Abstract
Species identification is important for epidemiological, clinical and treatment purposes. The aim of this study was to find out whether hippurate hydrolysis is a reliable test for differentiating between Campylobacter coli and Campylobacter jejuni. To achieve this, hippurate hydrolysis test was compared with multiplex Polymerase Chain Reaction (mPCR) for their ability to speciate C. coli and C. jejuni. Eighteen Campylobacter strains from poultry samples were used for this study. The results from 17 of the 18 strains were in agreement with both methods. Thus, the hippurate hydrolysis test can be used for distinguishing C. jejuni from C. coli although occasionally some strains of C. jejuni may be mis-identified as C. coli.
Keywords: C. coli, C. jejuni, Hippurate Hydrolysis, mPCR
Abstract
Pengenalpastian spesies penting untuk tujuan epidemiologikal, klinikal dan perubatan. Tujuan kajian ini adalah untuk mengetahui sama ada ujian hidrolisis hippurate berkesan untuk membezakan antara Campylobacter coli dan Campylobacter jejuni. Untuk mencapai ini, ujian hidrolis hippurate dibandingkan dengan multiplex Polymerase Chain Reaction (mPCR) untuk mengkaji kebolehan membezakan spesies C. coli dan C. jejuni. Lapan belas strain Campylobacter daripada sampel haiwan ternakan digunakan. Keputusan 17 daripada 18 strain menunjukkan keberkesanan menggunakan kedua-dua jenis ujian. Maka ujian hidrolisis hippurate berkesan untuk membezakan C. jejuni daripada C. coli, walaupun kadang-kadang terdapat strain C. jejuni yang diidentifikasi secara salah sebagai C. coli.
Campylobacters are Gram-negative, nonspore-forming, oxidase and catalase positive, curved spiral or rod shaped bacteria, that are microaerophilic in nature and unable to grow at 25°C (Corry et al. 2003). They are also motile, with either uni- or bi-polar flagella, 0.2–0.5 mm wide and 0.5–8 mm long (Corry et al. 2003; Moore et al. 2005). Campylobacters cannot ferment carbohydrate because they do not have the enzyme phosphofructokinase which is engaged in energy metabolism (Velayudhan & Kelly 2002) but obtain their energy from amino acids and/or tricarboxylic acid cycle intermediates (Vandamme 2000; EFSA 2005).
Campylobacters have been reported to be the most common cause of foodborne bacterial enteritis (Mead et al. 1999). The European Food Safety Authority (EFSA 2005) report also indicated that the number of campylobacteriosis cases per year have surpassed salmonellosis in the European Union (EU) nations. In most developing countries including Africa reliable data on foodborne illness cases is unavailable (Adzitey & Nurul 2011). The genus Campylobacter is made up of 17 species (On 2001) of which C. jejuni and C. coli are the most important in terms of food safety. C. jejuni is responsible for about 90% of all campylobacter infections, and most of the rest are caused by C. coli (EFSA 2005). C. jejuni infection can lead to serious autoimmune diseases such as Guillain-Barré syndrome and reactive arthritis.
Hippurate hydrolysis relies on the ability of the enzyme called hippurate hydrolase produced by microorganisms to hydrolyse sodium hippurate to benzoic acid and glycine. This test does not require the microorganisms to grow, but instead it detects the presence of already formed enzyme by testing for glycine, one of the end products of the hydrolysis. If glycine is present a blue or deep purple colour is formed. Hippurate hydrolysis has been successfully used to identify group B streptococci (Hwang & Ederer 1975; Mugg 1983). In recent years, several Polymerase Chain Reaction (PCR)-based techniques have been mentioned or described which are more specific, accurate and sensitive than phenotypic methods for distinguishing Campylobacter species (EFSA 2005; Didelot & Falush 2007; Ridley et al. 2008; Adzitey & Nurul 2011). However, most Campylobacter isolates from human cases or from poultry are either C. jejuni or C. coli, and it is important for clinical and treatment purposes to be able to distinguish between them by means of a simple and economical test.
