Abstract
A total of 13 Colletotrichum isolates were obtained from different banana cultivars (Musa spp.) with symptoms of anthracnose. Colletotrichum isolates from anthracnose of guava (Psidium guajava) and water apple (Syzygium aqueum) were also included in this study. Based on cultural and morphological characteristics, isolates from banana and guava were identified as Colletotrichum musae and from water apple as Colletotrichum gloeosporiodes. Isolates of C. musae from banana and guava had similar banding patterns in a randomly amplified polymorphic DNA (RAPD) analysis with four random primers, and they clustered together in a UPGMA analysis. C. gloeosporiodes from water apple was clustered in a separate cluster. Based on the present study, C. musae was frequently isolated from anthracnose of different banana cultivars and the RAPD banding patterns of C. musae isolates were highly similar but showed intraspecific variations.
Keywords: Colletotrichum, Banana, Anthracnose, RAPD
Abstract
Sebanyak 13 pencilan Colletotrichum telah diperolehi daripada gejala antraknos pada kultivar pisang (Musa spp.) yang berbeza. Pencilan Colletotrichum daripada antraknos jambu batu (Psidium guajava) dan jambu air (Syzygium aqueum) turut disertakan dalam kajian ini. Berdasarkan ciri-ciri kultur and morfologi pencilan-pencilan Colletotrichum daripada pisang dan jambu batu, Colletotrichum musae telah dikenalpasti dan Colletotrichum gloeosporiodes pula dipencilkan daripada jambu air. Daripada analisis polimorfisma DNA rawak teramplifikasi (RAPD) menggunakan empat pencetus menunjukkan corak jalur C. musae daripada pisang dan jambu batu adalah serupa dan analisis kluster mengelompokkan pencilan-pencilan C. musae dalam kelompok yang sama. C. gloeosporiodes daripada jambu batu dikelompokkan dalam kluster yang berasingan. Berdasarkan kajian yang telah dijalankan, C. musae kerap dipencilkan daripada gejala antraknos kultivar pisang yang berlainan dan corak jalur RAPD pencilan-pencilan C. musae adalah sangat serupa tetapi menunjukkan variasi intraspesifik.
Anthracnose of banana is caused by the Colletotrichum species and is one of the most serious diseases of ripe banana. Symptoms of anthracnose include black and sunken lesions with spore masses or acervuli in the lesion. Infection on the banana usually starts during the development of the fruit but remains quiescent until the fruit ripens; symptoms often manifest during storage and marketing (Prusky & Plumbley 1992). Anthracnose becomes severe when the banana fruits are wounded by scratches during handling and transportation, making the fruit unmarketable. In Malaysia, studies on the occurrence of Colletotrichum species associated with anthracnose of local banana cultivar are limited. Thus, the present preliminary study was conducted to isolate and identify Colletotrichum species that are associated with anthracnose on different banana cultivars in Pulau Pinang and to characterise the isolates using cultural, morphological and RAPD analysis.
Thirteen banana cultivars with symptoms of anthracnose were obtained from several markets in Pulau Pinang. The banana cultivars were pisang Mas, pisang berangan, pisang raja, pisang abu keling, pisang embun, pisang kelat, pisang nangka, pisang susu, pisang udang, pisang awak, pisang rastali and pisang abu. Guava and water apple with symptoms of anthracnose were also included in the study for comparison. Colletotrichum isolates were either isolated directly from an anthracnose lesion on the surface of the fruit or from pieces of infected tissues that were surface sterilised with 70% ethanol. Conidia from the conidia masses in the lesion and pieces of the infected tissues were plated on potato dextrose agar (PDA) supplemented with 100 μg/ml streptomycin and incubated at 27 ± 1°C for 5–7 days or until mycelial growth was visible. The mycelia that grew on the plates were then sub-cultured on new PDA plates. Single spore isolates were prepared for each isolate for morphological and molecular characterisation.
