverse range of glycans. In this unique niche, it would be a survival advantage if a microorganism can utilize host sialic acid. In this regard, it is not surprising that all three ‘red-complex’ bacteria possess neuraminidases. In addition, several lines of evidence have suggested that those neuraminidases (PG0352, TF0035, and Tdneu) have similar biochemical features. First, they are all dual-domain proteins (Li et al., 2012;Thompson et al., 2009). The neuraminidase domains are located at their C-terminal regions while the N-termini have different domains. PG0352 has a peptidoglycan-binding domain (aa 30 to 180). The N-terminus of Tdneu contains a Listeria-Bacteroides-Repeat domain that is often found in internalin families (CDD212296) of Listeria species and is implicated in cell adhesion and invasion (Dramsi et al., 1993). Second, they are active over a wide pH range (e.g., TF0035 is active from pH 4.5 to 8.0, with an optimum pH of 5.5) (Thompson et al., 2009). Tdneu is active in a similar pH range to TF0035 (Fig.1B). Third, they all show exo-neuraminidase activity. Similar to PG0352 and TF0035 (Li et al., 2012;Thompson et al., 2009), Tdneu is able to remove α2,3- and α2,6-linked sialic acid of AGP (Fig.1D). Finally, they all contain a signal peptide, suggesting they are most likely secreted and/or surface-localized. Previous studies have shown that TF0035 is both surface-localized and secreted (Honma et al., 2011). Similar to TF0035, Tdneu is also surface-exposed and secreted (Fig.3). These biochemical characteristics are compatible with the gingival crevice microenvironment – the pH of GCF in healthy individuals is about 6.9 (Eggert et al., 1991;Bickel and Cimasoni, 1985). In addition, α2,3- and α2,6-linked sialic acids are the most abundant and are ubiquitous at the termini of plasma sialoglycoconjugates (Ponnio et al., 1999;Crocker et al., 2007;Varki, 2008). These features may allow these enzymes to maintain their optimal activities in terms of withstanding pH fluctuation and acquiring sufficient substrates in the periodontal pockets.
Does T. denticola utilize sialic acid as a nutrient?
Several bacteria use sialic acid as an alternative nutrient when other carbon resources are limited (Vimr et al., 2004;Severi et al., 2007). The plasma in the gingival crevices contains a variety of sialoglycoconjugates that can be desialylated by Tdneu (Fig.4). The released free sialic acid presumably can be transported and metabolized within T. denticola cells. This can explain why Td35405 is able to grow in the serum medium but Tde471mut cannot (Fig.5). One intriguing question remaining to be answered is how T. denticola metabolizes sialic acid and converts it to other nutrients. In Escherichia coli, NanA first metabolizes Neu5Ac and then releases ManNAc and pyruvate. ManNAc is ultimately converted to fructose-6-phosphate and ammonia via a series of reactions catalyzed by NanK, NanE, NagB and NagA [for review, see (Vimr et al., 2004)]. However, the genes encoding these enzymes are absent in the genome of T. denticola (Seshadri et al., 2004), indicating that a similar catabolic pathway does not exist in the spirochete. The metabolism of T. denticola is poorly understood and the catabolism of sialic acid in anaerobes has not yet been documented. It is possible that T. denticola and other anaerobes may have evolved a novel catabolic pathway to metabolize sialic acid. Recent studies have suggested that T. forsythia might use Neu5Ac and N-acetyl-glucosamine (NAG) to produce N-acetylmuramic acid (NAM) (Roy et al., 2011), a key component of cell wall peptidoglycan. A similar scenario may also occur in T. denticola.
How does T. denticola escape complement killing?
In this report, we found that Tde471mut was more vulnerable to the serum killing (Fig.6). However, under the tested condition, approximately 39% of the mutant cells were still viable, indicating that other mechanisms are also involved in the complement resistance of T. denticola. Dentilisin, a surface protease of T. denticola, hydrolyzes C3b (Yamazaki et al., 2006) and cleaves FH (McDowell et al., 2011). Since C3b plays a key role in the complement cascade activation (Lambris et al., 2008;Ricklin et al., 2010), it has been previously postulated that dentilisin may contribute to the serum resistance of T. denticola (Yamazaki et al., 2006). However, McDowell et al recently reported that dentilisin has no impact on the serum resistance of T. denticola (McDowell et al., 2011). This observation is contradictory to its activity of cleaving C3b and FH; and more comprehensive studies are needed to further elucidate its role in the serum resistance of T. denticola. In addition, recent reports described that FhbB, an FH-binding protein, is involved in complement evasion (McDowell et al., 2011). Conceivably, these factors may constitute a complex and redundant network that protects T. denticola from the complement killing, ensuring that the spirochete can survive and thrive in the oral flora while facing the constant immune surveillance that is mediated by the complement system.