Correct differentiation of C. coli from C. jejuni is important particularly in the treatment of human illness, because the antibiotics employed will depend on the causative species. For example, erythromycin is commonly used to treat gastrointestinal infections caused by C. jejuni, while C. coli is more likely to be resistant to this antibiotic, rendering treatment ineffective if C. coli is the causative agent (Aarestrup et al. 1997). It is also important to differentiate between these two pathogenic species because C. jejuni infection is an important predisposal factor in the development of Guillian-Barré syndrome, as well as reactive arthritis and Reiter’s syndrome whereas C. coli is less strongly associated with these sequelae (Smith 1995). This paper compares the results of distinguishing C. jejuni and C. coli using the hippurate hydrolysis test and multiplex PCR (mPCR).
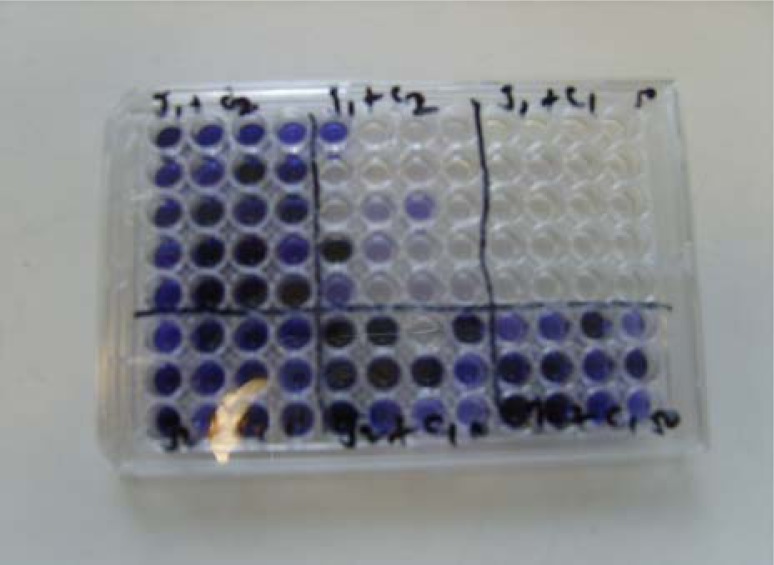
The study was conducted in the laboratory of the School of Veterinary Medicine, University of Bristol (UK) using 18 campylobacter isolates originating from carcasses, faeces and caecal contents taken from different poultry flocks and various poultry processing plants.The ability of the Campylobacter strains to hydrolyse hippurate was checked using a modification of the method of Hwang and Ederer (1975). 100 μl of 1% (w/v) sodium hippurate solution were dispensed into each well of a microtitre plate. A loopful (1 μl) of Campylobacter culture grown on modified cefaperazone charcoal deoxycholate agar (mCCDA) microaerobically for 48 h at 41.5°C was added, agitated with the loop to produce a suspension, and covered with cling film for incubation aerobically for 4 h at 37°C. After incubation, 50 μl of 3.5% (w/v) ninhydrin solution was added to each suspension, mixed well and incubation was continued at 37°C for 30 min before the results were checked. A deep purple (not medium or light purple) positive reaction was developed by all C. jejuni strains. Figure 1 shows a typical example of colour changes observed for the hippurate test.
Figure 1:
Representative results of hippurate hydrolysis test carried out on microtitre plate.
Note: Deep purple colour indicates C. jejuni while medium or no colour change indicates C. coli.
DNA templates were prepared by adding 10 μl loopful of Campylobacter culture from mCCDA plate to 500 μl peptone buffered saline (PBS) contained in an eppendorf tube and heated to 100°C for 10 min using a heating block. Primers were made to a concentration of 100 pmol according to manufacturer’s (MWG Operon, Eberberg, Germany), instructions. They were then diluted to a working concentration of 10 μM by adding 10 μl of concentrated (100 pmol) primer stocks into 90 μl nuclease free water. The primers used for the detection of C. jejuni and C. coli strains are listed in Table 1. The PCR mixture for one reaction contained 0.5 μl of each primer (10 μM concentration), 12.5 μl hotstart taq mastermix (Qiagen, West Sussex, UK), 2.75 μl nuclease free water, 0.75 μl of 50 mM magnesium chloride (Qiagen, West Sussex, UK) and 5 μl template DNA. The temperature cycling was performed at 95°C for 15 min, 30 cycles of denaturing at 94°C for 1 min, annealing at 60°C for 1 min and extension at 72°C for 1 min, with a final extension time of 72°C for 10 min.