In the morphological characterisation, the shapes and sizes of the conidia were examined under a light microscope after incubation at room temperature for seven days on PDA. For each isolate, the length and width of 25 conidia were measured. To induce germ tube and appresoria formation, a drop of conidial suspension (1 X 105 conidia/ml) was inoculated on cellophane membrane on a glass slide and incubated at room temperature for 24–48 h. The mean sizes of the conidia length and width and appresoria were calculated. Cultural characteristics and colony growth were observed after growth on PDA at room temperature for seven days; three replicates were prepared for each isolate. The mean colony growth for each isolate was calculated after seven days. The colour of the colony and conidial masses was also recorded after seven days. The description and determination of the Colletotrichum species was done according to Sutton and Waterston (1970) and Mordue (1971).
For DNA extraction, fungal isolates were grown on PDA plates for 5–7 days, after which the mycelial were harvested and lyophilised. DNA was extracted using DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instruction.
After screening and optimisation, four random primers, OPA01 (5′CAGGCCCTTC), OPB03 (5′AGTCAGCCAC), OPB07 (5′GGTGACGCAG) and OPB10 (5′CTGCTGGGAC) were used in the RAPD analysis. A polymerase chain reaction (PCR) reaction was performed in a 25 μl reaction volume, containing 2.5mM MgCl2, 1X PCR buffer, 1 unit Taq polymerase (Promega, Madison, WI), 0.7mM dNTP, 0.8mM of each primer and 25 ng DNA. PCR was run for 40 cycles in a PTC Peltier Thermal Cycler with an initial denaturation of 98°C for 4 min followed by 45 cycles of denaturation at 95°C for 1 min, annealing at 36°C for 1 min, extension at 72°C for 2 min and final extension at 72°C for 10 min to ensure complete extension of the amplification products. PCR products were separated by electrophoresis using a 1.7% agarose gel in Tris borate EDTA (TBE) buffer for 2 h 40 min at 5Vcm−1. A 1 kb DNA marker (GeneRuler, Fermentas, Lithuania) and 100 bp DNA ladder (Promega, Madison, WI) were used to estimate the sizes of the RAPD bands. Two markers were used because the preliminary results of RAPD banding patterns showed some bands with sizes of more than 4000 bp. The gel was stained with 0.5 μg/ml ethidium bromide, visualised under the ultraviolet (UV) light and photographed.
The Numerical Taxonomy System of Multivariate Program (NT-SYS) software package Version 2.0 (Rohlf 2000) was used to analyse the RAPD data. RAPD bands were scored based on the presence (1) or absence (0) of a particular band to create a binary matrix. A genetic similarity matrix was constructed from the binary data using a simple matching coefficient, and a dendrogram was constructed using an unweighted pair-group method with arithmetic (UPGMA) cluster analysis.
Thirteen Colletotrichum isolates were recovered from different banana cultivars and one isolate each from the guava and water apple. The colony characteristics of the Colletotrichum isolates from different banana cultivars and guava were different from those of the isolates from the water apple. Isolates from banana and guava were white to orange with fast growing mycelia and a mean colony diameter of 8.50 ± 0.02 cm after 5–6 days. The isolate from the water apple had white to greyish white colonies, mycelia that grew slower than the isolates from banana and guava and a mean colony diameter of 8.05 ± 0.04 cm after 7 days.
Based on morphological characteristics, Colletotrichum isolates from banana and guava were identified as C. musae according to the descriptions of Sutton and Waterston (1970). The isolate from water apple was identified as C. gloeosporioides, according to the description of Mordue (1971). A comparison of the morphological characteristics of the two Colletotrichum species is shown in Table 1. C. musae could be distinguished from C. gloeosporioides by the shape and size of the conidia; the conidia of C. musae were broader, whereas those of C. gloeosporioides were longer. Figure 1 shows the conidia of C. musae isolated from anthracnose of pisang Berangan. Appresoria of C. musae were formed from the mycelial and also directly from the germ tube. For C. gloeosporioides, appresoria were formed from the mycelial only. Figure 2 shows the appresoria of C. musae from pisang Mas.