How does Tdneu protect T. denticola from the serum killing?
Previous reports have shown that FhbB, an FH-binding protein, protects T. denticola from serum killing (McDowell et al., 2005; McDowell et al., 2011). In Neisseria gonorrhoeae, sialylation increases the binding activity of FH to LPS, which results in higher serum resistance (Ram et al., 1998b;Ram et al., 1998a). We sought to determine if a similar scenario accounts for the serum resistance of T. denticola (e.g., the free sialic acid released by Tdneu is utilized to modify FhbB, which results in increased binding of FH to FhbB). To test this hypothesis, an affinity-ligand-binding immunoblotting (ALBI) assay was carried out (Fig.S6). The results showed that FH bound to both Td35405 and Tde471mut in a similar pattern, but not to FhbBmut, a previously constructed fhbB isogenic mutant (McDowell et al., 2011). This indicates that the loss of Tdneu has no impact on FH binding to Tde471mut. Thus, it is unlikely that Tdneu affects the serum resistance of T. denticola by acting on FhbB.
Several pathogenic bacteria (e.g., Campylobacter jejuni, Haemophilus influenza, and Neisseria spp) acquire serum resistance by the incorporation of sialic acid into their surface macromolecules (i.e., LPS and capsule) (Figueira et al., 2007;Ram et al., 1998b;Madico et al., 2007;Avril et al., 2006;Vimr and Lichtensteiger, 2002;Severi et al., 2007). However, T. denticola lacks both LPS and capsule (Seshadri et al., 2004). If a similar mechanism exists in T. denticola, the organism must utilize other surface molecules rather than capsule and LPS. Proteomic analysis disclosed that at least four proteins had either changed expression levels or had altered molecular weights in Tde471mut (Li, et al, unpublished data). These proteins were Msp (TDE0405), oligopeptide-binding protein (TDE0985), OppA (TDE1071), and a putative outer-membrane protein (TDE2602). Msp and OppA are two virulence factors of T. denticola (Fenno et al., 1998;Fenno et al., 2000). These proteins could be potential candidates for modification by sialic acid. Consistent with this possibility, a recent proteomic analysis showed that Msp and OppA have different forms (Veith et al., 2009), suggesting that they are probably modified (e.g., glycosylation). Furthermore, T. denticola has a novel type of outer membrane lipid that contains lipoteichoic acids (Schultz et al., 1998), and genomic information suggests that it may produce surface polysaccharides (Seshadri et al., 2004). These two molecules are potential candidates for sialylation. We are currently employing different approaches to investigate if these candidate molecules are sialylated or not.
Summary
The evidence described in this report along with previous studies has demonstrated that ‘red-complex’ bacteria have evolved to employ neuraminidases to scavenge host sialic acid. This can benefit the pathogens at least in two ways: nutrient acquisition and immune evasion. Accumulating evidence has shown that neuraminidases play critical roles in the physiology and pathogenicity of ‘red-complex’ bacteria (Roy et al., 2011;Aruni et al., 2011;Li et al., 2012). These neuraminidases are surfaced-exposed and share similar biochemical features. Given these facts, the neuraminidases are ideal targets for developing new therapeutic agents for intervention and prevention of periodontitis (e.g., designing specific inhibitors against neuraminidases). In addition, the surfaces of host immune system cells contain a family of sialic-acid-binding immunoglobulin-like lectins (Siglecs), which play critical roles in regulating the functions of the cells in innate and adaptive immune systems via glycan recognition (Varki, 2007;Crocker et al., 2007;Varki, 2008). It is intriguing to consider the potential effects that neuraminidases could impose on innate and adaptive immunity regulated by the Siglecs. Furthermore, sialylation is essential for the anti-inflammatory activity of human plasma immunoglobulin G (also referred to as intravenous IgG, IVIG), which is often utilized to treat a variety of human hematological and immunological disorders (Nimmerjahn and Ravetch, 2008). Recent studies have shown that desialylation of IVIG not only depletes its anti-inflammatory activity but also potentially leads to generation of pathogenic antibodies (Kaneko et al., 2006). Our preliminary studies suggested that Tdneu is able to cleave sialic acid from IVIG (data not shown). It is even more intriguing to consider the potential impact of neuraminidases on IgG in the GCF and the role of IgG in the pathogenesis of periodontitis as well as systemic diseases that are associated with periodontitis.
EXPERIMENTAL PROCEDURES
Ethics statement
All animal experimentation was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Human serum samples were collected from healthy volunteers. The protocols for animal studies and the use of human serum were approved by the Institutional Animal Care and Use Committee (permit number: ORB 23068Y) and the Human Subjects Review Committee (permit number: ORB0321006E) of the State University of New York at Buffalo, respectively.