Table 1:
Primers used for the identification of C. jejuni and C. coli by mPCR.
| Species | Target gene | Reaction direction | Sequence (5’-3’) | Reference |
|---|---|---|---|---|
| C. jejuni | lpxA | forward | ACA ACT TGG TGA CGA TGT TGT A | Klena et al. (2004) |
| reverse lpxARKK2m |
CAA TCA TGD GCD ATA TGA SAA TAH GCC AT |
|||
| hipO | forward | ACT GCA AAA TTA GTG GCG | Bang et al. (2002) | |
| reverse | GAG CTT TTA GCA AAC CTT CC | |||
| C. coli | lpxA | forward | AGA CAA ATA AGA GAG AAT CAG | Klena et al. (2004) |
| reverse lpxARKK2m |
CAA TCA TGD GCD ATA TGA SAA TAH GCC AT |
|||
| glyA | forward | TCA AGG CGT TTA TGC TGC AC | Dingle et al. (2005) | |
| reverse | CCA TCA CTT ACA AGC TTA TAC |
The restriction fragments were separated by gel electrophoresis, on a 2% agarose gel prepared by adding 4 g agarose (Sigma-Aldrich, Dorset, UK) to 200 ml tris acetate EDTA (1xTAE) buffer (Sigma, Dorset, UK) containing 1 μg ml−1 ethidium bromide (Sigma, Dorset, UK), and visualised on an ultra violet transilluminator (UVP BioDoc-It™ imaging-system, Cambridge, UK). Hyperladder IV (Bioline, London, UK) was used as the molecular weight marker and band positions were determined by eye using the molecular weight marker.
Comparison of the 18 isolates tested for hippurate hydrolysis and mPCR assay demonstrated that the same species result was obtained for 17 isolates. Figure 1 shows a representative sample of how hippurate positive (C. jejuni) and hippurate negative (C. coli) Campylobacter species look on a microtitre plate. Deep purple coloured well indicates C. jejuni whilst medium, light or no colour changes is C. coli. A comparison between hippurate hydrolysis and mPCR for the identification of C. jejuni and C. coli is depicted in Table 2.
Table 2:
Comparison between hippurate hydrolysis and mPCR for the identification of C. jejuni and C. coli.
| Campylobacter isolate used | Colour | Hippurate results | Lane | mPCR results |
|---|---|---|---|---|
| C. coli | medium | C. coli | 1 | C. coli |
| C. jejuni | medium | C. coli | 2 | C. jejuni |
| C. jejuni | deep purple | C. jejuni | 3 | C. jejuni |
| C. jejuni | deep purple | C. jejuni | 4 | C. jejuni |
| C. coli | medium | C. coli | 5 | C. coli |
| C. coli | medium | C. coli | 6 | C. coli |
| C. coli | medium | C. coli | 7 | C. coli |
| C. coli | medium | C. coli | 8 | C. coli |
| C. coli | medium | C. coli | 9 | C. coli |
| C. coli | medium | C. coli | 10 | C. coli |
| C. coli | medium | C. coli | 11 | C. coli |
| C. coli | medium | C. coli | 12 | C. coli |
| C. coli | medium | C. coli | 13 | C. coli |
| C. coli | medium | C. coli | 14 | C. coli |
| C. jejuni | deep purple | C. jejuni | 15 | C. jejuni |
| C. jejuni | deep purple | C. jejuni | 16 | C. jejuni |
| C. jejuni | deep purple | C. jejuni | 17 | C. jejuni |
| C. jejuni | deep purple | C. jejuni | 18 | C. jejuni |
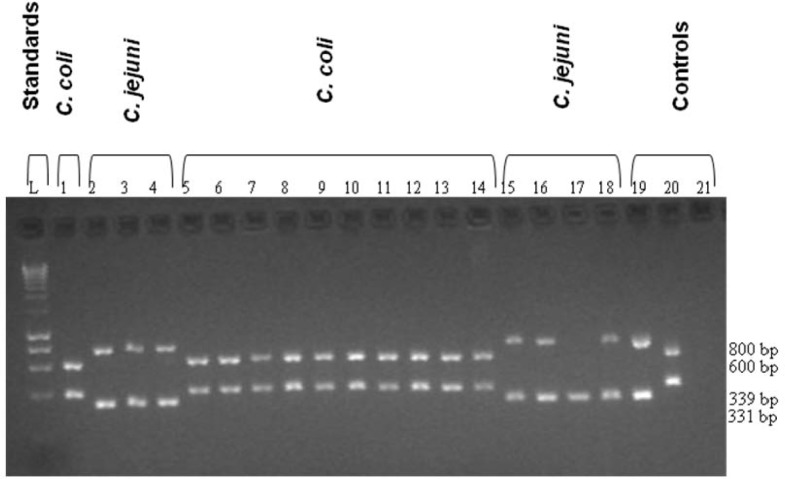
Hydrolysis of sodium hippurate by C. jejuni produces a deep purple colour, while C. coli strains produce medium or no colour change. mPCR amplification of the DNA from C. jejuni yielded two bands of approximately 331 and 800 bp, while amplification of C. coli DNA yielded bands of 391 and 600 bp. No amplification products were obtained from PCR analysis of the negative control (Fig. 2).
