Abstract
Contamination of paddy seeds (rice with husk) by Fusarium species can cause spoilage and subsequent production of mycotoxins, especially fumonisins that affect human and animal health. A mycological study was conducted to evaluate the natural occurrence of fumonisin B1 produced by Fusarium proliferatum on paddy grown in different geographic regions of Karnataka (India). A total of 65 isolates of F. proliferatum from paddy samples were analysed by polymerase chain reaction (PCR). One set of primers, Fp3-F and Fp4-R was employed to identify the species F. proliferatum, and another set of primers, FUM1 was employed to determine the fumonisin producing ability of the isolates. All 65 isolates of F. proliferatum scored positive with both set of primers, producing amplified products of the expected sizes. Furthermore, thin layer chromatography (TLC) analysis detected fumonisin B1 (FB1) in all of the PCR positive isolates of F. proliferatum.
Keywords: Fp3-F/Fp4-R, FUM1, PCR Diagnostics, TLC
Abstract
Pencemaran biji benih padi (beras bersama sekam/ sekam padi) oleh spesies Fusarium boleh mengakibatkan kerosakan dan diikuti dengan penghasilan mikotoksin, terutamanya fumonisin yang memberi kesan terhadap kesihatan manusia dan haiwan. Satu kajian mikologi telah dijalankan untuk menilai penghasilan fumonisin secara semulajadi oleh Fusarium proliferatum dalam padi yang tumbuh di kawasan-kawasan geografi berbeza di Karnataka (India). Sejumlah 65 sampel F. proliferatum diasingkan daripada padi dan dianalisa menggunakan polymerase chain reaction (PCR). Satu set primer-primer Fp3-F dan Fp4-R telah digunakan untuk mengenal pasti spesies F. proliferatum manakala satu lagi set primer-primer, FUM1 telah digunakan untuk mengenal pasti kebolehan menghasilkan fumonisin. Kesemua 65 sampel F. proliferatum memberi keputusan positif kepada kedua-dua set primer, dengan menghasilkan produk yang dibesarkan kepada saiz yang dijangka. Tambahan lagi, melalui analisa thin layer chromatography (TLC), fumonisin B1 (FB1) telah dikesan di kesemua sampel F. proliferatum yang memberi keputusan PCR yang positif.
INTRODUCTION
Paddy is an important worldwide food crop and is the staple food for most of Asia. Paddy is the basic food for the Indian population, as it is a rich source of dietary energy and a good source of amino acids, such as thiamine, riboflavin, glutamic acid and niacin (FAO 2004). However, paddy is highly susceptible to fungal infections, such as Fusarium species (Trung et al. 2001), F. proliferatum (Matsushima) Nirenberg and F. verticillioides (Sacc.) Nirenberg (synonym F. moniliforme Sheldon), which produce mycotoxins, such as trichothecenes, zearalenones and fumonisins (Abbas et al. 1998; Desjardins et al. 2000). Fumonisin B1 (FB1) and fumonisin B2 (FB2) are the major toxins of the family of mycotoxins produced by F. proliferatum and F. verticillioides. While fumonisin production by F. verticillioides has been well studied there is limited data for F. proliferatum (Rheeder et al. 2002).
Fusarium contamination of paddy has been reported in many parts of the world (Tonon et al. 1997; Pacin et al. 2002), but with less frequency compared to other cereals. Despite the importance of paddy as a staple food and the reported occurrence of mycoflora and fumonisins (Park et al. 2005; Hinojo et al. 2006), there is a scarcity of information on the incidence of Fusarium species and fumonisin contamination in paddy produced in the Indian state of Karnataka.
The International Agency for Research on Cancer (IARC) evaluated FB1 as a possible carcinogen to humans (Group 2B) (Domijan et al. 2005). It is important to detect fumonisins in order to prevent human and animal exposure to such toxic substances. Polymerase chain reaction (PCR) protocols based on intergenic spacer (IGS) sequences have been extensively used for the accurate detection of Fusarium species (Jurado et al. 2006). In the present investigation, species-specific primers Fp3-F and Fp4-R based on IGS sequences were used to detect F. proliferatum isolated from paddy. In addition, the fumonisin producing ability of these isolates was confirmed by PCR using a second set of primers, FUM1, and by thin layer chromatography (TLC) studies that detect FB1.