Table 1:
Morphological characteristics of C. musae and C. gloeosporioides.
| C. musae | C. gloeosporioides | |
|---|---|---|
| Conidia | ||
| - shape | cylindrical or ellipsoidal | cylindrical |
| - colour | brown | hyaline |
| - size (μm) | 11.8 – 19.1 x 3.2 – 8.5 | 13.4 – 24 x 4.0 – 5.9 |
| Appresoria | ||
| - shape | irregularly lobe | obovate to clavate |
| - colour | dark brown | brown |
| - size (μm) | 5.8 – 12.5 x 5.6 – 10.5 | 6.7 – 19.8 x 3.8 – 11.9 |
Figure 1:
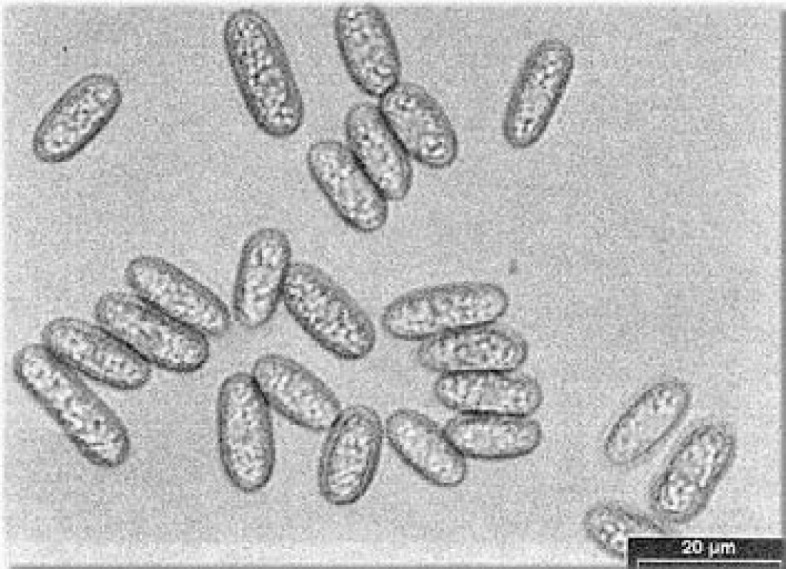
Conidia of C. musae isolated from anthracnose of pisang Berangan.
Figure 2:
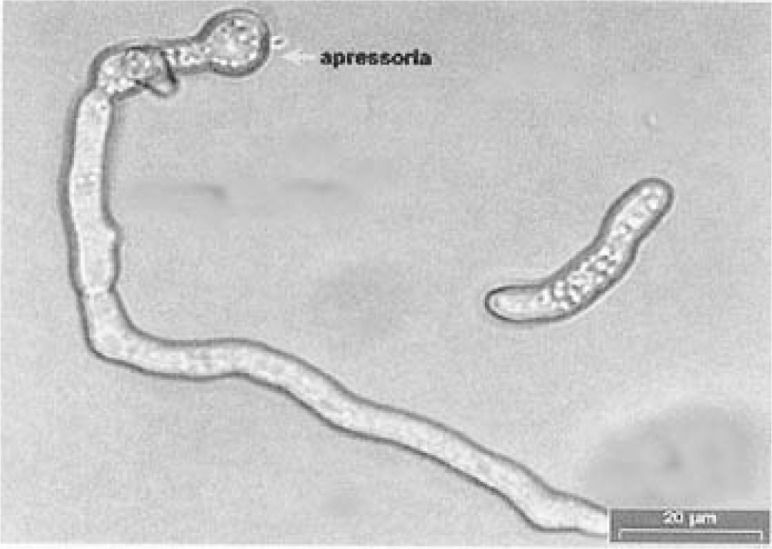
Appresorium formed from the germ tube of C. musae isolated from anthracnose of pisang Mas.