Figure 2:
A representative mPCR assay showing results for 18 Campylobacter isolates from poultry-related samples. Lane 1, C. coli; lanes 2–4, C. jejuni; lanes 5–14, C. coli; lanes 15–18, C. jejuni (lane 17, has a missing hipO band); lane 19, C. jejuni (11168) positive control; lane 20, C. coli (RM28) negative control; lane 21, negative control.
Notes: Lanes 1–2, were medium purple (C. coli); lanes 3–4, were deep purple (C. jejuni); lanes 5–14, were medium purple (C. coli); 15–18, were deep purple (C. jejuni) by hippurate hydrolysis test.
One isolate was identified as C. coli (medium purple) by a negative hippurate test but was identified as C. jejuni by the mPCR (Fig. 2, lane 2). The samples might have contained both C. coli and C. jejuni or contained a hippurase hydrolysis-negative C. jejuni strain (personal communication with Dr. Frieda Jørgenson). This could happen if the gene was present but not expressed. Rönner et al. (2004) reported that 5% of human Campylobacter isolates and 10% of chicken isolates were hippurase negative (presumptive C. coli isolates) but were actually C. jejuni. Similarly, Burnett et al. (2002) reported that the hippurate hydrolysis test was particularly unreliable since 28 of 29 hipO negative isolates, mostly of poultry origin were positive in this biochemical test. The PCR method offers more accurate results for species identification since the hippurate test could yield misleading reactions. This experiment agrees that hippurate hydrolysis test can be used to differentiate between C. jejuni and C. coli especially in areas where molecular equipments are unavailable.
A missing hipO band was observed with one C. jejuni strain (band 17). This is most likely because the DNA from the strain did not bind to the primers (as the strain was able to hydrolyse hippurate). To investigate this, the PCR has to be repeated using primers designed for a different region of the hipO gene (personal communication with Dr. Frieda Jørgenson). Slater and Owen (1997) reported that occasionally a typical strain of C. jejuni (less than 1%) may not produce hipO product due to the base pair substitution/deletion in the annealing site of these primers.
The results reported in this study confirm that the hippurate hydrolysis test is useful for distinguishing C. jejuni from C. coli although additional verification using methods such as mPCR is very useful. Sodium hippurate hydrolysis reagents are easily available in bacteriolological laboratories in Africa and other developing countries, and can easily be used to differentiate or identify hippurase positive and negative Campylobacters. Additionally, as C. jejuni (subsp. jejuni and subsp. doylei) is the only Campylobacter species positive for hippurase, this test is also a quick method for identifying any Campylobacter isolate as C. jejuni; assuming that it has been correctly identified as a Campylobacter. Presumptive identification of Campylobacter species is best determined from oxidase reaction (positive) and from typical spiral morphology on microscopic examination of a fresh culture, together with the inability to grow in aerobic atmosphere, so that if hippurase positive, it is almost certainly C. jejuni, as no other Campylobacter or Arcobacter species is hippurase positive (Corry et al. 2003).
Acknowledgments
The corresponding author acknowledges the support given by the Department for International Development, United Kingdom and the Institute of Postgraduate Studies, Universiti Sains Malaysia to pursue his Masters degree and PhD programme, respectively. The corresponding author also expresses his sincere gratitude to Dr. Janet Corry and Dr. Frieda Jørgenson for their guidance in this project.