MATERIALS AND METHODS
Chemicals and Reagents
The reagents for PCR, including primers, were procured from Bangalore Genei, Bangalore (India). All other chemicals used in the study were of reagent grade (Merck Ltd., Mumbai). FB1 standard was purchased from Sigma Aldrich Chemicals Pvt. Ltd., Bangalore, India.
Collection of Paddy Seed Samples
In total, 109 lots of different paddy seed samples were collected from farmer’s fields from 15 districts in the state of Karnataka during the month of June 2006. The seed samples (0.5 kg) were packed, appropriately labelled and used for further studies.
Isolation of Fusarium Species
Isolation of Fusarium species from the 109 paddy samples was accomplished by using modified malachite green agar medium containing malachite green oxalate (2.5 mg/l) (MGA 2.5) (Bragulat et al. 2004). Modified Czapek-Dox agar (CZA) and Spezieller Nährstoffarmer Agar media (SNA) were used (Leslie & Summerell 2006) to maintain cultures.
For mycological analysis, 109 paddy samples were subjected to sampling by the hand-halving method (Mathur & Kongsdal 2003). A total of 200 paddy seeds from each sample were surface sterilised by treatment with 1% sodium hypochlorite solution for 1 min and rinsed twice in sterile distilled water. The surface-sterilised seeds were plated (10 per plate) on MGA 2.5 containing chloramphenicol (50 mg/l). The plates were incubated at 26°C for 5 days. The developing fungal colonies were counted and Fusarium species were isolated and sub-cultured on potato dextrose agar medium. The cultures were identified using fungal keys and manuals (Booth 1977; Leslie & Summerell 2006).
The isolation frequency (Fr) (Gonzalez et al. 1995) of the Fusarium isolates was calculated as below:
DNA Extraction
Genomic DNA was extracted from isolates of F. proliferatum (65), F. verticillioides (27), F. semitectum Berkeley & Ravenel (12) and F. oxysporum Schlechtendahl emend Snyder & Hansen (10). Each fungal species was inoculated aseptically into 500 μl of sterile potato dextrose broth in 2 ml microfuge tubes. The tubes were incubated for 5 days at 28°C and DNA was isolated from each fungal sample (Zhang et al. 1998). Mycelia from the microfuge tubes were centrifuged at 5000 rpm (REMI C24 Cooling Centrifuge, Chennai) for 8 min and the supernatant was discarded. The mycelium was resuspended in cell lysis buffer (2% CTAB, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris-HCl, pH 8.0, preheated at 65°C) and heated at 65°C for 20 min in a water bath. An equal volume of phenol:chloroform (1:1) was added to each tube and centrifuged at 3000 rpm for 5 min. The supernatants were transferred to new microfuge tubes and an equal volume of isopropyl alcohol was added. The microfuge tubes were incubated at −20°C for 2 h and then centrifuged at 8000 rpm for 8 min to precipitate the DNA. The DNA pellet was air-dried and resuspended in 20 μl of nuclease free water and used directly for PCR analysis.
Primers for PCR
A pair of primers, Fp3-F (5′CGGCCACCAGAGGATGTG 3′) and Fp4-R (5′ CAACACGAATCGCTTCCTGAC 3′) (Jurado et al. 2006), specific to F. proliferatum was used for species identification. Another set of primers specific to the fum1 gene (involved in fumonisin biosynthesis), FUM1 forward (5′-CCATCACAGTGGGACACAGT-3′) and FUM1 reverse (5′-CGTATCGTCAG CATGATGTAGC-3′) (Bluhm et al. 2004), was used to determine the fumonisin producing ability of F. proliferatum. The expected amplicon sizes were 230 bp and 183 bp, respectively.