Bacterial strains and culture conditions
T. denticola ATCC 35405 (wild type) (Seshadri et al., 2004) was used in this report. Cells were grown either in tryptone-yeast extract-gelatin-volatile fatty acids-serum medium (TYGVS) (Ohta et al., 1986) or in 50% heat-inactivated (56°C for 30 min) rabbit serum diluted in PBS (phosphate buffered saline, pH 7.4) supplemented with 6 μg/ml thiamine pyrophosphate (TPP) (designated as serum growth medium) at 37°C with an atmosphere of 85% nitrogen, 5% carbon dioxide, and 10% hydrogen. T. denticola isogenic mutants were grown with appropriate antibiotic(s) for selective pressure as needed: erythromycin (70 μg/ml) and/or gentamicin (20 μg/ml). E. coli TOP10 strain (Invitrogen, Carlsbad, CA) was used for DNA cloning; BL21 Star™ (DE3) (Invitrogen) and M15 (Qiagen, Valencia, CA) strains were used for preparing recombinant proteins. The E. coli strains were cultured in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Preparation of recombinant TDE0471 and its antiserum
To prepare a recombinant protein for enzymatic analysis, the full-length TDE0471 gene was PCR amplified with primers P1/P2 using Vent DNA polymerase (New England Biolabs, Ipswich, MA). The obtained PCR product was cloned into pET101/D-TOPO vector (Invitrogen), which encodes a six-histidine tag at the C-terminus. The resulting plasmid was then transformed into BL21 Star (DE3) cells. The expression of TDE0471 was induced using 1 mM isopropyl β-D-1-thiogalactosidase (IPTG). The recombinant protein was purified using Ni-NTA agarose (Qiagen) under native conditions as previously described (Sze et al., 2011;Li et al., 2012). The purified protein was then dialyzed in a buffer containing 20 mM Tris-HCl at 4°C overnight using 6.0 kDa molecular weight cut-off Spectra/Por® dialysis bags (Spectrum Laboratories, Rancho Dominguez, CA). The concentration of purified protein was determined using a Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA). A similar method was used to prepare a truncated TDE0471 recombinant protein without the C-terminal catalytic domain (from aa 324 to 543). This protein was used either as an antigen for production of TDE0471antibody or as a control in the biochemical analyses. For antibody production, 5 mg purified truncated TDE0471 protein was used to immunize rats (2.5 mg for each animal) following a standard immunization procedure as previously described (Sze and Li, 2011). The primers for preparation of the recombinant proteins are listed in Table S1.
Electrophoresis and immunoblotting analyses
Sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) and immunoblotting using the enhanced chemiluminescent (ECL) detection system were carried out as described before (Sze and Li, 2011). T. denticola cells were harvested at the late-logarithmic-phase (~108 cells/ml). Equal amounts of whole cell lysates (10-50 μg) were separated on SDS-PAGE gels and then transferred to PVDF membranes (Bio-Rad). The immunoblots were probed with specific antibodies against various proteins and developed using horseradish peroxidase-coupled secondary antibody with an ECL luminol assay. The antibodies against the T. denticola flagellin protein (FlaA) and Msp were gifts from J. C. Fenno (University of Michigan). The polyclonal antibodies against T. denticola were raised in this study by immunizing rats twice with the wild-type whole cells. A monoclonal antibody against human FH was purchased from Thermo Scientific (Thermo Scientific, Rockford, IL). A polyclonal against C3 and monoclonal antibodies against C9 and C5b-9 complex were purchased from Abcam (Abcam Inc., Cambridge, MA).
Enzyme assays
A previously documented filter paper spot sialidase assay was used to detect the neuraminidase activity (Moncla and Braham, 1989;Li et al., 2012). Briefly, the recombinant TDE0471, the whole cell lysates, or the supernatants from the T. denticola cultures were incubated with 2’-4-methylumbelliferyl-α-D-N-acetyl-neuraminic acid (4-MUNANA) (Sigma-Aldrich), a fluorogenic neuraminidase substrate. Fluorescence was measured using a ChemiDoc XRS imaging system (Bio-Rad) with an excitation wavelength of 302 nm and an emission wavelength of 548 nm. To test the neuraminidase specificity, 2,3-dehydro-2-deoxy-N-acetyl-neuraminic acid (Neu5Ac2en), a neuraminidase inhibitor (Barrett et al., 2011), was included in the assay. To determine the optimal pH for the enzymatic activity, the filter paper spot assay was carried out at various pH values (ranging from 3.0 to 9.0). The neuraminidase kinetics analysis was carried out as previously described (Thompson et al., 2009;Gut et al., 2008). Briefly, 3.342 nM purified recombinant TDE0471 protein was incubated with various amounts of 4-MUNANA (ranging from 0.02 to 0.25 mM) in a reaction buffer containing 70 mM sodium citrate (pH 7.0) at 37°C for 15 minutes. The saturation curves were fitted to Michaelis-Menten kinetics using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA), and the Km and Vmax were calculated.