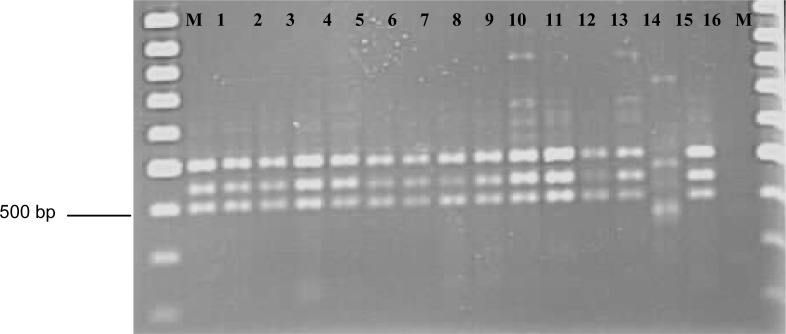
Depending on the primers, 3–7 bands were amplified with sizes ranging from 300–1150 bp. In general, the RAPD patterns of the C. musae isolates of the banana and guava were similar. The RAPD patterns of C. gloeosporioides from the water apple were different from those of the C. musae isolates from the banana and guava. Figure 3 shows the RAPD banding patterns of C. musae isolates from banana and guava and of C. gloeosporioides from the water apple using OPA7. RAPD patterns also revealed intraspecific variations within C. musae isolates from different banana cultivars.
Figure 3:
RAPD banding patterns of Colletotrichum isolates from banana, guava and water apple using the OPB07 primer. Lane 1–13: C. musae isolates from banana. Lane 14: C. gloeosporiodes from water apple. Lane 15: C. musae from guava. Lane 16: Control. M: markers.
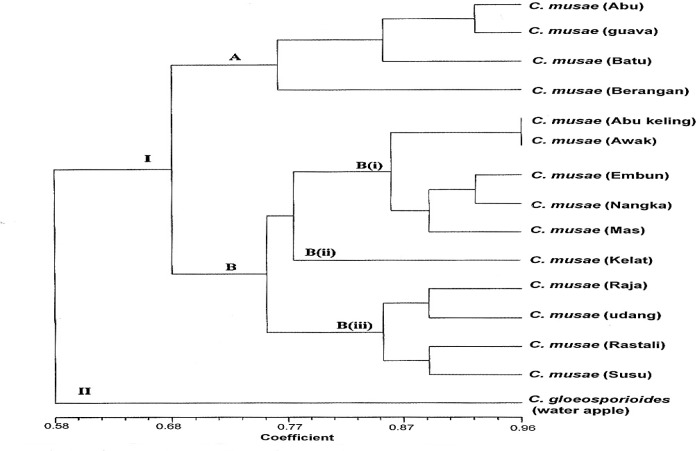
The dendrogram obtained from the UPGMA cluster analysis separated the C. musae isolates from banana into two separate clusters, A and B (Fig. 4). The similarity value within C. musae isolates ranged from 68%–100%. C. gloeosporioides isolated from water apple was clustered in a separate cluster, i.e., major cluster II.
Figure 4:
Dendogram from UPGMA cluster analysis using simple matching coefficient based on RAPD of C. musae from banana cultivars and guava, and C. gloeosporioides from water apple.
C. musae and C. gloeosporioides could be differentiated based on RAPD banding patterns and UPGMA cluster analysis, which clearly separated the two Colletorichum species into separate groups. The molecular characterisation agreed with the morphological characterisation of the two Colletorichum species.
Anthracnose is a post-harvest disease that develops during the storage and ripening of the banana. C. musae is the most common species associated with anthracnose of banana and has been widely accepted as the causal agent of the disease (Chakravarty 1957; Meredith 1960; Jeger et al. 1995; Jones & Slabaugh 1998). Besides causing anthracnose on banana fruits, C. musae can also infect the bracts, flowers, petioles and leaves of banana plants (Jones & Slabaugh 1998). Anthony et al. (2004) reported that C. musae was isolated from anthracnose lesions of three banana cultivars and was also associated with crown rot disease of banana in Sri Lanka.
In the present study, C. musae was also isolated from anthracnose of guava (P. guajava). In addition to Musa spp., C. musae has been reported to be pathogenic to apple (Malus pumila), mango (Mangifera indica), avocado (Persea americana), guava (P. guajava) and Vigna sp. (Sutton & Waterston 1970).
C. gloeosporioides was isolated from anthracnose of water apple (S. aqueum). In cultural and morphological studies and RAPD analysis, C. gloeosporioides appeared to be different from C. musae; C. gloeosporioides is considered to be a group species or species complex and is found on a wide variety of fruits, such as apple, avocado, citrus, papaya, peach, mango and strawberry (Freeman 2000).
