SUMMARY
EGFRvIII, a frequently occurring mutation in primary glioblastoma, results in a protein product that cannot bind ligand, but signals constitutively. Deducing how EGFRvIII causes transformation has been difficult because of autocrine and paracrine loops triggered by EGFRvIII alone or in heterodimers with wild-type EGFR. Here, we document co-expression of EGFR and EGFRvIII in primary human glioblastoma that drives transformation and tumorigenesis in a cell-intrinsic manner. We demonstrate enhancement of downstream STAT signaling triggered by EGFR-catalyzed phosphorylation of EGFRvIII, implicating EGFRvIII as a substrate for EGFR. Subsequent phosphorylation of STAT3 requires nuclear entry of EGFRvIII and formation of an EGFRvIII-STAT3 nuclear complex. Our findings clarify specific oncogenic signaling relationships between EGFR and EGFRvIII in glioblastoma.
INTRODUCTION
The epidermal growth factor receptor (EGFR) plays a prominent role in many tumors including glioblastoma, the most common primary brain tumor. Amplification and over-expression is observed in >50% of glioblastoma. Half of EGFR amplified tumors in-turn harbor the EGFRvIII mutant, an intragenic rearrangement generated by in-frame deletion of exons 2-7 from this receptor tyrosine kinase (RTK), which consequently signals constitutively in the absence of ligand (Huang et al., 1997; Sugawa et al., 1990; Wong et al., 1992). A number of studies noted that amplification and over-expression of both EGFR and EGFRvIII conferred a worse prognosis in glioma patients (Heimberger et al., 2005; Shinojima et al., 2003), with a clinical trial suggesting vaccination against EGFRvIII as a promising immunotherapy (Sampson et al., 2010). In contrast, a recent report failed to associate amplification of EGFR with outcome (Weller et al., 2009). Expression of EGFRvIII in glioblastoma is heterogeneous and is usually observed in a subpopulation of neoplastic cells (Nishikawa et al., 2004). Most antibodies against EGFR and EGFRvIII cross-react, complicating efforts to examine specific co-expression of EGFR or EGFRvIII in individual tumor cells within a glioblastoma.
Does cross-talk occur between EGFR and EGFRvIII signaling? EGFRvIII induces heparin binding EGF (HB-EGF) in glioma cells. A neutralizing antibody to HB-EGF blocked EGFRvIII-induced proliferation, raising the possibility of a EGFRvIII-HB-EGF-EGFR autocrine loop in glioblastoma (Inda et al., 2010; Ramnarain et al., 2006). Expression of EGFRvIII also induces secretion of interleukin 6 and leukemia inhibitory factor. These cytokines activate gp130, generating a paracrine loop that promotes activation of EGFR in neighboring cells (Inda et al., 2010). Physical interaction of EGFRvIII with EGFR has additionally been proposed, associated with phosphorylation of both EGFRvIII and EGFR (Luwor et al., 2004). Collectively, these studies suggest paracrine interactions between cells expressing EGFR or EGFRvIII, as well as physical interactions between EGFRvIII and EGFR within individual cells, as contributors to progression in glioma. Here, we analyze co-expression of EGFR and EGFRvIII in primary glioblastoma tumor cells from patients, and elucidate functional implications of these findings.
RESULTS
Coexpression of EGFR and EGFRvIII in Human Glioblastoma
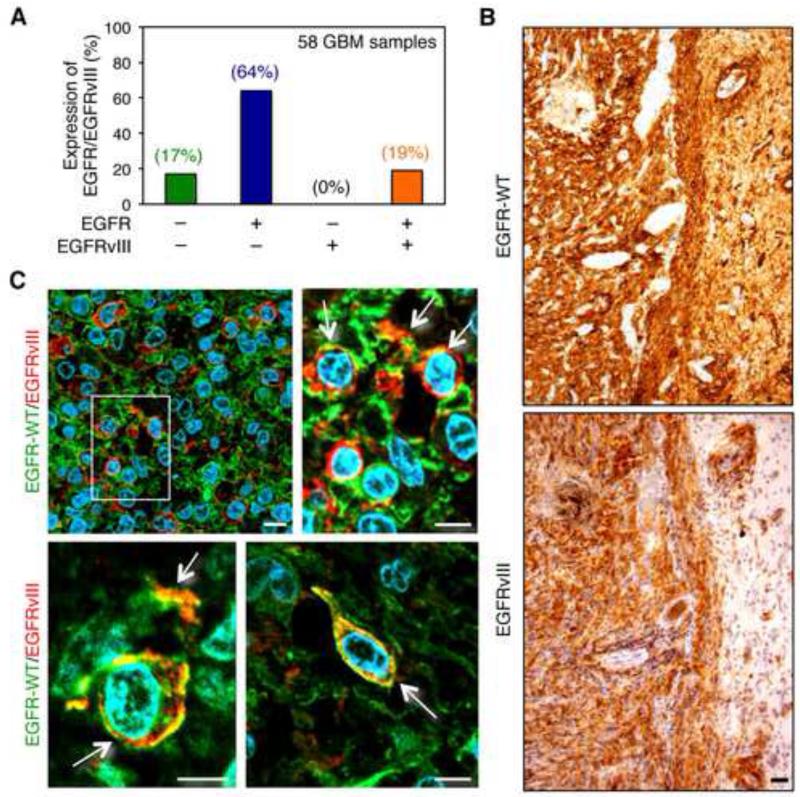
(Figure 1A and Table S1) demonstrate by immunohistochemistry, the expression status of EGFR and EGFRvIII across a series of human primary glioblastoma tissues. Among 58 tumors, 83% (48 of 58) stained for EGFR. Of these, 11 (19% of the total) were positive for EGFRvIII, with all EGFRvIII positive tumors also expressing EGFR. Although these data require that we subtract the EGFRvIII staining from the EGFR/EGFRvIII costained samples (a relatively imprecise process), these data are nevertheless consistent with findings by others (Biernat et al., 2004), and suggest that expression of EGFRvIII typically occurs in glioblastoma tumors that also over-express EGFR. Representative immunostaining is shown (Figure S1A-F).
Figure 1.
Detection of EGFR and EGFRvIII in primary human glioblastoma. (A) Graphical analysis of immunohistochemical data from Table S1. (B-C) Immunohistochemical staining of a primary human GBM with EGFR-specific (top panel) or EGFRvIII-specific antibody (bottom panel) on consecutive sections (brown, diaminobenzidine; light blue, nuclear counterstain with hemalum). Black scale bar corresponds to 50 μm. (C) Immunofluorescence double-staining of primary GBM tissue sections using EGFR-specific and EGFRvIII-specific antibodies. Cells double-positive for both EGFR-WT and EGFRvIII are indicated by arrows. An enlarged image of region marked with a white square (upper left panel in (C)) is shown on the right side. Lower panels demonstrate EGFR-WT/EGFRvIII-positive tumor cells in two additional distinct tumors; green fluorescence, EGFR-WT; red fluorescence, EGFRvIII; blue fluorescence, nuclei (4′,6-diamidino-2-phenylindole). White scale bar corresponds to 10 μm. Staining in A used pan EGFR antibody (Ventana 790-2988, clone 3C6) and EGFRvIII antibody (Duke University, L8A4). Staining in B and C used EGFR antibody (Dako DAK-H1-WT) and EGFRvIII antibody (Celldex polyclonal rabbit antiserum 6549). See also Figure S1 and Table S1.
The EGFR antibody used in (Figure 1A, Figure S1A-F, and Table S1) recognizes both full length EGFR and EGFRvIII. Therefore double-immunofluorescence staining experiments were performed using EGFR- and EGFRvIII-specific antibodies. We assessed co-expression of EGFR and EGFRvIII in individual tumor cells in glioblastoma tissue sections from 10 cases previously shown by immunohistochemistry to be positive for both proteins. Representative immunostaining is shown (Figure 1B). Antibody specificity is shown in (Figure S1G). The majority of cells within tumors co-expressing EGFR and EGFRvIII showed expression of a single RTK. In each sample however, individual tumor cells or groups of tumor cells were detected that over-expressed both proteins, with EGFR and EGFRvIII co-localized in tumor cells (Figure 1C). These results indicate that EGFR and EGFRvIII are jointly over-expressed within subsets of tumor cells in human primary glioblastoma tissue.
