Abstract
Prader-Willi Syndrome (PWS) is caused by the absence of paternally expressed, maternally silenced genes at 15q11-q13. We report four individuals with truncating mutations on the paternal allele of MAGEL2, a gene within the PWS domain. The first subject was ascertained by whole genome sequencing analysis for PWS features. Three additional subjects were identified by reviewing results of exome sequencing of 1248 cases in a clinical laboratory. All four subjects had autism spectrum disorder (ASD), intellectual disability (ID), and a varying degree of clinical and behavioral features of PWS. These findings suggest MAGEL2 is a novel gene causing complex ASDs, and MAGEL2loss of function can contribute to several aspects of the PWS phenotype.
Prader-Willi syndrome (MIM176270) is characterized by infantile hypotonia with poor suck and failure to thrive, followed by overeating and rapid weight gain during childhood, developmental delay, intellectual disability, hypogonadism, short stature, and a characteristic behavioral profile. Comprehensive diagnostic criteria have been established (Holm et al., 1993)1. PWS can result from paternal deletion of 15q11-q13 (65–75% of cases), maternal uniparentaldisomy 15 (20–30%), an imprinting defect (1–3%)2, or possibly rare deletions of the SNORD116 cluster as discussed below. Point mutations causing PWS have not been reported to date.
The Prader-Willi locus contains four paternally expressed genes with six open reading frames coding for five polypeptides (MKRN3, MAGEL2, NDN, NPAP1 and SNURF- SNRPN) and a family of six paternally expressed snoRNA genes or clusters. The search for candidate genes contributing to specific phenotypic components of PWS has not been entirely conclusive. Three patients with deletions of the SNORD116 snoRNA cluster manifested key characteristics of PWS3–5. Paternal deletion of MKRN3, MAGEL2 and NDN alone occurred in one patient, and was associated with obesity and intellectual disability, but not the typical PWS phenotype6 leading the authors to conclude that “deficiency of MKRN3, MAGEL2 and NDN is not sufficient to cause PWS. “While multiple PWS mouse models have been generated, including targeted mutations of Ndn, Magel2, Mkrn3, Snurf-Snrpn, and Snord1167, no single model recapitulates early growth deficiency and hyperphagia leading to subsequent obesity. Numerous publications from Wevrick and colleagues argue that Magel2 null mice have selected biological findings similar to PWS, including neonatal growth retardation, excessive weight gain after weaning, impaired hypothalamic regulation, and reduced fertility8–10. In conjunction, human and animal data suggest that several genes and snoRNAs in the PWS locus may contribute to its clinical phenotype, but whether PWS truly represents a “contiguous gene syndrome” remains elusive.
Here, we report the first individuals with point mutations in a protein-coding gene within the PWS domain. Subject 1 was enrolled in a whole genome sequencing research study, and subjects 2–4 were identified through clinical whole exome sequencing. In both instances, samples are submitted without pre-screening criteria. Submission is based on the referring provider’s determination that the affected individual likely has genetic cause to his disease.
Subject 1was ascertained at age 13 years with a history signficiant for aPWS-like phenotype, ASD, and mild ID. Using highly accurate whole genome sequencing with 60X coverage (developed by Drmanac et al.)11, he was found to carry a heterozygous de novo c.1652delT (p.V551fs) mutation in MAGEL2(NM_019066.4), one of the protein-coding genes in the PWS domain. Given that MAGEL2 is only expressed from the paternal allele, we investigated whether the mutation was present on the maternal or paternal copy of chromosome 15. We performed long fragment analysis (in part as previously reported by Peters et al.)12, which in conjunction with parental SNP genotypes in proximity to the mutated locus determined that the MAGEL2 mutation was on the paternal allele (Online Methods).
Based on the findings in subject 1, the Baylor College of Medicine Whole Genome Laboratory clinical whole exome sequencing (WES) database was searched for probable pathogenic mutations in MAGEL2. Three additional subjects with nonsense of frameshifting indel mutations were ascertained. A total of 1248 WES cases were reviewed.