Polymerase Chain Reaction
Genomic DNA was subjected to PCR analysis using Advanced Primus 25 Thermocycler (Peqlab, Germany). The PCR mixture for primer Fp3-F and Fp4-R contained 100 ng of genomic DNA, 1.25 μl of each primer (20 pmol), 0.5 μl of Taq DNA polymerase (3 U/μl), 2.5 μl of 10X PCR buffer, 1.0 μl of MgCl2 (25 mM) and 1 μl of 2 mM dNTPs. The final volume was brought up to 25 μl with nuclease free water.
PCR conditions were set at 94°C for 4 min for the initial denaturation, followed by 35 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 40 s, primer extension at 72°C for 1 min and the final extension at 72°C for 5 min.
The PCR mixture for FUM1 primers contained 2 μl of genomic DNA, 1 μl of each FUM1 primers (20 pmol), 0.5 μl of Taq DNA polymerase (3 U/μl), 2.5 μl of 10X PCR buffer, 2.5 μl of MgCl2 (25 mM) and 1 μl of 2 mM dNTPs. The final volume was made up to 25 μl with nuclease free water.
The PCR conditions were 94°C for 4 min for the initial denaturation, followed by 35 cycles of denaturation at 94°C for 1 min, primer annealing at 60°C for 1 min, primer extension at 72°C for 1 min and the final extension at 72°C for 5 min. Amplified products were analysed on a 1.5% agarose gel in 1X TAE buffer (40 mM Tris acetate and 1.0 mM EDTA) and documented with a gel documentation system (UTP-Bio Doc, USA).
Sample Preparation for Thin Layer Chromatography
PCR positive isolates of F. proliferatum (25) were cultured on rice medium for FB1 detection. About 30 g of polished rice in 9 ml of sterile distilled water were autoclaved in a 250 ml conical flask. A conidial suspension (106/ml) of each F. proliferatum isolate was prepared by adding 7 ml of sterile water to a Petri dish containing a 14-day-old fungal culture. Each flask containing the rice medium was inoculated with 5 ml of conidial suspension, mixed well and incubated at 28°C for 28 days in the dark. Rice cultures were oven-dried overnight at 60°C and finely ground in a laboratory mill. The powdered samples were stored at −20°C until further analysis (Desjardins et al. 2000).
Extraction and Cleanup
FB1 was extracted from the finely ground sample using a protocol developed by Rice et al. (1995). 10 g samples were placed into a 250 ml conical flask containing 50 ml of acetonitrile:water (ACN:water, 50+50, v/v) and the flask was covered and shaken for 30 min. 15 ml of the supernatant was filtered through a Whatman No. 4 filter paper. A C18 Sep Pak solid phase extraction (SPE) clean up column (Waters, USA) was preconditioned by rinsing with 2 ml ACN followed by 2 ml of 1% aqueous potassium chloride (KCl). The filtered extract (2 ml) and 1% KCl (6 ml) were mixed in a vial and applied to the column.
The solution flowed through the SPE column at a flow rate of 4 ml/min. The column was rinsed with 2 ml of 1% KCl followed by 2 ml of ACN:water (15+85, v/v). The rinse was discarded and air was forced through the column to expel all the rinse solution. FB1 was eluted from the column with 2 ml of ACN:water (70+30, v/v).
Following extraction and clean up the samples were analysed by TLC (Bailly et al. 2005). Sample extracts (5 μl) and FB1 standards (200 ng/ml and 400 ng/ml) were spotted on TLC plates (AluGram SIL G/UV 254, Machery Nagel, Germany). Separation was carried out in a mixture of 1-butanol:acetic acid:water (20+10+10, v/v/v). After drying, plates were sprayed with a solution containing methanol:0.5% p-anisaldehyde:acetic acid:sulphuric acid (85:0.5:10.5 v/v) and heated for 10 min at 110°C. The plates were observed under daylight conditions for the presence of purple FB1 spots.
RESULTS
In total, 109 paddy samples screened on MGA 2.5 containing chloramphenicol yielded 10 different Fusarium species namely, F. solani (Martius) Appel & Wollenweber emend (63.3%), F. anthophilum (A. Braun) Wollenweber (59.63%), F. oxysporum (55%), F. semitectum (54.12%), F. sporotrichioides Sherbakoff (50.45%), F. proliferatum (40.36%), F. verticillioides (24.77%), F. graminearum Schwabe (23.85%), F. lateritium Nees (16.51%) and F. poae (Peck) Wollenweber (14.67%). F. proliferatum isolates produced aerial mycelium with purple violet pigmentation on the agar medium. Many polyphialides were observed with abundant club shaped microconidia in long chains. The macroconidia were slender, thin walled, curved and 5-septate. Chlamydospores were absent.