EGFR and EGFRvIII Cooperate to Promote Tumor Growth in vitro and in vivo
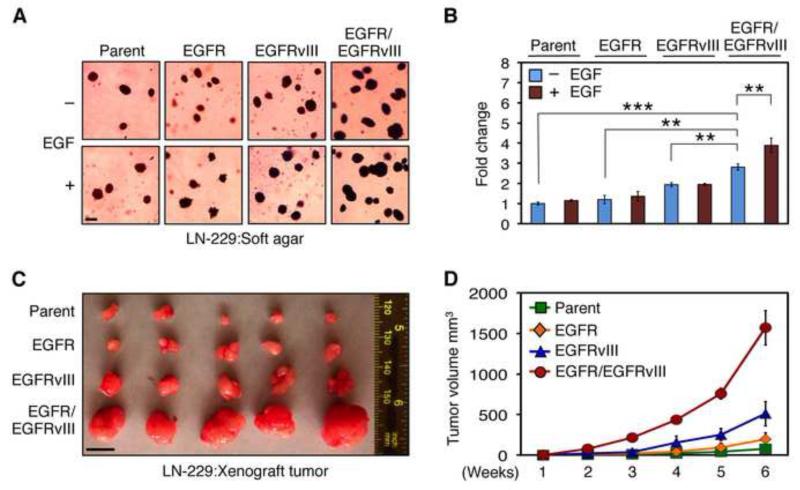
Both EGFR and EGFRvIII amplicons are rapidly lost upon culturing primary glioblastoma tumors (Pandita et al., 2004). To recapitulate co-expression, we therefore transduced EGFR, EGFRvIII, or both in human glioma cell lines LN-229 and U87MG. Because endogenous EGFR is expressed at low levels in these lines, we also examined mouse NIH3T3 fibroblast cells, which show little to no expression of EGFR (Bishayee et al., 1999). EGFR and EGFRvIII cooperated in transformation, forming both significantly more and larger colonies, as compared with parent, EGFR, or EGFRvIII; when expressed in LN-229 cells in the absence of EGF (Figure 2A and B, p<0.0001 by Student’s t test-281% increase in colony number with LN-229:EGFR/EGFRvIII versus LN-229:parent cells; p=0.0003 by Student’s t test-234% increase for LN-229:EGFR/EGFRvIII versus LN-229:EGFR cells; p=0.0011 by Student’s t test-145% increase for LN-229:EGFR/EGFRvIII versus LN-229:EGFRvIII cells). Addition of EGF led to increased colony numbers in LN-229:EGFR/EGFRvIII cells (p=0.0083 by Student’s t test-139% increase in the presence EGF when compared with absence of EGF) with little effect on cells expressing vector, EGFR, or EGFRvIII alone. LN-229:EGFR/EGFRvIII cells were transformed to modest levels without EGF treatment, perhaps due to EGFR ligands present in the fetal calf serum used in this assay (Figure 2A and B). Similar results were obtained in U87MG cells (Figure S2). Importantly, to exclude the possibility that increased transformation was related to doubling of total levels of EGFR/EGFRvIII kinase activity, we also analyzed untransformed NIH3T3 cells. In these cells (Figure S2), EGFR and EGFRvIII synergize in transformation, suggesting a greater than additive effect. Similar results were observed for LN229 cells in vivo, where EGFR and EGFRvIII led to a greater than additive effects (compared to LN229:EGFR or LN229:EGFRvIII cells) in driving tumor size (Figure 2C and D).
Figure 2.
Co-expression of EGFR and EGFRvIII enhances malignancy. (A, B) Anchorage-independent growth of LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, or LN-229:EGFR/EGFRvIII cells was measured by colony formation in soft agar, in the presence or absence of EGF (50 ng/ml). (A) Representative colonies were photographed after 3 weeks. Scale bar corresponds to 50 μm. (B) The number of colonies on 6-well plates in triplicate, normalized to parental cells without EGF, was quantified after 3 weeks. Data are presented as mean ± SE obtained from three 6-well plates. **, p < 0.05, ***, p < 0.0001. (C, D) LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, and LN-229:EGFR/EGFRvIII cells (106) were injected subcutaneously and separately in BALB/c nu/nu animals, 5 mice/group. (C) Representative tumors after 6 weeks. Scale bar corresponds to 10 mm. (D) Each point represents mean tumor volume ± SE obtained from five mice. See also Figure S2.
To further evaluate the oncogenic potential of cells co-expressing EGFR and EGFRvIII in vivo, we established xenografts from cell lines in Figure 2A and B. Growth of EGFR/EGFRvIII tumor xenografts was increased markedly (Figure 2C and D) as compared with tumors driven by parent, EGFR, or EGFRvIII (p=0.0003, Student’s t test-198% increase in LN-229:EGFR/EGFRvIII cells compared with LN-229:parent cells; p=0.0005, Student’s t test-82% increase when compared with LN-229:EGFR cells; and p= 0.0028, Student’s t test-31% increase when compared with LN-229:EGFRvIII cells).
Co-expression of EGFR and EGFRvIII phosphorylates STAT proteins in vitro and in vivo
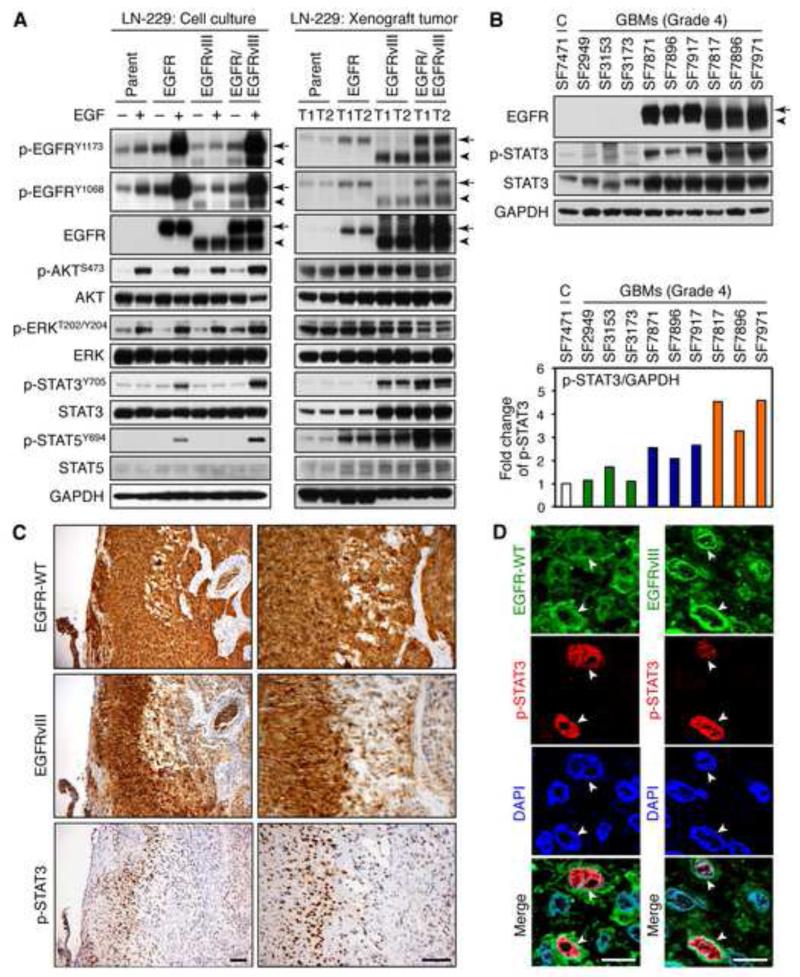
No significant differences were observed in the abundance of phosphorylated AKT (p-AKT) and p-ERK among EGF-treated LN-229:EGFR/EGFRvIII, LN-229:EGFR, and LN-229:EGFRvIII lines (Figure 3A). STAT signals downstream of EGFR, is activated in 60% of glioblastoma patients, drives progression in animal models of glioma, and correlates inversely with survival (Birner et al., 2010; Darnell et al., 1994; Doucette et al., 2012; Shao et al., 2003), prompting us to analyze this pathway. Addition of EGF led to a statistically significant increase in the abundance of p-STAT3 and p-STAT5 in EGFR/EGFRvIII cells, as compared with parent, EGFR, and EGFRvIII cells (Figure 3A and Figure S3). Induction of p-STAT3 and p-STAT5 in cells co-expressing EGFR/EGFRvIII was also observed in U87MG human glioma, and mouse fibroblast NIH3T3 cells (Figure S3). Higher levels of p-STAT3 and p-STAT5 were also observed in xenografted tumors from LN-229:EGFR/EGFRvIII cells, as compared with parent, EGFR, or EGFRvIII tumors (Figure 3A). Interestingly, although levels of p-STAT3 and p-STAT5 were very low in cultured LN-229:EGFRvIII cells, both STAT3 and STAT5 were phosphorylated to a moderate degree in xenografted EGFRvIII tumors (perhaps due to re-expression of EGFR) albeit to lower levels than observed in EGFR/EGFRvIII tumors (Figure 3A).