Subject 2 is an 8 year-old boy with classic PWS, meeting the 1993 diagnostic criteria. He carries a heterozygous c.1802delC (p.P601fs) mutation, which is not maternally inherited (father unavailable). Subject 3 is a 5 year-old boy with a de novo heterozygous c.3181_3182del (p.I1061fs) mutation. While he has a history of feeding difficulties as an infant requiring tube feeding, followed by excessive weight gain during childhood, cryptorchidism, short stature, some of the typical behavioral phenotypes of PWS, and ID, he does not meet full clinical criteria for the condition. This is similar to subject 4, who meets four of the major diagnostic criteria (five necessary): neonatal hypotonia, feeding difficulties requiring tube feeding, followed by hyperphagia and absence of satiety, and ID. He has a de novoc.3124C>T (p.Q1024X) mutation. Clinical phenotypes are summarized in Table 1. Full clinical descriptions are provided in Supplementary Note 1, and patient photographs in Supplementary Fig. 1. Of note, all four probands had normal methylation testing for PWS, and normal chromosome microarray analysis.
Table 1.
Molecular and clinical phenotypes of four individuals with truncating MAGEL2 mutations.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Summary | |
|---|---|---|---|---|---|
| Sex | M | M | M | M | 4 M |
| Age at time of diagnosis | 12 years | 8 years | 5 years | 19 years | average: 11 years |
| Mutation | c.1652delT p.V551fs | c.1802delC p.P601fs | c.3181_3182del p.I1061fs | c.3124C>T p.Q1024X | 1 nonsense, 3 frameshift indels |
| Inheritance | de novo | not maternal | de novo | de novo | 3 de novo, 1 not maternal |
| Affected allele | paternal | paternal | paternal | paternal | 4 paternal |
| PWS major criteria* | |||||
| Nenonatalhypotonia, poor suck | + | + | − | + | 3/4 |
| Feeding problems in infancy, with need for special feeding technique | − | + | + | + | 3/4 |
| Excessive weight gain before age 6 years | + | + | + | − | 3/4 |
| Hyperphagia, lack of satiety | − | + | − | + | 2/4 |
| Developmental delay, Intellectual disability | + | + | + | + | 4/4 |
| PWS characteristic facial features | − | + | − | − | 1/4 |
| Hypogonadism | + | + | + | − | 3/4 |
| PWS minor criteria* | |||||
| Infantile lethargy, weak cry | + | − | + | + | 3/4 |
| Short stature | − | − | + | + | 2/4 |
| Small hands | − | − | − | + | 1/4 |
| Narrow hands | − | − | − | + | 1/4 |
| Eye abnormalities | − | + | + | + | 3/4 |
| Hypopigmentation | − | − | − | − | 0/4 |
| Thick saliva | − | − | − | − | 0/4 |
| Characteristic behavior (temper tantrums, violent outbursts, oppositional behavior, etc) | − | − | + | + | 2/4 |
| Speech articulation defects | − | + | + | + | 3/4 |
| Skin picking | − | − | + | + | 2/4 |
| Sleep apnea | − | + | − | + | 2/4 |
| Other symptoms (non-PWS criteria) | |||||
| Autism spectrum disorder | + | + | + | + | 4/4 |
| Contractures of the proximal and distal interphalangeal joints | − | − | + | + | 2/4 |
M, male; +, present; −, not present;
, based on Holm et al1.