Lectin blot analysis
A previously described lectin blot analysis (Gut et al., 2008) was used to determine the substrate specificity of TDE0471. Briefly, 50 μl aliquots containing 1 μg human α-1 acid glycoprotein (AGP), 0.7 nM recombinant TDE0471 (0.7 nM) or 0.0064 U Clostridium perfringens neuraminidase NanH (Sigma-Aldrich), and 10 mM potassium phosphate (pH 7.0) were incubated overnight at 37°C. After the incubation, the samples were separated on SDS-PAGE gels, and then transferred to PVDF membranes (Bio-Rad). The resultant blots were probed with biotin-labeled lectins (Vector Laboratories, Inc., Burlingame, CA) from three plants: Datura stramonium (DSA), Sambucus nigra (SNA), and Maackia amurensis (MAA) (Crowley et al., 1984). These three lectins specifically recognize different carbohydrates – SNA binds to α2,6-linked sialic acid, MAA to α2,3-linked sialic acid, and DSA to galactose-linked GlcNAc. The final blots were incubated with streptavidin-horseradish peroxidase conjugate and developed with the ECL luminol assay (Thermo Scientific).
Treatment of human serum with recombinant TDE0471
Human serum samples were collected from healthy laboratory donors. Freshly drawn blood was allowed to clot and separated by centrifugation at 2,000 × g for 20 min. The serum samples were aliquoted in small volumes and stored at -80°C. Each aliquot was thawed only once. To determine if TDE0471cleaves sialic acid from sialoglycoconjugates present in the serum, 0.15% (final concentration in the reactions) or 1.5% normal human serum (NHS) was incubated with various amounts of recombinant TDE0471 in a reaction buffer (10 mM sodium phosphate, pH 5.0) for 1 to 3 hrs at 37°C. After incubation, the treated samples were then subjected to SDS-PAGE and immunoblotting analyses. The blots were first blocked in 1× Carbo-Free blocking solution (Vector Laboratories) containing 0.05% Tween-20 and then incubated with biotinylated lectins in an incubation buffer (0.2 × Carbo-Free solution, 0.05% Tween-20) for 1 h at room temperature. The lectins used here include SNA, MAA (Vector Laboratories), and Concanavalin A (ConA, Sigma-Aldrich). The resulting blots were washed four times with PBS-T buffer (PBS, 0.05% Tween-20) and then incubated with streptavidin-horseradish peroxidase conjugate. After incubation, the blots were washed four times with PBS-T buffer and developed with the ECL luminol assay.
Constructing a TDE0471 isogenic mutant and its cognate complemented strain
The construct TDE0471∷ermR (Fig.S3A) was used to inactivate TDE0471 via allelic exchange recombination. The construct ErmR∷TDE0471 (Fig.S3B) was used for cis-complementation of TDE0471 mutant. To construct TDE0471∷ermR, a previously described erythromycin resistant cassette (ermR) (Li et al., 1996) and a DNA fragment containing the entire TDE0471 gene were amplified by PCR using primers P5/P6 and P7/P8, respectively. The resulting amplicons were cloned into pGEM-T vector. The cloned ermR cassette was then released by EcoRI from the plasmid and inserted into TDE0471 at the same cleavage site, yielding TDE0471∷ermR. ErmR∷TDE0471 was constructed by two step PCR and DNA cloning. The 5’-portion of ermR and a previously constructed gentamicin resistance cassette (aacC1) (Bian et al., 2012) were PCR amplified with primers P9/P10 and P11/P12, respectively, and then fused together with primers P9/P12, generating Fragment 1 (Fig.S3B). The 3’-portion of ermR and a DNA fragment containing the full length of TDE0471 and its upstream promoter sequence were PCR amplified with primers P15/P16 and P13/P14, respectively, and then fused together by PCR using primers P13/P16, generating Fragment 2 (Fig.S3). The primers used are listed in Table S1. The two obtained DNA fragments were cloned into the pGEM-T easy vector and then fused together at the engineered XbaI cleavage site. Preparation of T. denticola competent cells, electroporation, and cell plating were carried out as previously described (Li et al., 1996;Bian et al., 2011). The resultant antibiotic-resistant clones were first characterized by PCR and the selected positive clones were further confirmed by immunoblotting analysis.