Figure 3.
Co-expression of EGFR and EGFRvIII is associated with phosphorylation of STAT proteins. (A) LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, or LN-229:EGFR/EGFRvIII cells were serum-staved for 24 hr then treated with or without EGF (50 ng/ml) for 15 min prior to harvest, lysis, and analysis by immunoblot using antisera indicated (left panels). This same panel of cells (106) was injected subcutaneously in BALB/c nu/nu mice, and animals were sacrificed after 6 weeks. Two representative tumors in each group were lysed, and analyzed by immunoblot (right panels). (B) Control (autopsy specimen) or human GBMs from the Brain Tumor Research Center at UCSF were lysed and analyzed by immunoblot with the antisera indicated. EGFR and EGFRvIII status were pre-confirmed by immunohistochemical staining. Samples were lysed and immunoblotted. In EGFR immunoblot, the top band (arrow) has mobility of wild-type EGFR, whereas the lower band (arrowhead) has mobility of EGFRvIII. The intensity of p-STAT3, quantified by densitometry using Silver Fast Scanner and ImageJ software, is shown below each immunoblot as fold increase relative to normal brain, normalized to GAPDH (bottom panel). (C and D) Immunohistochemical staining of adjacent sections (left) and immunofluorescent costaining of primary glioblastoma tissue using EGFR, EGFRvIII, and p-STAT3 (Tyr705, mouse monoclonal antibody 3E2, Cell Signaling) specific antibodies (right). Arrows indicate position of the nucleus and identify tumor cells positive for both EGFR (green) and p-STAT3 (red); or for both EGFRvIII (green) and p-STAT3 (red). Black scale bar corresponds to 100 μm and white bar to 10 μm. See also Figure S3.
We evaluated whether STAT signaling correlated with co-expression of EGFR and EGFRvIII in human tumors. Analysis of 58 human glioblastoma tumors demonstrated that expression of EGFRvIII was limited to tumors that expressed EGFR (Figure 1A and Table S1). We therefore further analyzed primary human glioblastoma tumors negative for expression of both EGFR and EGFRvIII, positive for EGFR alone, or positive for both EGFR and EGFRvIII. Figure 3B demonstrates that p-STAT3 was expressed at highest levels in primary human tumors that co-expressed EGFR and EGFRvIII, and at lower levels in tumors that expressed either EGFR alone, or neither kinase. Analysis of nine additional human glioblastoma tumors demonstrated general alignment among levels of EGFR, EGFRvIII and p-STAT proteins (Figure S3E-G). Immunohistochemical staining of 10 EGFR/EGFRvIII-positive primary human glioblastoma tissue sections demonstrated subsets of tumors cells with strong nuclear expression of p-STAT3, that overlapped regionally with both EGFR and EGFRvIII in every case (representative staining shown in Figure 3C); with immunofluorescence suggesting some nuclear or perinuclear expression of EGFRvIII (Figure S3H) and co-localization of p-STAT3 with EGFRvIII or with EGFR within individual cells (Figure 3D).
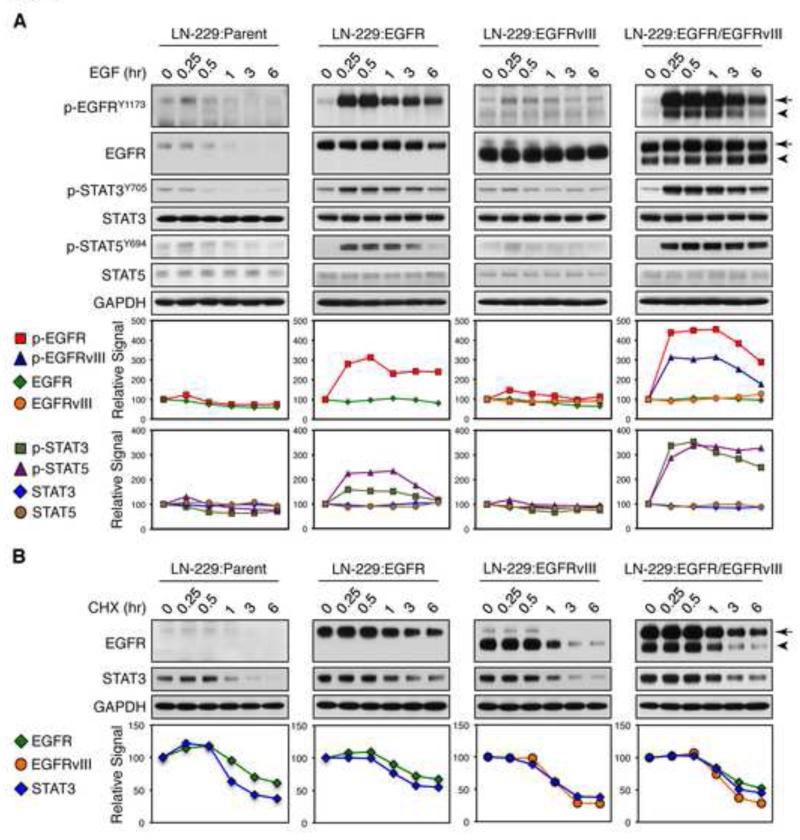
We next examined the kinetics through which EGFR and EGFRvIII could enhance STAT signaling. EGF treatment of LN-229:EGFR/EGFRvIII cells led to prolonged phosphorylation of both EGFR and EGFRvIII, starting at 15 min, continuing through 60 min; and correlating with sustained phosphorylation of STAT3 and STAT5 (Figure 4A). In contrast, phosphorylation of EGFR in LN-229:EGFR cells peaked at ~30 min, then declined more rapidly, with less robust peak and temporal phosphorylation of STAT signaling. In LN-229:parent and LN-229:EGFRvIII cells, EGF treatment had little effect on phosphorylation of EGFRvIII, STAT3, or STAT5. Using the protein synthesis inhibitor cycloheximide, we showed similar half lives for EGFR, EGFRvIII, and STAT3 proteins in LN-229:EGFR/EGFRvIII cells, as compared with parent, EGFR, and EGFRvIII cells (Figure 4B), indicating that prolonged temporal phosphorylation of STAT3 in LN-229:EGFR/EGFRvIII cells was independent of receptors and STAT3 stability. These data suggest that EGFR and EGFRvIII coordinately drive enhanced and prolonged STAT phosphorylation.
Figure 4.
EGF treatment of LN-229:EGFR/EGFRvIII cells leads to prolonged phosphorylation of EGFR, EGFRvIII, and STAT3/5. (A) LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, or LN-229:EGFR/EGFRvIII cells were serum-starved for 24 hr then treated with EGF (50 ng/ml) for times shown, prior to harvest, lysis, and analysis by immunoblot using antisera indicated (Top panel). (B) Cells were grown in 10% FBS then treated with protein synthesis inhibitor cycloheximide (CHX, 50 μg/ml) for times shown, prior to harvest, lysis, and analysis by immunoblot using antisera indicated (Top panel). EGFR is indicated by arrow, whereas EGFRvIII is indicated by arrowhead. (A, B) The intensity of each protein, quantified by densitometry using Silver Fast Scanner and ImageJ software, is shown below each immunoblot--as fold increase relative to untreated samples (EGF 0 hr or cycloheximide 0 hr) after normalization to GAPDH (Bottom panel). See also Figure S4.
To address biological effects of EGFRvIII-STAT signaling in human tumors, we queried TCGA data for key expression differences in EGFR/EGFRvIII co-amplified tumors, comparing these to tumors with amplification of EGFR in the absence of EGFRvIII. These data (Figure S4), demonstrate 33 genes that were differentially expressed between the EGFR-EGFRVIII and EGFR-amplified samples. Of note, these genes converge on PKC and PLC, both of which have been previously demonstrated to interact with STAT3(Lo et al., 2010; McBeth et al., 2013; Parsons et al., 2013)
EGFR Phosphorylates vIII, Cooperating in Transformation
Surprisingly, the abundance of phosphorylated EGFRvIII was strongly increased when LN-229:EGFR/EGFRvIII cells were treated with EGF (Figure 3A), evident at Y1068 and Y1173 tyrosine residues (Figure S3). EGFRvIII is unable to bind ligand, and demonstrates constitutive albeit low activity, whereas EGFR shows ligand-dependent signaling. Accordingly, EGF-induced increase in phosphorylation of EGFRvIII in LN-229:EGFR/EGFRvIII cells suggests that EGFR cross-phosphorylates EGFRvIII in response to EGF.