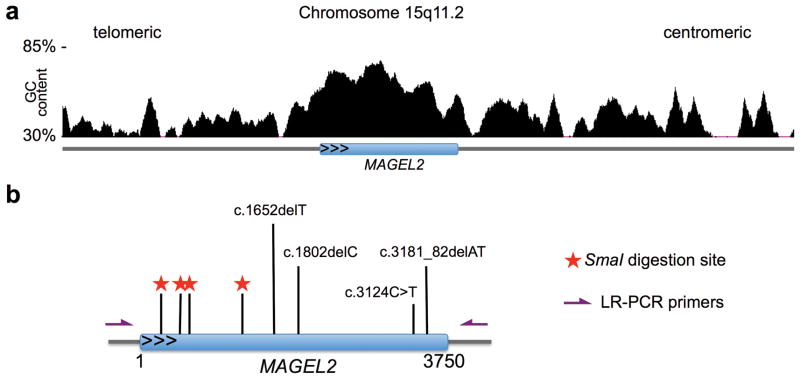
We then developed a test allowing determination of parental origin of de novo MAGEL2 mutations independent of SNP genotypes. MAGEL2is a relatively GC-rich, single-exon gene (Fig. 1a) with a CpGs methylated on the maternal chromosome 15, and exclusively expressed from the unmethylated paternal allele. The MAGEL2 coding sequence contains four restriction sites (5′-CCCGGG-3′) for the methylation sensitive restriction endonuclease SmaI (Fig. 1b). The unmethylated allele is digested by SmaI, while the methylated allele remains intact. PCR amplification following SmaI digestion using oligonucleotide primers flanking one or more of the digestion sites, followed by Sanger sequencing, detects only the maternal allele (Online Methods). The detection of known MAGEL2 mutations following SmaI digestion suggests that they are located on the maternal (i.e. inactive) allele. In contrast, a mutation undetectable by Sanger sequencing following SmaI digest is located on the paternal allele and potentially pathogenic.
Figure 1.
Truncating mutations on the paternal allele of MAGEL2. (a) GC content of MAGEL2 and flanking sequence on 15q11.2 (based on UCSC genome browser, hg19). (b) Truncating MAGEL2 mutations reported in this manuscript are indicated relative to their position in the coding sequence of this single-exon gene. For phasing of MAGEL2 mutations, genomic DNA was digested with the methylation-sensitive restriction endonuclease SmaI, which leaves only the methylated maternal MAGEL2 allele intact. Digestion is followed by long-range PCR. Red stars indicate SmaI digestion sites within the MAGEL2 sequence. Purple arrows indicate the position of oligonucleotide primers used for long-range PCR.
Using this approach, we showed that the MAGEL2 mutations in all four subjects are on the paternal allele (Table 2). Given the functional hemizygosity for this gene, a truncating mutation on the paternal allele leaves affected individuals without functional, expressed MAGEL2, making it potentially pathogenic.
Table 2. Phasing of MAGEL2 mutations.
The methylation-sensitive restriction endonuclease SmaI cuts at four sites within the paternal (unmethylated) allele of MAGEL2. The inability to detect a respective mutation in MAGEL2 by Sanger sequencing following SmaI restriction and long-range PCR suggests its presence on the paternal allele.
| Sequence at mutation site without digestion | Sequence at mutation site after SmaI digestion | Conclusion | |
|---|---|---|---|
| Subject 1 | T/del | T | Mutation on pat allele |
| Subject 2 | C/del | C | Mutation on pat allele |
| Subject 3 | AT/del | AT | Mutation on pat allele |
| Subject 4 | C/T | C | Mutation on pat allele |
In summary, this is the first report of point mutations in the imprinted MAGEL2 gene in the 15q11-q13 domain, causing classic PWS (subject 2) and PWS-like phenotype (subjects 1,3, and 4). This is particularly interesting, given previous reports3–5 of three individuals with small deletions of the SNORD116 snoRNA cluster, two of which meet full diagnostic criteria for PWS. There could be genetic heterogeneity of PWS, or the phenotypic overlap between SNORD116 deletion cases and MAGEL2 mutation cases might be caused by changes in higher-order chromatin structure at the 15q11-q13 locus13. Many speculations could be offered, but the answer is unknown at present. At this time, there does not seem to be a genotype-phenotype correlation (Supplementary Table 1).