REFERENCES
- Aarestrup FM, Nielsen EM, Madsen M, Engberg J. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from human, pigs, cattle and broilers in Denmark. Antimicrobial Agents and Chemotherapy. 1997;41(10):2244–2250. doi: 10.1128/aac.41.10.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzitey F, Nurul H. Campylobacter in poultry: Incidences and possible control measures. Research Journal of Microbiology. 2011;6(2):182–192. [Google Scholar]
- Bang DD, Wedderkopp A, Pedersen K, Madsen M. Rapid PCR using nested primers of the 16S rRNA and hippuricase (hip O) genes to detect Campylobacter jejuni and Campylobacter coli in environmental samples. Molecular and Cellular Probes. 2002;16(5):359–369. doi: 10.1006/mcpr.2002.0434. [DOI] [PubMed] [Google Scholar]
- Burnett TA, Hornitzky MA, Kuhnert P, Djordjevic SP. Speciating Campylobacter jejuni and Campylobacter coli isolates from poultry and humans using six PCR-based assays. FEMS Microbiology Letters. 2002;216(2):201–209. doi: 10.1111/j.1574-6968.2002.tb11436.x. [DOI] [PubMed] [Google Scholar]
- Corry JEL, Atabay HI, Forsythe SJ, Mansfield LP. Culture media for the isolation of campylobacters, helicobacters and arcobacters. In: Corry JEL, Curtis GDW, Baird RM, editors. Handbook of culture media for food microbiology. 2nd ed. Amsterdam: Elsevier Science; 2003. pp. 271–316. [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Falush D, Maiden MCJ. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. Journal of Clinical Microbiology. 2005;43(1):340–347. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Scientific report of the scientific panel on biological hazards on the request from the Commission related to Campylobacter in animals and foodstuffs. EFSA Journal. 2005;173:1–105. [Google Scholar]
- Hwang MN, Ederer GM. Rapid hippurate hydrolysis method for presumptive identification of group B streptococci. Journal of Clinical Microbiolology. 1975;1(1):114–115. doi: 10.1128/jcm.1.1.114-115.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klena JD, Parker CT, Knibb K, Ibbitt JC, Devane PML, Horn ST, Miller WG, Konkel ME. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. Journal of Clinical Microbiology. 2004;42(12):5549–5557. doi: 10.1128/JCM.42.12.5549-5557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerging Infectious Diseases. 1999;5(5):607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, Matsuda M, McDowell DA, et al. Review article: Campylobacter. Veterinary Research. 2005;36(3):351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- Mugg P. A rapid hippurate hydrolysis test for the presumptive identification of group B streptococci. Pathology. 1983;15(3):251–252. doi: 10.3109/00313028309083502. [DOI] [PubMed] [Google Scholar]
- On SLW. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: Current status, future prospects and immediate concerns. Journal of Applied Microbiology. 2001;90(56):1S–15S. doi: 10.1046/j.1365-2672.2001.01349.x. [DOI] [PubMed] [Google Scholar]
- Ridley AM, Allen VM, Sharma M, Harris JA, Newell DG. Real-time PCR approach for detection of environmental sources of campylobacter strains colonizing broiler flocks. Applied and Environmental Microbiology. 2008;74(8):2492–2504. doi: 10.1128/AEM.01242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönner AC, Engvall EO, Andersson L, Kaijser B. Species identification by genotyping and determination of antibiotic resistance in Campylobacter jejuni and Campylobacter coli from humans and chickens in Sweden. International Journal of Food Microbiolology. 2004;96(2):173–179. doi: 10.1016/j.ijfoodmicro.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Slater ER, Owen RJ. Restriction fragment length polymorphism analysis shows that the hippuricase gene of Campylobacter jejuni is highly conserved. Letters in Applied Microbiolology. 1997;25(4):274–278. doi: 10.1046/j.1472-765x.1997.00218.x. [DOI] [PubMed] [Google Scholar]
- Smith JL. Arthritis, Gullain-Barré syndrome and other sequelae of Campylobacter jejuni enteritis. Journal of Food Protection. 1995;58(10):1153–1170. doi: 10.4315/0362-028X-58.10.1153. [DOI] [PubMed] [Google Scholar]
- Vandamme P. Microbiology of campylobacter infections: Taxonomy of the family Campylobacteraceae. In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington DC: ASM Press; 2000. pp. 3–26. [Google Scholar]
- Velayudhan J, Kelly DJ. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: An essential role for phosphoenolpyruvate carboxykinase. Microbiology. 2002;148:685–694. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]