Preparations of proteinase K-treated T. denticola cells and culture supernatants
The proteinase K treatment was performed as previously described (Brooks et al., 2006). Briefly, 10 ml of stationary-phase T. denticola cultures (5 × 108 cells/ml) were harvested by centrifugation at 2,000 × g for 15 min. The harvested cells were washed three times in PBS. They were then resuspended in 2.5 ml PBS containing 600 μg of proteinase K (Sigma-Aldrich) and incubated at 37°C for 1 h. The reactions were stopped by adding 10 μl of 10 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich), followed by centrifugation at 2,000 × g for 15 min. The obtained cell pellets were washed twice in PBS and subjected to SDS-PAGE and immunoblotting analyses. T. denticola culture supernatants were prepared as previously described (Rosen et al., 1995). Briefly, 400 ml of the late-logarithmic-phase T. denticola cultures (108 cells/ml) were centrifuged and the obtained culture supernatants were filtered through 0.2 μm-pore-size filters to further remove the remaining cells. The filtered supernatants were then dialyzed in 4 liters of 50 mM Tris-HCl buffer (pH 7.8) at 4°C overnight using 3.5 kDa molecular weight cut-off Spectra/Por® dialysis membranes and further concentrated to 4 ml.
Immunofluorescence assay (IFA)
IFA was conducted to determine the surface location of TDE0471 as previously described with slight modifications (Xu et al., 2011). Briefly, 1.5 ml of T. denticola cultures were harvested, washed one time with PBS buffer, and then treated with methanol at -20°C for 1 hour. The resultant cells were then placed on poly-L-lysine-coated cover slips, and allowed to fully air dry. The obtained cover slips were first incubated in a blocking solution (2% BSA in PBS, pH 7.5) for 1 hour, followed by incubation in the blocking solution containing 1:100 diluted αTdneu for 1 hour at room temperature. Finally, the cover slips were washed five times with PBS, incubated with the secondary goat anti-mouse Texas red antibody (Invitrogen) for 1 hour at room temperature, washed with PBS, and mounted in 40% glycerol for image processing as described previously (Xu et al., 2011).
Measuring T. denticola growth rates
To measure the growth rates, 5 μl of the late-logarithmic-phase T. denticola cultures (108 cells/ml) were inoculated into 5 ml of the TYGVS medium, the serum growth medium, or the serum medium supplemented with either 100 or 500 μg/ml of N-acetyl-neuraminic acid (Neu5Ac). T. denticola cells in the cultures were enumerated every 24 h or 48 h using a Petroff Hausser counting chamber (Hausser Scientific, Horsham, PA). Each growth curve is representative of at least three independent cultures, and the results are represented as the mean of cell numbers ± standard error of the mean (SEM).
Serum killing assay
The serum killing assay was conducted as previously described (McDowell et al., 2011;Li et al., 2012). Briefly, 75 μl of T. denticola cultures (containing 106 spirochete cells) were first mixed with 25 μl of either normal human serum (NHS) or heat-inactivated serum (HIS) and then incubated at 37°C in the anaerobic chamber for 1 h. After incubation, the number of living spirochete cells was enumerated as described above. The living cells were determined by whether they were motile during the observation time (at least 1 min). The average survival rates (the number of living cells in NHS / the number of living cells in HIS) were calculated from at least three independent experiments, and the results are represented as the mean of survival rates ± standard error of the mean (SEM).
Complement deposition assay
The complement deposition assay was carried out as previously described (Barnes and Weiss, 2001). For this assay, 75 μl of T. denticola cultures (containing 106 cells from the late-logarithmic-phase cultures) were mixed with 25 μl of NHS and then incubated for 20 min at 37°C in the anaerobic chamber. As controls, 10 mM ethylenediamine-tetraacetic acid (EDTA) and 10 mM MgCl2-ethylene glycol-bis (2-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (Mg-EGTA) were included in the assay either to block activation of the complement pathways or to selectively activate the alternative pathway. After the incubation, the reactions were incubated on ice for 1 min and then ice-cold PBS containing 10 mM EDTA was added to stop complement activation. The cells were collected by centrifugation. The cell pellets were washed three times with ice-cold PBS and resuspended in 50 μl PBS. After adding 20 μl Laemmli sample buffer, the resultant samples were boiled for 5 min, adjusted to 100 μl, and finally 5 μl of samples were subjected to SDS-PAGE and immunoblotting analyses.
Affinity-ligand-binding immunoblotting (ALBI) assay
The ALBI assay was carried out as previously described (McDowell et al., 2005). Briefly, the late-logarithmic-phase T. denticola cultures were harvested, separated on 15% SDS-PAGE gels, and transferred to PVDF membranes. The blots were incubated with purified human factor H (FH) (Complement Technology Inc., Tyler, TX). The bound FH was detected by a monoclonal antibody against FH, following the standard immunoblotting procedure described above.