All four subjects reported here have a diagnosis of ASDs, based on DSM-IV criteria and expert clinical impression. This suggests MAGEL2 as an additional gene in the ever-growing list of autism susceptibility genes (https://gene.sfari.org/). In a previous study investigating the co-morbidity of PWS with ASDs 19% of individuals with PWS met diagnostic criteria for ASDs14. With that, ASDs are over-represented in our cohort of individuals with truncating MAGEL2 mutations, but additional individuals need to be ascertained before drawing conclusions. Nonsense or frameshifting mutations in MAGEL2 have not been reported in exome sequencing studies of individuals with autism15–22. The GC richness of the gene may impair exon capture, as well as subsequent sequencing.
The phenotypes of MAGEL2loss-of-function mutations reported herein appear consistent with data from Magel2 knockout mouse models, which predominantly manifest poor suckling, neonatal growth retardation, excessive weight gain after weaning, impaired hypothalamic regulation, and delayed onset of puberty as well as reduced fertility23. Learning and memory were found to be normal in the Magel2-null mice, which led to the interpretation that other genes had to be responsible for these features. There are no reports of Magel2-null mice being tested for autism-like behaviors. The latter should probably be considered given the high prevalence of autism spectrum disorders among individuals with MAGEL2 loss-of-function at least in this first report.
Based on our data, were commend considering MAGEL2 sequencing or exome sequencing in complex autism, especially in individuals with a history of neonatal hypotonia, feeding difficulties, or hypogonadism.
The identification of neurological disorders caused by loss-of-function mutations in imprinted genes is particularly important, as novel therapeutic approaches might be envisioned. For another neurodevelopmental disorder, Angelman syndrome, caused by deletion or mutation of the paternally imprinted gene UBE3A, topoisomerase inhibitors were able to unsilence the dormant UBE3A allele in mouse neurons24. Also, antisense RNAs have the potential to activate the inactive, methylated allele of imprinted genes25. We hope that our report will generate new research efforts to investigate the function and clinical importance of this gene, to ultimately benefit individuals with its associated disorders.
Online Methods
Human subjects
Patient 1 and his parents were enrolled in a whole genome sequencing study, approved by the Institutional Review Board of Baylor College of Medicine (BCM), Houston, USA. Enrollment in this study is not based on a particular phenotype, but rather on the referring physician’s determination that the enrolled subject likely has a genetic change in the DNA that has led to genetic disease. Patients 2–4 were referred to the Medical Genetics Laboratories at BCM for clinical whole exome sequencing. Whole exome sequencing (WES) has been offered as a clinical test at the Baylor College of Medicine Whole Genome Laboratory since October 2011. These are consecutive, unrelated samples without pre-screening criteria. The clinical WES test is not a designed study. Among the cases referred for clinical WES, 91.2% were pediatric, 8.1% were adult, and 0.7% were prenatal. 78% of the referred subjects had a history of developmental delay and/or intellectual disability, and 12.2% had a history of autism spectrum disorder.
Following the identification of truncating MAGEL2 mutations, Patients 2–4 and their respective parents were subsequently enrolled in a research study investigating variants of unknown significance, approved by the Institutional Review Board of BCM. Informed consent for all study participants was obtained. For individuals 1, 2, and 3, for whom clinical photographs are shown in Supplementary Note 1, consent was obtained specifically stating the agreement to publish these photographs in medical publications, even if the individual displayed in the picture can be recognized.