In the RAPD analysis, variation was observed within C. musae isolates. This variation may be explained by the presence of a perfect stage of C. musae, i.e., Gloeosporium musarum. The variation may also be attributed to the adaptation of the species to a non-specific, broad host range (Freeman et al. 1998). For Colletotrichum species, it is common for single hosts to become infected by a single species or for multiple hosts to be infected by a single species of the pathogen (Freeman 2000). Infection of multiple hosts by C. musae has been reported by Sutton and Waterston (1970).
The data from this study provide a basic understanding of the population structure and genetic diversity of C. musae isolates associated with anthracnose of different banana cultivars; we hope these data will help us develop effective control methods for the disease. The management and control of banana anthracnose currently includes the application of fungicides and the use of resistant banana cultivars. The population structure and genetic diversity of a pathogen is likely to affect the pathogen’s ability to evolve in response to these control measures (Abang et al. 2003). Therefore, knowledge of the genetic diversity of the pathogen population is a prerequisite for implementing effective programs for screening banana cultivars for resistance to anthracnose and to avoid fungicide resistance.
In conclusion, the present study used cultural and morphological characteristics and RAPD analysis to identify C. musae as the most common species associated with anthracnose of different banana cultivars.
REFERENCES
- Abang MM, Winter S, Mignouna HD, Green KR, Asiedu R. Molecular taxonomic, epidemiological and population genetic approaches to understanding yam anthracnose disease. African Journal of Biotechnology. 2003;2(12):486–496. [Google Scholar]
- Anthony S, Abeywickrama K, Dayananda R, Wijeratnam SW, Arambewela L. Fungal pathogens associated with banana fruits in Sri Lanka and their treatment with essential oils. Mycopathologia. 2004;157:91–97. doi: 10.1023/b:myco.0000012226.95628.99. [DOI] [PubMed] [Google Scholar]
- Chakravarty T. Anthracnose of banana (Gloeosporium musarum cooke and massae) with special reference to latent infection in storage. Transactions of the British Mycological Society. 1957;40:337–345. [Google Scholar]
- Freeman S. Genetic diversity and host specificity of Colletotrichum species on various fruits. In: Prusky D, Freeman S, Dickman MB, editors. Colletotrichum: host specificity, pathology and host pathogen interaction. Minnessota, USA: American Pathological Society Press; 2000. pp. 131–144. [Google Scholar]
- Freeman S, Katan T, Shabi E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Disease. 1998;82:596–605. doi: 10.1094/PDIS.1998.82.6.596. [DOI] [PubMed] [Google Scholar]
- Jeger MJ, Eden-Green S, Thresh JM, Johannson A, Waller JM, Brown AE. Banana diseases. In: Gowen SR, editor. Banana and plantains. London: Chapman and Hall; 1995. pp. 3117–3381. [Google Scholar]
- Jones DR, Slabaugh WR. Anthracnose and fungal scald. In: Ploetz RC, Zentmyer GA, Nishijima WT, Rohrbach KG, Ohr HD, editors. Compendium of tropical fruit diseases. Minnessota, USA: American Pathological Society Press; 1998. pp. 4–5. [Google Scholar]
- Meredith DS. Studies on Gloeosporium musarum (Gke and massee) causing storage rots of Jamaican bananas. Anthracnose and its chemical control. Annals of Applied Biology. 1960;48(2):279–290. [Google Scholar]
- Mordue JEM. CMI description of pathogenic fungi and bacteria, No. 315. London: Eastern Press; 1971. Glomerella cingulata. [Google Scholar]
- Prusky D, Plumbley RA. Quiescent infections of Colletotrichum in tropical and subtropical fruits. In: Bailey JA, Jeger MJ, editors. Colletotrichum: Biology, pathology and control. Wallingford, UK: CAB International; 1992. pp. 289–307. [Google Scholar]
- Rohlf FJ. NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2.1. Setauket, New York: Exeter Publishing Ltd; 2000. [Google Scholar]
- Sutton BC, Waterston NW. CMI Description of pathogenic fungi and bacteria, No. 222. London: Eastern Press; 1970. Colletotrichum musae. [Google Scholar]