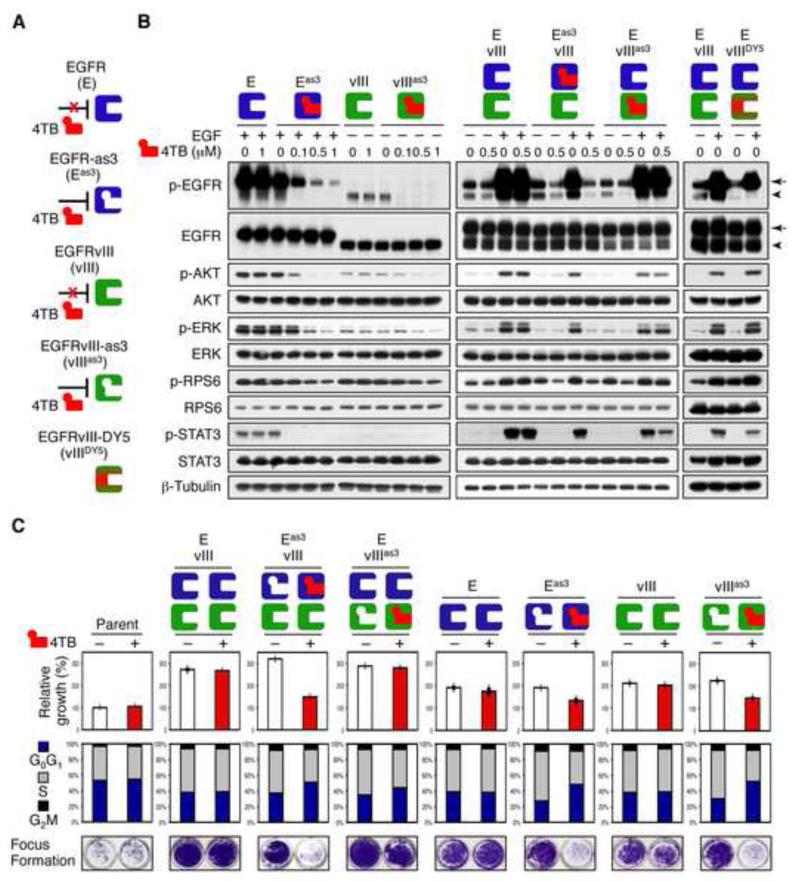
To explore mechanisms through which EGFR could phosphorylate EGFRvIII, we tested analogue-sensitive (as) alleles of EGFR, engineered to accept analogs of ATP not efficiently used by wild-type kinases (Bishop et al., 2000; Blair et al., 2007). NIH3T3 cells, which have low or undetectable levels of endogenous EGFR (Bishayee et al., 1999), were stably transduced either with wild-type or analogue sensitive alleles of EGFR (EGFRas3) or EGFRvIII (vIIIas3), individually and in combination. Cells transduced with EGFRas3 demonstrated EGF-dependent phosphorylation of EGFRas3; whereas cells transduced with EGFRvIIIas3 showed baseline phosphorylation of vIIIas3, suggesting retention of basal and EGF-driven kinase activities (Figure 5A and B). Treatment with the bulky covalent ATP-analogue 4TB [N-(4-(4-tert-butylphenylamino)quinazolin-6-yl)acrylamide (Blair et al., 2007)] blocked phosphorylation of EGFRas3, EGFRvIIIas3, and downstream targets. Under the same conditions, as expected, 4TB had no effect on phosphorylation of downstream signaling in cells transduced with either wild type EGFR or EGFRvIII.
Figure 5.
Unidirectional phosphorylation of EGFRvIII by EGFR correlates with proliferation and transformation. (A) Chemical genetic approach: as-allele-selective irreversible inhibitor 4TB did not affect EGFR or EGFRvIII, while as-alleles of these kinases (EGFRas3, EGFRvIIIas3) were inhibited. EGFRvIIIDY5 represents an allele of EGFRvIII with tyrosine mutated to phenylalanine at codons 992, 1068, 1086, 1148, and 1173. (B) Immunoblot of NIH3T3 cells transduced with EGFR, EGFRvIII, EGFRas3, EGFRvIIIas3, EGFR/EGFRvIII, EGFRas3/EGFRvIII, EGFR/EGFRvIIIas3, or EGFR/EGFRvIIIDY5. Cells were grown in 10% FBS and treated with or without indicated doses of 4TB for 24 hr. EGF (50 ng/ml) was added to cells 15 min before harvest, and lysates immunoblotted using antisera indicated. EGFR is indicated by arrow, whereas EGFRvIII is indicated by arrowhead. (C) NIH3T3 cells stably transduced with the indicated retroviral constructs were grown in 10% FBS and treated with or without 0.5 μM of 4TB. Cell proliferation (WST-1 assay, top panel), flow cytometry (middle panel), and focus formation analyses (bottom panel). Data are mean ± SD of triplicate measurements. See also Figure S5.
Having confirmed that EGFRas3 and EGFRvIIIas3 could be blocked by 4TB, we next analyzed transphosphorylation. In EGFRas3/EGFRvIII cells, 4TB blocked phosphorylation of EGFRas3, EGFRvIII, and STAT3, consistent with EGFRvIII phosphorylation by EGFR (Figure 5A and B). In EGFR/EGFRvIIIas3 cells in the absence of EGF, 4TB potently blocked phosphorylation of vIIIas3 (likely related to the low intrinsic activity of EGFRvIII), with no impact on phosphorylation of EGFR. In the absence of EGF, p-STAT3 was undetectable in these cells. In the presence of EGF however, 4TB was unable to block phosphorylation of EGFR in EGFR/EGFRvIIIas3 cells with modest effects on phosphorylation of both EGFRvIIIas3 and p-STAT3. Notably, in response to EGF, the abundance of p-STAT3 in EGFR/EGFRvIIIDY5 cells [which contain five non-phosphorylatable phenylalanine residues in place of tyrosine in the C-terminus of EGFRvIII (Huang et al., 1997)] was much lower than that in EGFR/EGFRvIII cells. Consistent with EGFR/EGFRvIIIDY5 result, treatment of LN-229:EGFR/EGFRvIII cells with EGFRvIII siRNA (Fan and Weiss, 2004) led to decreased phosphorylation of STAT3 (Figure S5A-C).
We used densitometry to quantify the relative levels of p-STAT3 in LN-229:EGFR/EGFRvIII and NIH3T3:EGFR/EGFRvIIIDY5 cells. The relative intensity of p-STAT3 in NIH3T3:EGFR/EGFRvIII cells (after addition of EGF, 15 min, and normalization to β-Tubulin) was set to 100%. The relative intensity of p-STAT3 dropped to 67% in NIH3T3:EGFR/EGFRvIIIDY5 cells (Figure 5B). Similarly, in Figure S5, we set the relative intensity of p-STAT3 in LN-229:EGFR/EGFRvIII cells treated with control siRNA to 100% (after addition of EGF, 15 min and normalization to GAPDH). The relative intensity of p-STAT3 dropped to 59% in response to EGFRvIII siRNA in Figure S5B and to 55% in Figure S5C respectively. These data suggest that phosphorylation of EGFRvIII contributes to STAT activation.
We next addressed functional effects of selective EGFR and EGFRvIII inhibition. Consistent with our immunoblot results (Figure 5A and B), treatment of EGFRas3/EGFRvIII cells with 4TB led to decreases in both proliferation and focus formation, inducing arrest at G1. In contrast, 4TB had a modest effect on EGFR/EGFRvIII3as3 or had little effect on EGFR/EGFRvIII cells, with control EGFRas3 and EGFRvIIIas3 cells showing expected responses (Figure 5C). Collectively, data in Figure 5 suggest that EGFR phosphorylates EGFRvIII, and that EGFR and EGFRvIII converge to phosphorylate STAT proteins, thereby driving transformation.