PCR Amplification
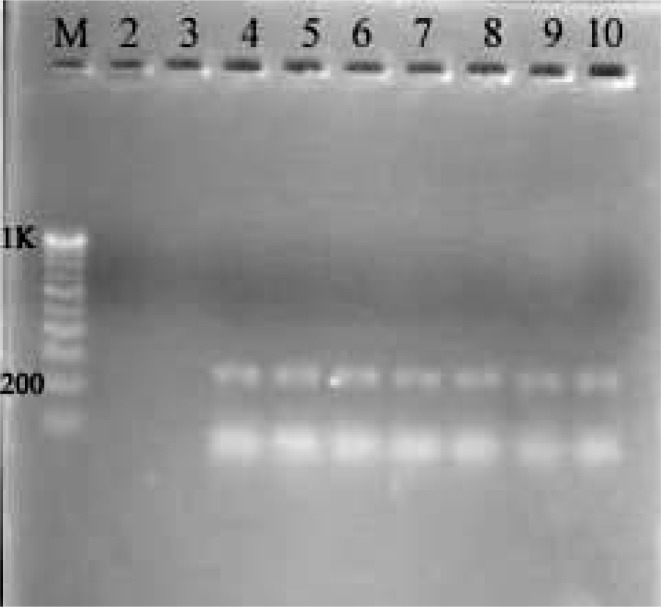
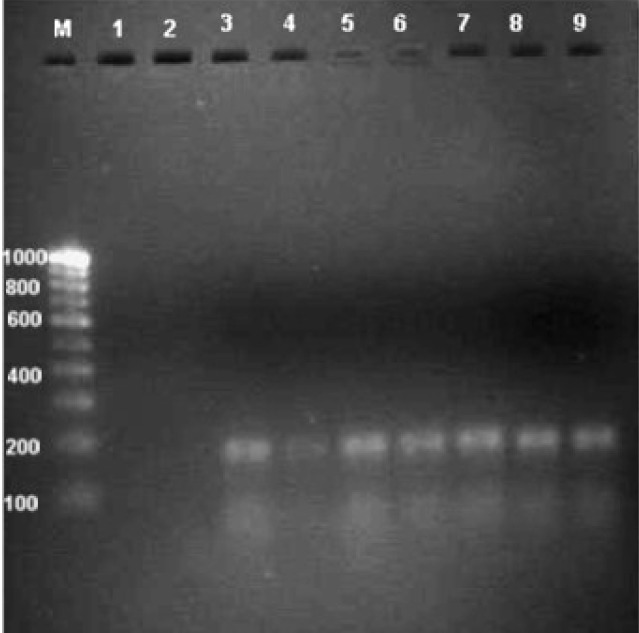
The species-specific primers Fp3-F and Fp4-R used in this study amplified a 230 bp PCR products in all 65 isolates of F. proliferatum; such amplified products were not detected in other Fusarium species tested (Fig. 1). Furthermore, all 65 isolates of F. proliferatum and 27 isolates of F. verticillioides scored positive with the FUM1 set of primers by producing a 183 bp product, indicating their potential fumonisin producing ability. No such amplification was detected in other Fusarium species tested (Fig. 2).
Figure 1:
Agarose gel (1.5%) showing PCR products using the Fp3-F/Fp4-R (230 bp) set of primers specific to F. proliferatum. Lane (M): (100 bp) DNA ladder; lane (2): F. semitectum; lane (3): F. verticillioides; lane (4–10): F. proliferatum.
Figure 2:
Agarose gel (1.5%) showing the amplified products of fumonisin producing F. proliferatum and F. verticillioides with the FUM1 (183 bp) set of primers. Lane (M): (100 bp) DNA ladder; Lane (1): F. semitectum; lane (2): F. oxysporum; lane (3–4); F. verticillioides; lane (5–9): F. proliferatum.