Animal studies
A previously documented mouse skin abscess model was used to assess the virulence of T. denticola (Kesavalu et al., 1997). For the animal studies, ~6-8 week-old BALB/C mice, complement-deficient mice (B6.129S4-C3tm1Crr/J) (Wessels et al., 1995) and its parental wild-type stain C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were included. Each mouse (at least 4 mice per bacterial strain) received a single subcutaneous injection on its posterior dorsolateral surface with 200 μl PBS containing 1 × 109 bacterial cells. After infection, mice were monitored for symptoms of infection on a daily basis for a total of ten days. The diameters of the observed abscesses were measured with a caliper gauge. Each abscess was measured at least three times from different angles and average sizes were calculated (area = πr2).
Supplementary Material
Acknowledgments
We thank C. J. Fenno for providing antibodies against FlaA and Msp, R. Marconi for providing the FhbB mutant strain, and S. Ruhl for providing Concanavalin A. This research was supported by Public Health Service Grants DE019667 and DE023080 to C. Li.
Reference List
- Achyuthan KE, Achyuthan AM. Comparative enzymology, biochemistry and pathophysiology of human exo-alpha-sialidases (neuraminidases) Comp Biochem Physiol B Biochem Mol Biol. 2001;129:29–64. doi: 10.1016/s1096-4959(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Aruni W, Vanterpool E, Osbourne D, Roy F, Muthiah A, Dou Y, Fletcher HM. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect Immun. 2011;79:2779–2791. doi: 10.1128/IAI.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avril T, Wagner ER, Willison HJ, Crocker PR. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect Immun. 2006;74:4133–4141. doi: 10.1128/IAI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MG, Weiss AA. BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition. Infect Immun. 2001;69:3067–3072. doi: 10.1128/IAI.69.5.3067-3072.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S, Mohr PG, Schmidt PM, McKimm-Breschkin JL. Real time enzyme inhibition assays provide insights into differences in binding of neuraminidase inhibitors to wild type and mutant influenza viruses. PLoS One. 2011;6:e23627. doi: 10.1371/journal.pone.0023627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben NA, Klimpel GR. Subversion of complement activation at the bacterial surface promotes serum resistance and opsonophagocytosis of Francisella tularensis. J Leukoc Biol. 2008;84:77–85. doi: 10.1189/jlb.0807526. [DOI] [PubMed] [Google Scholar]
- Berger M, Birx DL, Wetzler EM, O’Shea JJ, Brown EJ, Cross AS. Calcium requirements for increased complement receptor expression during neutrophil activation. J Immunol. 1985;135:1342–1348. [PubMed] [Google Scholar]
- Bian J, Fenno JC, Li C. Development of a modified gentamicin resistance cassette for genetic manipulation of the oral spirochete Treponema denticola. Appl Environ Microbiol. 2012;78:2059–2062. doi: 10.1128/AEM.07461-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Shen H, Tu Y, Yu A, Li C. The riboswitch regulates a thiamine pyrophosphate ABC transporter of the oral spirochete Treponema denticola. J Bacteriol. 2011;193:3912–3922. doi: 10.1128/JB.00386-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian XL, Wang HT, Ning Y, Lee SY, Fenno JC. Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect Immun. 2005;73:1252–1255. doi: 10.1128/IAI.73.2.1252-1255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel M, Cimasoni G. The pH of human crevicular fluid measured by a new microanalytical technique. J Periodontal Res. 1985;20:35–40. doi: 10.1111/j.1600-0765.1985.tb00408.x. [DOI] [PubMed] [Google Scholar]
- Brooks CS, Vuppala SR, Jett AM, Akins DR. Identification of Borrelia burgdorferi outer surface proteins. Infect Immun. 2006;74:296–304. doi: 10.1128/IAI.74.1.296-304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacalano G, Kays M, Saiman L, Prince A. Production of the Pseudomonas aeruginosa neuraminidase is increased under hyperosmolar conditions and is regulated by genes involved in alginate expression. J Clin Invest. 1992;89:1866–1874. doi: 10.1172/JCI115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JT, Jourdian GW, Roseman S. The sialic acids. VI Purification and properties of sialidase from Clostridium perfringens. J Biol Chem. 1965;240:3501–3506. [PubMed] [Google Scholar]
- Chien CH, Shann YJ, Sheu SY. Site-directed mutations of the catalytic and conserved amino acids of the neuraminidase gene, nanH, of Clostridium perfringens ATCC 10543. Enzyme Microb Technol. 1996;19:267–276. doi: 10.1016/0141-0229(95)00245-6. [DOI] [PubMed] [Google Scholar]
- Comb DG, Roseman S. The sialic acids. I. The structure and enzymatic synthesis of N-acetylneuraminic acid. J Biol Chem. 1960;235:2529–2537. [PubMed] [Google Scholar]
- Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Copley RR, Russell RB, Ponting CP. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Protein Sci. 2001;10:285–292. doi: 10.1110/ps.31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell SJ, Garman EF, Laver WG, Vimr ER, Taylor GL. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc Natl Acad Sci U S A. 1993;90:9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Crowley JF, Goldstein IJ, Arnarp J, Lonngren J. Carbohydrate binding studies on the lectin from Datura stramonium seeds. Arch Biochem Biophys. 1984;231:524–533. doi: 10.1016/0003-9861(84)90417-x. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Kocks C, Forestier C, Cossart P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenes is regulated by the bacterial growth state, temperature and the pleiotropic activator prfA. Mol Microbiol. 1993;9:931–941. doi: 10.1111/j.1365-2958.1993.tb01223.x. [DOI] [PubMed] [Google Scholar]
- Eggert FM, Drewell L, Bigelow JA, Speck JE, Goldner M. The pH of gingival crevices and periodontal pockets in children, teenagers and adults. Arch Oral Biol. 1991;36:233–238. doi: 10.1016/0003-9969(91)90091-8. [DOI] [PubMed] [Google Scholar]
- Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol 2000. 2005;38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto VJ, McBride BC. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66:1869–1877. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Tamura M, Hannam PM, Wong GW, Chan RA, McBride BC. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect Immun. 2000;68:1884–1892. doi: 10.1128/iai.68.4.1884-1892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira MA, Ram S, Goldstein R, Hood DW, Moxon ER, Pelton SI. Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect Immun. 2007;75:325–333. doi: 10.1128/IAI.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen JE, Ketley JM, Fasano A, Richardson SH, Wasserman SS, Kaper JB. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut H, King SJ, Walsh MA. Structural and functional studies of Streptococcus pneumoniae neuraminidase B: An intramolecular trans-sialidase. FEBS Lett. 2008;582:3348–3352. doi: 10.1016/j.febslet.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Complement and periodontitis. Biochem Pharmacol. 2010;80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012a;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol. 2012b;946:69–85. doi: 10.1007/978-1-4614-0106-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Honma K, Mishima E, Sharma A. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect Immun. 2011;79:393–401. doi: 10.1128/IAI.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- Kesavalu L, Walker SG, Holt SC, Crawley RR, Ebersole JL. Virulence characteristics of oral treponemes in a murine model. Infect Immun. 1997;65:5096–5102. doi: 10.1128/iai.65.12.5096-5102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15:341–349. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- Leprat R, Michel-Briand Y. Extracellular neuraminidase production by a strain of Pseudomonas aeruginosa isolated from cystic fibrosis. Ann Microbiol (Paris) 1980;131B:209–222. [PubMed] [Google Scholar]
- Li C, Kurniyati, Hu B, Bian J, Sun J, Zhang W, et al. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect Immun. 2012;80:3–13. doi: 10.1128/IAI.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madico G, Ngampasutadol J, Gulati S, Vogel U, Rice PA, Ram S. Factor H binding and function in sialylated pathogenic neisseriae is influenced by gonococcal, but not meningococcal, porin. J Immunol. 2007;178:4489–4497. doi: 10.4049/jimmunol.178.7.4489. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Frederick J, Miller DP, Goetting-Minesky MP, Goodman H, Fenno JC, Marconi RT. Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola. Mol Oral Microbiol. 2011;26:140–149. doi: 10.1111/j.2041-1014.2010.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417–1425. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Lankford J, Stamm L, Sadlon T, Gordon DL, Marconi RT. Demonstration of factor H-like protein 1 binding to Treponema denticola, a pathogen associated with periodontal disease in humans. Infect Immun. 2005;73:7126–7132. doi: 10.1128/IAI.73.11.7126-7132.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncla BJ, Braham P. Detection of sialidase (neuraminidase) activity in Actinomyces species by using 2’-(4-methylumbelliferyl)alpha-D-N-acetylneuraminic acid in a filter paper spot test. J Clin Microbiol. 1989;27:182–184. doi: 10.1128/jcm.27.1.182-184.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncla BJ, Braham P, Hillier SL. Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacteria. J Clin Microbiol. 1990;28:422–425. doi: 10.1128/jcm.28.3.422-425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Anti-inflammatory actions of intravenous immunoglobulin. Annu Rev Immunol. 2008;26:513–533. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- Ohta K, Makinen KK, Loesche WJ. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect Immun. 1986;53:213–220. doi: 10.1128/iai.53.1.213-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Ponnio M, Alho H, Nikkari ST, Olsson U, Rydberg U, Sillanaukee P. Serum sialic acid in a random sample of the general population. Clin Chem. 1999;45:1842–1849. [PubMed] [Google Scholar]
- Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- Powell LD, Varki AP. Sialidases. Curr Protoc Mol Biol. 2001;Chapter 17(Unit17) doi: 10.1002/0471142727.mb1712s27. [DOI] [PubMed] [Google Scholar]
- Ram S, McQuillen DP, Gulati S, Elkins C, Pangburn MK, Rice PA. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J Exp Med. 1998a;188:671–680. doi: 10.1084/jem.188.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med. 1998b;187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie GE, Moffatt BE, Sim RB, Morgan BP, Dwek RA, Rudd PM. Glycosylation and the complement system. Chem Rev. 2002;102:305–319. doi: 10.1021/cr990294a. [DOI] [PubMed] [Google Scholar]
- Roggentin P, Schauer R, Hoyer LL, Vimr ER. The sialidase superfamily and its spread by horizontal gene transfer. Mol Microbiol. 1993;9:915–921. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- Rosen G, Naor R, Rahamim E, Yishai R, Sela MN. Proteases of Treponema denticola outer sheath and extracellular vesicles. Infect Immun. 1995;63:3973–3979. doi: 10.1128/iai.63.10.3973-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Honma K, Douglas CW, Sharma A, Stafford GP. Role of sialidase in glycoprotein utilization by Tannerella forsythia. Microbiology. 2011;157:3195–3202. doi: 10.1099/mic.0.052498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkein HA, Fletcher HM, Bodnar M, Macrina FL. Increased opsonization of a prtH-defective mutant of Porphyromonas gingivalis W83 is caused by reduced degradation of complement-derived opsonins. J Immunol. 1995;154:5331–5337. [PubMed] [Google Scholar]
- Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. I. Quantitative study of immunoglobulins, complement components, and other plasma proteins. J Periodontol. 1977a;48:772–777. doi: 10.1902/jop.1977.48.12.772. [DOI] [PubMed] [Google Scholar]
- Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. II. Evidence for cleavage of complement components C3, C3 proactivator (factor B) and C4 in gingival fluid. J Periodontol. 1977b;48:778–784. doi: 10.1902/jop.1977.48.12.778. [DOI] [PubMed] [Google Scholar]
- Schultz CP, Wolf V, Lange R, Mertens E, Wecke J, Naumann D, Zahringer U. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. Functioning permeation barrier without lipopolysaccharides. J Biol Chem. 1998;273:15661–15666. doi: 10.1074/jbc.273.25.15661. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- Sillanaukee P, Ponnio M, Jaaskelainen IP. Occurrence of sialic acids in healthy humans and different disorders. Eur J Clin Invest. 1999;29:413–425. doi: 10.1046/j.1365-2362.1999.00485.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Li C. Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi. Infect Immun. 2011;79:1270–1279. doi: 10.1128/IAI.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Morado DR, Liu J, Charon NW, Xu H, Li C. Carbon storage regulator A (CsrA(Bb)) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol. 2011;82:851–864. doi: 10.1111/j.1365-2958.2011.07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- Thompson H, Homer KA, Rao S, Booth V, Hosie AH. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J Bacteriol. 2009;191:3623–3628. doi: 10.1128/JB.01618-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, et al. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis. 2009;199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- Uchiyama S, Carlin AF, Khosravi A, Weiman S, Banerjee A, Quach D, et al. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J Exp Med. 2009;206:1845–1852. doi: 10.1084/jem.20090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Dashper SG, O’Brien-Simpson NM, Paolini RA, Orth R, Walsh KA, Reynolds EC. Major proteins and antigens of Treponema denticola. Biochim Biophys Acta. 2009;1794:1421–1432. doi: 10.1016/j.bbapap.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10:254–257. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- Vimr ER. Microbial sialidases: does bigger always mean better? Trends Microbiol. 1994;2:271–277. doi: 10.1016/0966-842x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels MR, Butko P, Ma M, Warren HB, Lage AL, Carroll MC. Studies of group B streptococcal infection in mice deficient in complement component C3 or C4 demonstrate an essential role for complement in both innate and acquired immunity. Proc Natl Acad Sci U S A. 1995;92:11490–11494. doi: 10.1073/pnas.92.25.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C, Moter A, Choi BK, Dewhirst FE, Xue Y, Schupbach P, et al. Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol. 2004;54:1117–1122. doi: 10.1099/ijs.0.02806-0. [DOI] [PubMed] [Google Scholar]
- Xu H, Raddi G, Liu J, Charon NW, Li C. Chemoreceptors and flagellar motors are subterminally located in close proximity at the two cell poles in spirochetes. J Bacteriol. 2011;193:2652–2656. doi: 10.1128/JB.01530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Miyamoto M, Yamada S, Okuda K, Ishihara K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006;8:1758–1763. doi: 10.1016/j.micinf.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