EGFRvIII is a Substrate for EGFR
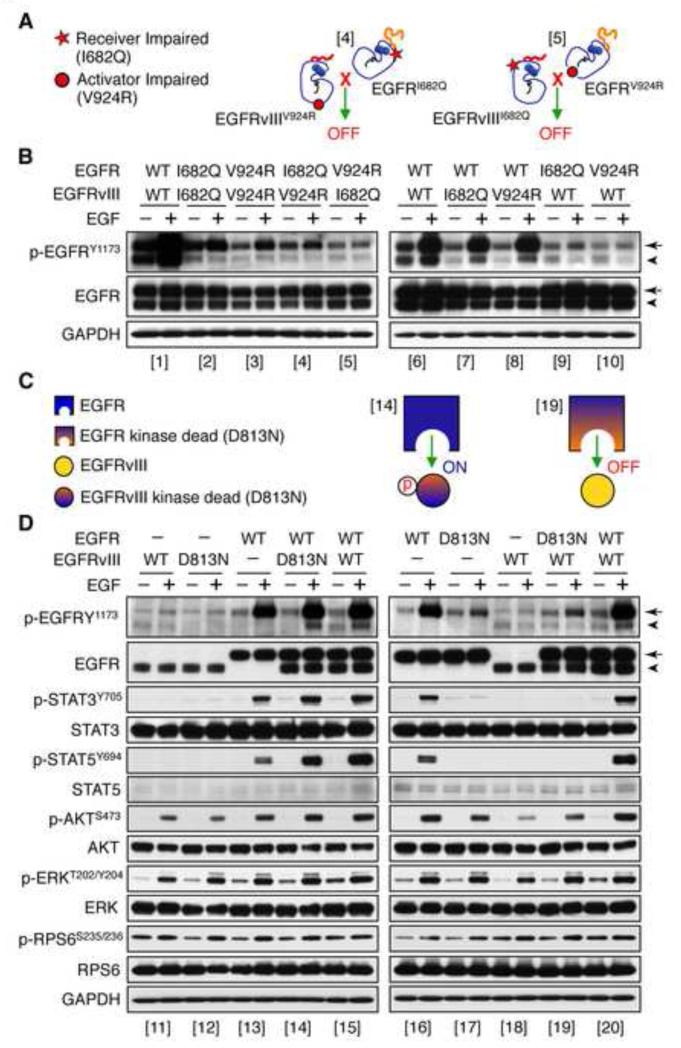
EGFR family members signal through allosteric interactions between monomers, which form an asymmetric dimer. In this dimer, only one kinase is catalytically activated (the receiver kinase) whereas the other functions as the allosteric activator (the activator kinase). To determine whether EGFR and EGFRvIII signal as an asymmetric heterodimer, we generated receiver-impaired (I682Q) and activator-impaired (V924R) mutations in both EGFR and EGFRvIII (Jura et al., 2009). LN-229 and NIH3T3 cells were stably transduced either with single or double retroviral constructs as shown in (Figure 6A and Figure S6). As expected, co-expression of EGFRI682Q and EGFRV924R restored significant signaling activity, as compared with cells transduced individually with either activator-impaired EGFRV924R or receiver-impaired EGFRI682Q (Figure S6).
Figure 6.
EGFRvIII serves as a substrate for EGFR. LN-229 cells stably transduced with retroviral constructs indicated were serum-starved for 24 hr then treated with or without EGF (50 ng/ml) for 15 min prior to harvest. Lysates were immunoblotted using antisera indicated. (A) Cartoon of receiver impaired (I682Q) or activator impaired (V924R) mutations. Bracketed numbers correspond to those in (B). (B) LN-229 cells co-transduced with EGFRI682Q/EGFRvIIIV924R or with EGFRV92R/EGFRvIIII682Q. EGFR is indicated by arrow. EGFRvIII is indicated by arrowhead. (C) Cartoon of EGFR, EGFR kinase dead (D813N), EGFRvIII, EGFRvIII kinase dead (D813N) and combinations. Bracketed numbers correspond to those in (D). (D) LN-229 cells co-transduced with EGFRWT, EGFRD813N, EGFRvIII, EGFRvIIID813N or combinations. EGFR is indicated by arrow. EGFRvIII is indicated by arrowhead. See also Figure S6.
To address whether EGFR and EGFRvIII signal as a heterodimer, we next generated EGFRI682Q/EGFRvIIIV924R cells, in which EGFR is an obligate activator, and EGFRvIII an obligate receiver. This combination (as well as the reciprocal EGFRV924R/EGFRvIIII682Q) failed to restore signaling. Co-immunoprecipitation results also failed to demonstrate a complex between EGFR and EGFRvIII (Figure S6I and J). In contrast when EGFRwt was co-transduced with EGFRvIIII682Q or EGFRvIIIV924R, downstream signaling from EGFRvIII was restored (Figure 6B). Collectively, these data suggest that EGFR and EGFRvIII signal together through a heterodimerization-independent mechanism.
We next asked whether EGFRvIII could serve as a substrate for EGFR. Kinase dead alleles EGFRD813N and EGFRvIIID813N showed low kinase activities as compared with EGFRwt and EGFRvIIIwt alleles (Figure 6C and D). The EGFRvIIID813N protein was fully phosphorylated by EGFRwt, whereas EGFRvIIIwt was unable to phosphorylate kinase dead EGFRD813N (Figure 6C and D). These EGFRvIIID813N data, in conjunction with our EGFRvIIIDY5 experiments (Figure 5B), suggest EGFRvIII is a substrate of EGFR, with phosphorylation of both EGFR and EGFRvIII promoting increased and sustained levels of phosphotyrosine in the tails of these two RTKs, phosphorylating STAT proteins, and driving progression in glioblastoma.
EGFR and EGFRvIII cooperate to phosphorylate STAT in nucleus
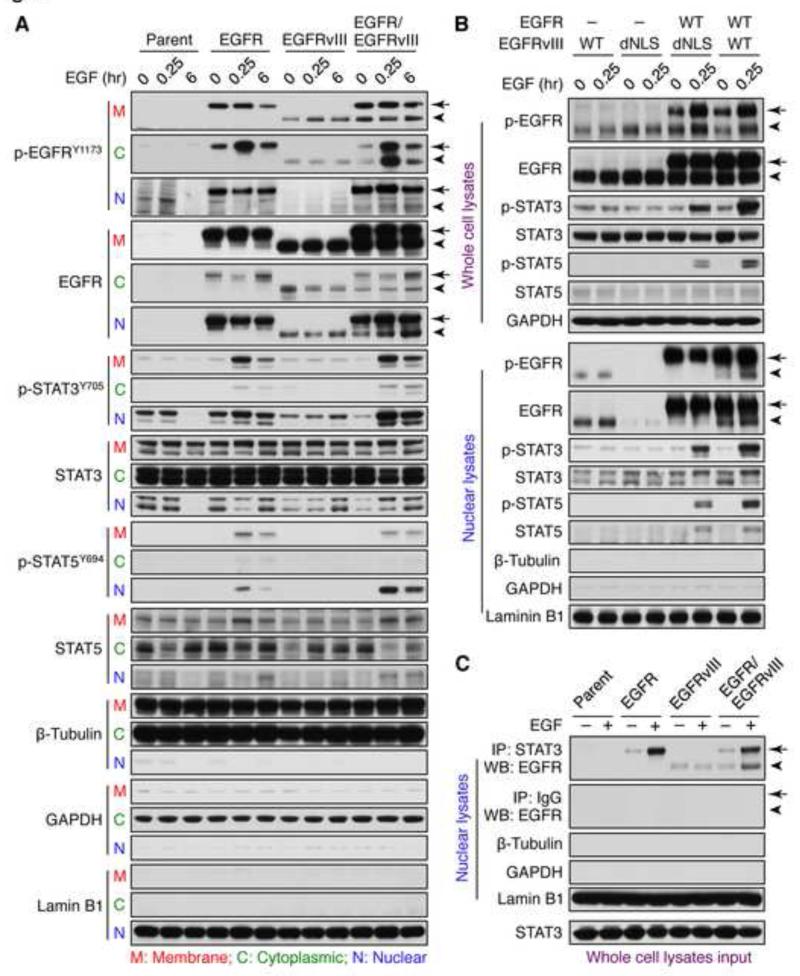
To address how EGFR and EGFRvIII converge on STAT signaling, we analyzed subcellular fractions. In the absence of EGF, both EGFR and EGFRvIII were detected in membrane, cytoplasmic, and nuclear extracts in LN-229:EGFR, LN-229:EGFRvIII, and LN-229:EGFR/EGFRvIII cells (Figure 7A). EGF treatment of LN-229:EGFR/EGFRvIII cells resulted in phosphorylation of EGFRvIII in both cytoplasmic and nuclear protein extracts (Figure 7A), associated with increased expression of EGFRvIII in the nucleus (Figure 7A), and with sustained phosphorylation of STAT3 and STAT5 in the nucleus. In contrast, EGF treatment of LN-229:EGFRvIII cells had little effect on expression of nuclear EGFRvIII, phosphorylation of EGFRvIII, or STAT signaling. EGF treatment only transiently phosphorylated nuclear STAT proteins in LN-229:EGFR cells, and had little effect in LN-229:parental cells (Figure 7A).