TLC Analysis
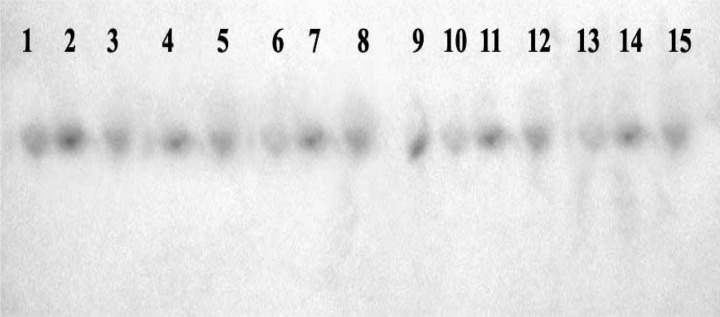
TLC revealed the occurrence of purple-coloured FB1 spots having relative front (Rf) values of 0.75, in all the 25 isolates of F. proliferatum (Fig. 3).
Figure 3:
TLC plate showing purple coloured spots after spraying with a mixture of 0.5% p-anisaldehyde. Lane (1–2): FB1 standards (200 ng and 400 ng, respectively); lane (3–15): culture extracts of FB1 from F. proliferatum isolates.
DISCUSSION
Mycotoxicological surveys of cereals are very important, especially in tropical countries, where temperature and relative humidity are favourable for fungal growth. Like all other cereals, paddy is susceptible to damage by mycotoxigenic fungi. After harvest, it is subjected to traditional sun drying for 3 to 5 days prior to being sent to the mills where it is stored for human and animal consumption. Due to this post harvest practice, paddy is susceptible to damage by fungi, especially the Fusarium species. Fusariums produce mycotoxins, such as fumonisins. Mycological analysis of 109 paddy seed samples revealed the presence of 10 different species of Fusarium. In terms of frequency, F. solani ranked the highest (63.3%). However, in terms of fumonisin producing ability, F. proliferatum (40.36%) ranked first. F. proliferatum is one of the dominant species on paddy and would probably be the main source of contamination by fumonisins. This finding is in agreement with earlier studies (Tonon et al. 1997; Pacin et al. 2002), which reported the Fusarium species (especially F. proliferatum) to frequently contaminate paddy. In a study by Abbas et al. (1998), paddy samples infected with F. proliferatum (40%) were found positive for FB1 at levels of 4.3 μg/g. Infection of paddy with F. proliferatum in Korean polished rice revealed the presence of FB1 at a concentration of 4.4–7.1 μg/g (Park et al. 2005).
Epidemiological studies have linked the human esophageal carcinoma to the consumption of maize with high incidences of F. verticillioides contamination (Marasas et al. 1981; Yoshizawa et al. 1994). Hence, there is a need for a rapid detection of the toxin producing fungi for the effective management of fumonisins in cereals. Molecular methods such as PCR have been developed for the detection of fumonisins to replace traditional methods based on molecular probes (Jurado et al. 2005). Unlike methods involving culturing, PCR does not require the presence of viable organisms for detection and may be performed even with small amounts of sample.
We employed diagnostic methods based on PCR to accurately detect fumonisin producers. F. proliferatum were identified using species-specific primers based on IGS sequences. Additionally, the use of the FUM1 primers, designed based on the fum1 gene, for the detection of fumonisin producing strains proved to be highly useful for identifying toxigenic Fusarium isolates. All 65 isolates tested scored positive with the species-specific primers. Furthermore, all 65 isolates of F. proliferatum were positive using the FUM1 primer set, indicating that all 65 isolates have the potential ability to produce fumonisin. Apart from the F. proliferatum species, 27 isolates of F. verticillioides also scored positive with the FUM1 set of primers.
The IGS region, which is commonly used for identification purposes in taxonomic studies, was found to be appropriate for the molecular detection of F. proliferatum in this study. The IGS regions contain high levels of sequence variability among the species of the same genus and allow differentiation of genetically related species (Edel et al. 2000; Kim et al. 2001; Konietzny & Greiner 2003; González-Jaén et al. 2004).