Whole genome sequencing analysis
Family 1 comprises two healthy parents and an affected son. Their DNA was sent to Complete Genomics, Inc. for whole genome sequence analysis. The genomics data was analyzed under a de novo model where we identified two high-quality private missense mutations in the affected individual, which are homozygous-reference genotypes in both parents (data obtained from the masterVar file from each genomic data set http://www.completegenomics.com/customer-support/documentation/100357139.html); both mutations were absent in the 1,000 Genomes project, dbSNP and in the ESP 6,500 project. The first de novo mutation in gene MYO1H altersaminoacid (aa) 83 of protein NP_001094891.3 from an Alanine to Valine, however there are no known human disorders associated with aa changes in MYO1H (according to OMIM). The second de novo mutation, in gene MAGEL2, generates a 1-base frameshift in protein NP_061939.3 (1,249 aa) starting at aa 551 and continuing until the new frame reaches a termination codon at aa 701. The MAGEL2 mutation was validated by Sanger sequencing using PCR reactions with primers Val1_Fw and Val1_Re (Supplementary Table 2). The change in the protein is biologically significant and given the fact that MAGEL2 is located in a maternally imprinted region 15q11.2, we proceeded to determine the phase of the mutation.
Phasing de novo mutation in MAGEL2 (also see Supplementary Figure 2)
Patient 1’s purified DNA was aliquoted at ~0.1 genome equivalents per well across a 384-well plate and subjected to the multiple displacement amplification (MDA) method of whole-genome amplification as previously described12. At this concentration, there is a 4% probability that any two linked loci from separate DNA molecules will be aliquoted to the same well. Furthermore, there is a 2% probability that two linked loci of different parental origin will be aliquoted to the same well. The MDA reaction was incubated at 37°C for 19 hours.
After heat inactivation of the MDA reaction, each well was diluted 13.4-fold with water. An aliquot of each well was diluted a further 5-fold with water. 1 μL of this 67-fold diluted DNA was used as template for qPCR to identify wells containing SNP regions linked to the de novo deletion – one paternally inherited SNP located 12,785 bp upstream of the deletion, and three maternally inherited SNPs located 506 bp upstream, 422 bp upstream, and 8,032 bp downstream of the deletion. Primers used were pSNP-12785qPCRseqFw, pSNP-12785qPCRseqRe, mSNP-506-422qPCRseqFw, mSNP-506-422qPCRseqRe, mSNP+8032qPCRseqFw, and mSNP+8032qPCRseqRe (Supplementary Table 2). Fast SYBR Green Master Mix (Applied Biosystems, Grand Island, NY) was used for qPCR. qPCR products of flanking SNPs were used as template for PCR with PfuTurboCx polymerase (Agilent Technologies, Santa Clara, CA), because the presence of UTP and uracil-N-glycosylase in the SYBR Green Master mix makes the qPCR products unstable.
Wells in which SNP regions were amplified by qPCR were assumed to contain DNA fragments spanning the amplified SNP and the de novo deletion. A subset of these wells were chosen for PCR with primers magel11f and magel11r (Supplementary Table 2) using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Woburn, MA) according to the manufacturer’s instructions and the cycling conditions described above to amplify the de novo deletion region. ~40 ng of MDA product was used as template for the KAPA PCRs.
KAPA HiFi and Pfu PCR products were gel purified (GeneJET Gel Extraction Kit, Fermentas, Waltham, MA) and submitted to Elim Biopharmaceuticals (Hayward, CA) for Sanger sequencing.
Copy number variation and methylation
We verified the absence of any copy number variation in the PWS region (15q11.2-q13) by analysis of the CNV calls generated by Complete Genomics in the CNV report files (http://www.completegenomics.com/customer-support/documentation/100357139.html) and by clinical chromosome microarray analysis26.
Methylations-sensitive digestion of MAGEL2 followed by Sanger sequencing
Patient DNA was digested with restriction endonuclease SmaI (New England Biosystems, Ipswich, MA, USA), followed by long-range PCR with DNA primers, LR_magel2_for and LR_magel2_rev (Supplementary Table 2). Specific mutation loci were amplified by nested-PCR and further analyzed by capillary electrophoresis sequencing. The following pairs of DNA primers were used for nested PCRs: Nested_PCR1_for, Nested_PCR1_rev, and Nested_PCR2_for, Nested_PCR2_rev (Supplementary Table 2).