Figure 7.
EGFR and EGFRvIII cooperate to active STAT in the nucleus. (A) LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, or LN-229:EGFR/EGFRvIII cells were serum-starved for 24 hr then treated with EGF (50 ng/ml) for times shown. Samples were harvested, subject to subcellular fractionation to obtain membrane (M), cytoplasmic (C), and nuclear (N) extracts, and analyzed by immunoblot using antisera indicated. EGFR is indicated by arrow, whereas EGFRvIII is indicated by arrowhead. (B) LN-229:EGFRvIII, LN-229:EGFRvIIIdNLS, LN-229:EGFR/EGFRvIIIdNLS, or LN-229:EGFR/EGFRvIII were serum-starved for 24 hr then treated with EGF (50 ng/ml) for 0 and 15 min, whole cell lysates or nuclear extracts were analyzed by immunoblot using antisera indicated. (C) LN229:parent, LN-229:EGFR, LN-229:EGFRvIII, or LN-229:EGFR/EGFRvIII cells were serum-starved for 24 hr then treated with EGF (50 ng/ml) for 0 and 15 min, and fractionated to obtain nuclear extracts. Nuclear STAT3 was immunoprecipitated using a mouse monoclonal STAT3 antibody, and immunoprecipitates analyzed by immunoblot to detect EGFR and EGFRvIII (using a rabbit polyclonal EGFR antibody, which recognizes both EGFR and EGFRvIII). Efficacy of subcellular fractionation in A, B, and C is indicated by membrane and cytoplasmic marker protein β-Tubulin, cytoplasmic marker protein GAPDH, and nuclear marker protein Lamin B1. See also Figure S7.
We used densitometry to quantify the relative levels of EGFRvIII phosphorylation in each fraction in LN-229:EGFR/EGFRvIII cells. The relative intensity of p-EGFRvIII in membrane fractions without EGF stimulation was set to 100% after normalization to β-Tubulin from membrane fractions. The relative intensity dropped to 86% after EGF stimulation for 15 min, and to 90% after EGF stimulation for 6h. Again, we set the relative intensity of p-EGFRvIII in cytoplasm fractions without EGF stimulation to 100%, after normalization to β-Tubulin from cytoplasmic fractions. The relative intensity then increased to 353% after EGF stimulation for 15 min, and to 340% after EGF stimulation for 6 h. In nuclear fractions, we again set the relative intensity of p-EGFRvIII in nuclear fractions without EGF to 100% after normalization to Lamin B1 from nuclear fractions. The relative intensity increased to 126% after EGF stimulation for 15 min, and to 142% after EGF stimulation for 6 h. These data suggest that EGF treatment of EGFR/EGFRvIII cells increased phosphorylation of EGFRvIII in cytoplasmic and nuclear fractions, but not in membrane fractions (Figure 7A).
To clarify whether EGFRvIII undergoes nuclear translocation to phosphorylate STAT proteins, we transfected LN-229 cells with EGFRvIIIdNLS, an allele of EGFRvIII defective for nuclear entry (Lo et al., 2010), and established stable LN-229:EGFRvIIIdNLS and LN-229:EGFR/EGFRvIIIdNLS cell lines. LN-229:EGFRvIIIdNLS and LN-229:EGFR/EGFRvIIIdNLS cells expressed levels of EGFRvIII equivalent to those in LN-229:EGFRvIII and LN-229:EGFR/EGFRvIII cells. EGF treated LN-229:EGFR/EGFRvIII cells had higher levels of phosphorylated STAT proteins, as compared to levels in LN-229:EGFR/EGFRvIIIdNLS cells (Figure 7B). Consistent with our immunoblot results using whole cell lysates (Figure 7B top panel) EGF treatment of LN-229:EGFR/EGFRvIIIdNLS cells resulted in decreased phosphorylation of STAT in the nuclear fraction (Figure 7B bottom panel). These data suggest that EGF treatment of EGFR/EGFRvIII cells enhances both nuclear transport of EGFRvIII, and phosphorylation of STAT in the nucleus.
We next asked whether nuclear EGFR and EGFRvIII could complex with STAT3 in the nucleus. Nuclear STAT3 was immunoprecipitated, and immunoblots analyzed to detect nuclear EGFR and EGFRvIII (Figure 7C). Nuclear lysates, whole cell lysates (input) and nuclear immunoprecipitations are shown in (Figure 7C). In LN-229:EGFR/EGFRvIII cells, the levels of nuclear EGFRvIII complexed to STAT3 were enhanced following 15 min of EGF treatment. In contrast, EGF treatment had little effect on the EGFRvIII/STAT3 complex in LN-229:vIII cells. As expected, mouse IgG did not pull down STAT3 or EGFR/EGFRvIII. Collectively, these data suggest that EGFR phosphorylates EGFRvIII, leading to increased nuclear translocation of EGFRvIII, and enhanced binding of EGFRvIII to STAT3 in the nucleus.
High STAT3 activity may contribute to resistance of GBM patients to EGFR inhibitors (Mellinghoff et al., 2005; Reardon et al., 2006). Might inhibition of STAT3 cooperate with inhibition of EGFR in glioma? To partially address this issue, we showed that cells treated with the STAT3 tool inhibitor Stattic in combination with the EGFR inhibitor erlotinib showed differential induction of apoptosis (Fig S7). Baseline levels of apoptosis differ among the four lines, consistent with EGFR and EGFRvIII independently modestly blocking basal apoptosis, with a more prominent effect of EGFR/EGFRvIII in combination. Thus, LN-229:EGFR/EGFRvIII cells at baseline showed 2.1% apoptosis, compared to 44.8% in response to Stattic and erlotinib (~21 fold change). LN-229:EGFRvIII cells at baseline showed 3.4% apoptosis, compared to 46.9% in response to Stattic and erlotinib (~14 fold change). By comparison, LN-229:EGFR cells at baseline showed 4.3% apoptosis, compared to 40.6% in response to Stattic and erlotinib (~9 fold change); while LN-229:parent cells at baseline showed 6.8% apoptosis, compared to 42.1% in response to Stattic and erlotinib (~6 fold change). These data suggest cooperative blockde of EGFR and STAT3 as a developmental therapeutic strategy in EGFR/EGFRvIII coamplified glioma.
DISCUSSION
Amplification and over-expression of EGFR represent striking features of primary glioblastoma, with frequent co-amplification of EGFR and its deletion mutant, EGFRvIII (Huang et al., 1997; Sugawa et al., 1990; Wong et al., 1992). Using immunofluorescence double staining with EGFR-and EGFRvIII-specific antibodies, we demonstrate that EGFR and EGFRvIII co-localize within individual tumor cells in glioblastoma. We further highlight cooperation between EGFR and EGFRvIII in transformation in vivo, with cell-based experiments showing that EGF treatment of cells expressing both EGFR and EGFRvIII resulted in phosphorylation of both kinases. This result was unexpected, as EGFRvIII is unable to bind or be activated in response to EGF. Using chemical genetic approaches, we found that EGFR promoted unidirectional EGFRvIII signaling in glioblastoma cells, driving further phosphorylation of STAT proteins, and enhanced malignancy. It is intriguing that enhanced STAT signaling is so selectively affected by EGFR/EGFRvIII cross talk, while signaling through PI3K and MAPK is less prominently affected. In addition, STAT signaling has been suggested to feature prominently in glioma stem cells (Dubuc et al., 2013). That rare cells within GBM tumors co-amplify EGFR and EGFRvIII, and phosphorylate STAT3 could be consistent with a role for STAT3 activation within the stem cell compartment of GBM.