TLC, an economical analytical procedure, was employed for the detection of fumonisin produced by F. proliferatum isolates. TLC remains an important tool for mycotoxin detection in countries that often produce and export agricultural commodities but do not have expensive equipment at their disposal. As opposed to labour intensive and time consuming advanced high pressure liquid chromatography (HPLC) screening methods (Rottinghaus et al. 1992), TLC is a relatively easy and useful technique.
CONCLUSION
The presence of Fusarium species in food and animal feed can be a threat to the food chain, as fumonisins are known to survive heat processing. The present study revealed the natural occurrence of fumonisin producing F. proliferatum species on paddy seeds produced in Karnataka (India). The risks associated with F. proliferatum contaminated paddy warrants the need for further analysis of this basic food that is intended for human and animal consumption.
REFERENCES
- Abbas HK, Cartwright RD, Shier WT, Abouzied MM, Bird CB, Rice LG, Ross PF, Sciumbato GL, Meredith FI. Natural occurrence of fumonisins in rice with Fusarium sheath rot disease. Plant Disease. 1998;82:22–25. doi: 10.1094/PDIS.1998.82.1.22. [DOI] [PubMed] [Google Scholar]
- Bailly JD, Querin A, Tardieu D, Guerre P. Production and purification of fumonisins from a highly toxigenic Fusarium verticillioides strain. Revue de Médecine Vétérinaire. 2005;156(11):547–554. [Google Scholar]
- Bluhm BH, Cousin MA, Woloshuk CP. Multiplex real time PCR detection of fumonisin producing and trichothecenes producing groups of Fusarium species. Journal of Food Protection. 2004;67(3):536–543. doi: 10.4315/0362-028x-67.3.536. [DOI] [PubMed] [Google Scholar]
- Booth C. Kew, England: Commonwealth Mycological Institute; 1977. Fusarium: Laboratory guide to the identification of major species. [Google Scholar]
- Bragulat MR, Martinez E, Castella G, Cabanes FJ. Selective efficacy of culture media recommended for isolation and enumeration of Fusarium species. Journal of Food Protection. 2004;67:207–211. doi: 10.4315/0362-028x-67.1.207. [DOI] [PubMed] [Google Scholar]
- Desjardins AE, Manandhar HK, Plattner RD, Manandhar GG, Poling SM, Maragos CM. Fusarium species from Nepalese rice and production of mycotoxins and gibberellic acid by selected species. Applied and Environmental Microbiology. 2000;66(3):1020–1025. doi: 10.1128/aem.66.3.1020-1025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domijan A, Peraica M, Jurjevic Z, Ivic D, Cvjetkovi B. Fumonisin B1, fumonisin B2, zearalenone and ochratoxin A contamination of maize in Croatia. Food Additives and Contaminants. 2005;22(7):677–680. doi: 10.1080/02652030500132927. [DOI] [PubMed] [Google Scholar]
- Edel V, Steinberg C, Gautheron N, Alabouvette C. Ribosomal DNA-targeted oligonucleotide probe and PCR assay specific for Fusarium oxysporum. Mycological Research. 2000;104(5):518–526. [Google Scholar]
- FAO. Food and Nutrition Division Food and Agriculture Organization of the United Nations Viale delle Terme di Caracalla, Rome 00100 Italy. 2004. http://www.fao.org/rice2004/en/f-sheet/factsheet3.pdf (accessed on 12 February 2004).