Supplementary Material
Acknowledgments
We are indebted to the patients and their families for their willingness to participate in our research study. We thank P. Zimmerman and E. Austin for clinical assistance. Dr Schaaf ‘s work is generously supported by the Joan and Stanford Alexander Family. Dr. Schaaf is a recipient of a Clinical Scientist Development Award by the Doris Duke Charitable Foundation. Dr. Gonzalez-Garay’s and Dr. Caskey’s work are generously supported by the Cullen Foundation for Higher Education and the Houston Foundation. Dr. Beaudet’s work is supported by NIH grant HD037283.
Footnotes
Conflict of Interest
Drs Schaaf, Beaudet, Caskey, and Yang are faculty members of the Department of Molecular and Human Genetics at Baylor College of Medicine, which derives revenue from whole exome sequencing analysis offered in the Medical Genetics Laboratory. Drs. Peters, McElwain, and Drmanac are employees of Complete Genomics, a company that derives revenue from whole genome sequencing analysis. Complete Genomics has filed several patents on sequencing technology. The remaining authors declare no conflict of interest.
Author Contributions
M.G.-G. and M.McE. performed whole genome sequencing and phase determination on subject 1. M.G.-G., M.McE., B.P., R.D., and C.T. designed and analyzed the experiments for subject 1. F.X. and Y.Y. performed whole exome sequencing and phase determination on subjects 2–4. C.S., F.X., Y.Y., B.Z., A.B., and Y.Y. designed and analyzed the experiments for subjects 2–4. L.P. and K.G. contributed patients, and provided detailed physical examinations. C.S. conceived the overall study, coordinated enrollment, supervised the experiments, wrote the manuscript, and generated the figures and tables. All authors participated in the discussion and interpretation of data and results, and all participated in editing and revising the manuscript.
URLs. Leiden Open Variation Database (LOVD), www.lovd.nl; MAGEL2 specific variations listed in the Leiden Open Variation Database, http://databases.lovd.nl/shared/genes/MAGEL2; Simons Foundation for Autism Research Modular Database for Autism Research (SFARI GENE), https://gene.sfari.org; Complete Genomics Data File Formats, http://www.completegenomics.com/customer-support/documentation/100357139.html; 1000 Genomes project, http://www.1000genomes.org/; dbSNP, http://www.1000genomes.org/; Exome Variant Server, http://evs.gs.washington.edu/EVS/.
References
- 1.Holm VA, et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics. 1993;91:398–402. [PMC free article] [PubMed] [Google Scholar]
- 2.Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med. 2012;14:10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- 3.Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Smith AJ, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18:3257–65. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duker AL, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanber D, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–90. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resnick JL, Nicholls RD, Wevrick R. Recommendations for the investigation of animal models of Prader-Willi syndrome. Mamm Genome. 2013;24:165–78. doi: 10.1007/s00335-013-9454-2. [DOI] [PubMed] [Google Scholar]
- 8.Tennese AA, Wevrick R. Impaired hypothalamic regulation of endocrine function and delayed counterregulatory response to hypoglycemia in Magel2-null mice. Endocrinology. 2011;152:967–78. doi: 10.1210/en.2010-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer RE, Wevrick R. Loss of magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLoS One. 2009;4:e4291. doi: 10.1371/journal.pone.0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–9. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- 11.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 12.Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–5. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum Mol Genet. 2009;18:4227–38. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Descheemaeker MJ, Govers V, Vermeulen P, Fryns JP. Pervasive developmental disorders in Prader-Willi syndrome: the Leuven experience in 59 subjects and controls. Am J Med Genet A. 2006;140:1136–42. doi: 10.1002/ajmg.a.31235. [DOI] [PubMed] [Google Scholar]
- 15.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–42. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu TW, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–73. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bervini S, Herzog H. Mouse models of Prader-Willi Syndrome: a systematic review. Front Neuroendocrinol. 2013;34:107–19. doi: 10.1016/j.yfrne.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Huang HS, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–9. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berteaux N, et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–45. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boone PM, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–42. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.