How does EGFR activate EGFRvIII? While others have demonstrated physical binding of these two kinases, our co-immunoprecipitation experiments failed to demonstrate this interaction. We were similarly unable to reconstitute signaling in a heterodimeric complex where EGFR was an obligate allosteric activator, and EGFRvIII an obligate receiver. Kinase dead EGFRvIII was readily phosphorylated by EGFR, suggesting EGFRvIII as a substrate of EGFR. Phosphorylation of both EGFR and EGFRvIII were required to fully phosphorylate STAT proteins. EGFR, when expressed alone, led to lower level and shorter duration of STAT phosphorylation, as compared to levels and duration observed in cells co-expressing both EGFR and EGFRvIII. Further, co-expression of EGFR with EGFRvIIIDY5, a mutant in which tyrosines in the tail of EGFRvIII were replaced with phenylalanines, blunted phosphorylation of STAT3 protein.
Analysis of human glioblastoma tumors demonstrated alignment among EGFR, EGFRvIII and STAT signaling in tumors, supporting a model in which EGFR activation of EGFRvIII leads to transphosphorylation of both kinases, converging on STAT signaling. Using subcellular fractionation, we further demonstrated that EGFR phosphorylation of EGFRvIII led to nuclear transport of EGFRvIII, and enhanced the formation of a complex between EGFRvIII and STAT3 in the nucleus. These data suggest that EGFR and EGFRvIII coordinately drive enhanced and prolonged STAT activity in the nucleus. It remains possible however, that very high levels of EGFR could subserve this role even in the absence of EGFRvIII.
In this study, we identify an EGFR-EGFRvIII-STAT signaling axis in a subset of glioblastomas that co-amplify EGFR and EGFRvIII within individual tumor cells. Given that coexpressed EGFR and EGFRvIII, and high levels of STAT signaling may confer both more aggressive behavior in glioblastoma (Abou-Ghazal et al., 2008; Birner et al., 2010; Shinojima et al., 2003), our findings suggest targeting EGFR in conjunction with STAT signaling as a therapeutic strategy for patients with EGFRvIII-positive glioblastoma.
EXPERIMENTAL PROCEDURES
Tumor Samples
Primary tumor samples were obtained in accordance with research ethics board approval from UCSF and the Heinrich Heine University. Informed consent was obtained from all UCSF patients. Archival samples from Dusseldorf were investigated in an anonymized manner, approved by the ethics committee of the Medical Faculty of Heinrich Heine University.
Construction of EGFR and EGFRvIII mutants
Retroviral-based pWLZ-hygro-EGFR-as3 was described previously (Blair et al., 2007). pWLZ-hygro-EGFRas3 plasmid was digested with BstXI and ligated into a similarly digested pLRNL-neo-EGFRvIII plasmid, generating retroviral-based pLRNL-neo-EGFRvIII-as3, an analogue-sensitive allele of EGFRvIII. pLRNL-neo-EGFRvIII-DY5 (Johns et al., 2007) and pCMV-tag5A-EGFRvIIIdNLS (Lo et al., 2010) were generously provided by Dr. Frank Furnari and Hui-Wen Lo, respectively. Point mutations (I682Q and V924R) were engineered into the pWLZ-hygro-EGFR or into the pLRNL-neo-EGFRvIII by site-directed mutagenesis (QuickChange Mutagenesis kit, Stratagene). All primer sequences are in (Thiel and Carpenter, 2007). Point mutation pcDNA-neo-EGFR (D813N) plasmid was from Natalia Jura, UCSF; digested with BstXI, ligated into a similarly digested vector and used to generate either pWLZ-hygro-EGFR (D813N) or pLRNL-neo-EGFRvIII (D813N). All mutants were sequence validated.
Cell lines, reagents, transfection, and transduction
Human glioma cell lines LN-229 and U87MG were obtained from the Brain Tumor Research Center at UCSF. NIH3T3 mouse fibroblasts were from ATCC. These and derivate lines were grown in 0.5% or 10% FBS. To generate retrovirus, the packaging cell line 293T was co-transfected with plasmids gag/pol and VSVg, using Effectene-transfection reagent (Qiagen). High-titer virus was collected at 48 hr and used to infect cells as previously described(Fan et al., 2007). pCMV-tag5A-EGFRvIIIdNLS plasmid was generously provided by Dr. Hui-Wen Lo, Duke University, and transfected into LN229:parent or LN229:EGFR cells. Transfected and transduced cells were selected as pools with G418 (800 μg/ml) or hygromycin (500 μg/ml) for 2 weeks. EGF was from (Roche). Cyclohexmide and Stattic were from (Sigma). 4TB [N-(4-(4-tert-butylphenylamino)quinazolin-6-yl)acrylamide were synthesized as described (Blair et al., 2007).
Immunohistochemical analyses
Immunohistochemical staining was performed by the UCSF Neurosurgery Tissue Core. Paraffin-embedded sections (5 μm) of human GBM were immunostained on the Benchmark XT automated stainer (Ventana Medical System, Tucson, AZ). Antibodies were detected with the Ventana iVIEW DAB detection kit (yielding a brown reaction product). The EGFR antibody utilized in this study was a mouse monoclonal antibody obtained from Ventana 790-2988 (clone 3C6) which recognizes the extracellular domain of both full length and the EGFRvIII variant of EGFR and was supplied as part of an FDA validated clinical diagnostic kit for EGFR abundance, used clinically to assess EGFR status. The EGFRvIII mouse monoclonal antibody was from Duke University (L8A4). Slides were counterstained with hematoxylin. Immunostaining results were graded in a semi-quantitative manner by determining the intensity of staining of each section and grading from 0 (no staining), 1 (1-25% immunoreactivity of cells), 2 ((26-75% immunoreactivity) or 3 (>75% immunoreactivity). Human glioblastoma tissue samples were routinely fixed in 4% buffered formaldehyde and embedded in paraffin. Five μm thick tissue sections were deparaffinized in xylene and rehydrated over a graded ethanol series. For antigen retrieval, rehydrated sections were treated in 10 mM citrate buffer at pH 6.0 (for EGFRvIII and p-STAT3) or at pH 9.0 (for EGFR) for 20 min in a steamer. Sections were immunostained with antibodies against EGFR (mouse monoclonal DAK-H1-WT, Dako, diluted 1:200), EGFRvIII (rabbit polyclonal 6549, Celldex, diluted 1:5000), p-STAT3 (Tyr705) (rabbit monoclonal antibody D3A7 or mouse monoclonal antibody 3E2, Cell Signaling, each diluted 1:50). Immunohistochemistry was performed on the Dako Autostainer Plus automated slide processing system using the Ultravision LP Large Volume Detection System HRP Polymer (Thermo Scientific) for detection of antibody binding according to the manufacturer’s protocol. 3,3-Diaminobenzidine was used as substrate for the peroxidase reaction. Slides were counterstained with hemalum, dehydrated and mounted in DePeX (Serva) mounting medium.
Double-immunofluorescence analyses
Glioma cells grown as monolayer cultures on chamber slides were fixed with methanol. Formalin-fixed paraffin sections from primary tumor tissues were deparaffinized in xylene and rehydrated over a graded ethanol series, followed by treatment in 10 mM citrate buffer at pH 6.0 for 20 minutes in a steamer for antigen retrieval. Rehydrated fixed cells and tissue sections were blocked 5 min with Ultra V Block (Thermo Scientific) and incubated overnight at 4°C with primary antibodies against EGFR (mouse monoclonal DAK-H1-WT, Dako) and EGFRvIII (rabbit polyclonal 6549, Celldex) diluted 1:50. Additional double-labeling experiments were performed with antibodies against EGFR and p-STAT3 (Tyr705) antibody (rabbit monoclonal antibody D3A7, Cell Signaling), or antibodies against EGFRvIII and p-STAT3 (Tyr705) antibody (mouse monoclonal antibody 3E2, Cell Signaling). Three washing steps were followed by incubation at room temperature for 1 hr with Alexa Fluor 488/594 secondary antibodies (Invitrogen) diluted 1:500. Following repeated washing, stained sections and cells were mounted in ProLong® Gold antifade reagent (Invitrogen) with 4′,6-diamidino-2-phenylindole (DAPI).