- González-Jaén MT, Mirete S, Patińo B, Lopez-Errasquín E, Vázquez C. Genetic markers for the analysis of variability and for production of specific diagnostic sequences in fumonisin-producing strains of Fusarium verticillioides. European Journal of Plant Pathology. 2004;110:525–532. [Google Scholar]
- Gonzalez HH, Resnik SL, Boca RT, Marasas WFO. Mycoflora of Argentinian corn harvested in the main production area in 1990. Mycopathologia. 1995;130:29–36. doi: 10.1007/BF01104346. [DOI] [PubMed] [Google Scholar]
- Hinojo MJ, Medina A, Valle-algarra FM, Gimeno-adelantado JV, Jiménez M, Mateo R. Fumonisin production in rice cultures of Fusarium verticillioides under different incubation conditions using an optimized analytical method. Food Microbiology. 2006;23:119–127. doi: 10.1016/j.fm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Jurado M, Vázquez C, Patińo B, González-Jaén MT. PCR detection assays for the trichothecene-producing species Fusarium graminearum, Fusarium culmorum, Fusarium poae, Fusarium equiseti and Fusarium sporotrichioides. Systematic and Applied Microbiology. 2005;28(6):562–568. doi: 10.1016/j.syapm.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Jurado M, Vázquez C, Marín S, Sanchis V, González-Jaén MT. PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in maize. Systematic and Applied Microbiology. 2006;29(8):681–689. doi: 10.1016/j.syapm.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Choi YK, Min BR. Variation of the Intergenic Spacer (IGS) region of ribosomal DNA among Fusarium oxysporum formae speciales. The Journal of Microbiology. 2001;39(4):265–272. [Google Scholar]
- Konietzny U, Greiner R. The application of PCR in the detection of mycotoxigenic fungi in foods. Brazilian Journal of Microbiology. 2003;34:283–300. [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium laboratory manual. 1st ed. Ames, Iowa: Blackwell Publishing; 2006. [Google Scholar]
- Marasas WFO, Wehner FC, Van Rensburg SJ, Van Schalkwyk DJ. Mycoflora of corn produced in human esophageal cancer areas in Transkei, South Africa. Phytopathology. 1981;71:792–796. [Google Scholar]
- Mathur SB, Kongsdal O. Common laboratory seed health testing methods for detecting fungi. Switzerland: International Seed Testing Association (ISTA); 2003. [Google Scholar]
- Pacin AM, Gonzalez HHL, Etcheverry M, Resnik SL, Vivas Land, Espin S. Fungi associated with food and feed commodities from Ecuador. Mycopathologia. 2002;156:87–92. doi: 10.1023/a:1022941304447. [DOI] [PubMed] [Google Scholar]
- Park JW, Choi SY, Hwang HJ, Kim YB. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. International Journal of Food Microbiology. 2005;103:305–314. doi: 10.1016/j.ijfoodmicro.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Rheeder JP, Marasas WFO, Vismer HF. Production of fumonisin analogs by Fusarium species. Applied and Environmental Microbiology. 2002;68(5):2101–2105. doi: 10.1128/AEM.68.5.2101-2105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LG, Ross PF, Dejong J, Plattner RD, Coats JR. Evaluation of a liquid chromatographic method for the determination of fumonisins in corn, poultry feed and Fusarium culture material. Journal of AOAC International. 1995;78:1002–1009. [PubMed] [Google Scholar]
- Rottinghaus GE, Coatney CE, Minor HC. A rapid, sensitive thin layer chromatography procedure for the detection of fumonisin B1 and B2. Journal of Veterinary Diagnostic Investigation. 1992;4:326–329. doi: 10.1177/104063879200400316. [DOI] [PubMed] [Google Scholar]
- Tonon SA, Marucci RS, Jerke G, Garcia A. Mycoflora of paddy and milled rice produced in the Northeastern Argentina and Southern Paraguay. International Journal of Food Microbiology. 1997;37:231–235. doi: 10.1016/s0168-1605(97)00066-4. [DOI] [PubMed] [Google Scholar]
- Trung TS, Bailly JD, Querin A, Le Bras P, Guerre P. Fungal contamination of rice from South Vietnam, mycotoxinogenesis of selected strains and residues in rice. Revue de Médecine Véterinaire. 2001;152(7):555–560. [Google Scholar]
- Yoshizawa T, Yamashita A, Luo Y. Fumonisin occurrence in corn from high- and low-risk areas for human esophageal cancer in China. Applied and Environmental Microbiology. 1994;60(5):1626–1629. doi: 10.1128/aem.60.5.1626-1629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YP, Uyemoto JK, Kirkpatrick BC. A small-scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. Journal of Virological Methods. 1998;71:45–50. doi: 10.1016/s0166-0934(97)00190-0. [DOI] [PubMed] [Google Scholar]