In Vitro and In Vivo growth assays
We used the Cell Transformation Detection Assay kit (Millipore) to evaluate colony formation on soft agar. Briefly, plates were pre-coated with 0.7% agarose as the bottom layer. Cells were seeded at a density of 1 × 104 cells per 6-well in triplicate for each cell line and cultured in 0.35% agarose as the top layer in DMEM (without phenol red) plus 10% FBS at 37°C for 3 weeks. The cells were kept wet by adding a small amount of culture media. EGF (50 ng/ml) was added every 5 days. Colonies were stained overnight at 37°C (Cell transformation detection assay kit, Millipore). Colony numbers in the entire well were counted under the microscope. For nude-mice, LN-229:parent, LN-229:EGFR, LN-229:EGFRvIII, and LN-229:EGFR/EGFRvIII cells (106) were injected subcutaneously just caudal to the left forelimb in 4- to 6 week-old BALB/c nu/nu mice (Harlan Sprague-Dawley). Tumor diameters were measures with calipers at 7-day intervals, and volumes calculated from five mice per data point (mm3= width2 × Length/2). UCSF’s Institutional Animal Care and Use Committee approved all experiments. Each value represents mean tumor volume +/− standard error obtained from five mice.
Immunoblotting
Membranes were blotted with p-EGFRY845, p-EGFRY992, p-EGFRY1045, p-EGFRY1068, p-AKTS473, AKT, p-S6 ribosomal proteinS235/236, S6 ribosomal protein, p-ERKT202/Y204, p-STAT3 (Tyr705), STAT3, STAT5, Lamin B1 (all from Cell Signaling), p-EGFRY1173, EGFR, ERK, normal mouse IgG (Santa Cruz Biotechnology), p-STAT5Y694 (BD Transduction Laboratories), GAPDH, or β-Tubulin (Upstate Biotechnology). Bound antibodies were detected with HRP-linked anti-mouse or anti-rabbit IgG (Calbiochem), followed by ECL (Amersham).
Subcellular fractionation and Immunoprecipitation (IP)
For multi-compartmental fractionation of cells we used the subcellular protein fractionation kit (Thermo Scientific Pierce) according to the manufacturer’s instructions. For immunoprecipitation, 200 μg nuclear protein were incubated with 1 μg anti-STAT3 mouse monoclonal antibody (Cell Signaling) or control mouse IgG at 4°C overnight with gentle agitation. Following addition of 20 μl protein G-agarose and incubation for 1 hr at 4°C, the immunocomplexes were pelleted, washed for multiple cycles at 4°C, and then subjected to SDS-PAGE and Western blotting analysis probing for EGFR (rabbit antibody that recognize both EGFR and EGFRvIII, Santa Cruz Biotechnology).
Supplementary Material
Highlights.
Rare cells within human glioblastomas express both EGFR and tumor-derived EGFRvIII.
Co-expression of EGFR and EGFRvIII show enhanced transformation in vitro and in vivo.
EGFR promotes unidirectional activation of EGFRvIII, converging to activate STAT3.
Co-expression of EGFR and EGFRvIII leads to a nuclear complex between vIII and STAT3.
Significance.
EGFR is commonly amplified and mutated in primary glioblastoma, a highly-malignant brain tumor. The most commonly observed mutant variant, EGFRvIII, signals via potential autocrine and paracrine loops. Our inability to fully elucidate and target this complex signaling has contributed to failed clinical trials in patients with few options for therapy. We document co-expression of EGFR and EGFRvIII in human tumors, identify a cell intrinsic role for co-expression in vitro and in vivo, and demonstrate that EGFR and EGFRvIII cooperate to phosphorylate STAT proteins, promoting malignant progression. Our findings elucidate signaling interactions between EGFR and EGFRvIII and suggest combinatorial targeting of the EGFR-EGFRvIII-STAT axis as a therapeutic approach to treat EGFRvIII-mutant glioblastoma.
ACKNOWLEDGMENTS
Supported by K08NS079485 (W.C.G), U54CA163155 and U01CA176287 (W.A.W), Howard Hughes Medical Institute (K.M.S), and the Samuel Waxman Cancer Research Foundation (W.A.W., K.M.S). We thank Albert Baldwin, Lew Cantley, Peter Jackson, Mark Lemmon, and Frank McCormick for useful discussions, Miller Huang, Justin Meyerowitz and Theo Nicolaides for critical review, Frank Furnari for plasmid pLRNL-neo-vIII-DY5, Hui-Wen Lo for plasmid pCMV-tag5A-vIIIdNLS, and Cynthia Cowdrey and King Chiu for immunohistochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures, one table, and Supplemental Experimental Procedures and can be found at a single PDF file.
REFERENCES
- Abou-Ghazal M, Yang DS, Qiao W, Reina-Ortiz C, Wei J, Kong LY, Fuller GN, Hiraoka N, Priebe W, Sawaya R, Heimberger AB. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14:131–136. doi: 10.1111/j.1750-3639.2004.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. Journal of neuro-oncology. 2010;100:339–343. doi: 10.1007/s11060-010-0195-8. [DOI] [PubMed] [Google Scholar]
- Bishayee A, Beguinot L, Bishayee S. Phosphorylation of tyrosine 992, 1068, and 1086 is required for conformational change of the human epidermal growth factor receptor c-terminal tail. Molecular biology of the cell. 1999;10:525–536. doi: 10.1091/mbc.10.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Blair JA, Rauh D, Kung C, Yun CH, Fan QW, Rode H, Zhang C, Eck MJ, Weiss WA, Shokat KM. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–238. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Doucette TA, Kong LY, Yang Y, Ferguson SD, Yang J, Wei J, Qiao W, Fuller GN, Bhat KP, Aldape K, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro-oncology. 2012 doi: 10.1093/neuonc/nos139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, Kool M, Jones DT, Unterberger A, Morrissy AS, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta neuropathologica. 2013;125:373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Cheng CK, Nicolaides TP, Hackett CS, Knight ZA, Shokat KM, Weiss WA. A Dual Phosphoinositide-3-Kinase {alpha}/mTOR Inhibitor Cooperates with Blockade of Epidermal Growth Factor Receptor in PTEN-Mutant Glioma. Cancer research. 2007;67:7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Weiss WA. RNA interference against a glioma-derived allele of EGFR induces blockade at G(2)M. Oncogene. 2004 doi: 10.1038/sj.onc.1208227. [DOI] [PubMed] [Google Scholar]
- Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. The Journal of biological chemistry. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns TG, Perera RM, Vernes SC, Vitali AA, Cao DX, Cavenee WK, Scott AM, Furnari FB. The efficacy of epidermal growth factor receptor-specific antibodies against glioma xenografts is influenced by receptor levels, activation status, and heterodimerization. Clin Cancer Res. 2007;13:1911–1925. doi: 10.1158/1078-0432.CCR-06-1453. [DOI] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Molecular cancer research: MCR. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luwor RB, Zhu HJ, Walker F, Vitali AA, Perera RM, Burgess AW, Scott AM, Johns TG. The tumor-specific de2-7 epidermal growth factor receptor (EGFR) promotes cells survival and heterodimerizes with the wild-type EGFR. Oncogene. 2004;23:6095–6104. doi: 10.1038/sj.onc.1207870. [DOI] [PubMed] [Google Scholar]
- McBeth LR, St-Pierre NR, Shoemaker DE, Weiss WP. Effects of transient changes in silage dry matter concentration on lactating dairy cows. Journal of dairy science. 2013 doi: 10.3168/jds.2012-6330. [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain tumor pathology. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes, chromosomes & cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Kukula M, Jackson P, Pulse M, Simecka JW, Valtierra D, Weiss WJ, Kaplan N, Rock CO. Perturbation of Staphylococcus aureus Gene Expression by the Enoyl-Acyl Carrier Protein Reductase Inhibitor AFN-1252. Antimicrobial agents and chemotherapy. 2013 doi: 10.1128/AAC.02307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- Reardon DA, Quinn JA, Vredenburgh JJ, Gururangan S, Friedman AH, Desjardins A, Sathornsumetee S, Herndon JE, 2nd, Dowell JM, McLendon RE, et al. Phase 1 trial of gefitinib plus sirolimus in adults with recurrent malignant glioma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:860–868. doi: 10.1158/1078-0432.CCR-05-2215. [DOI] [PubMed] [Google Scholar]
- Sampson JH, Heimberger AB, Archer GE, Aldape KD, Friedman AH, Friedman HS, Gilbert MR, Herndon JE, 2nd, McLendon RE, Mitchell DA, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Cheng HY, Cook RG, Tweardy DJ. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63:3923–3930. [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.