Abstract
Animals comprise dynamic three-dimensional arrays of cells that express gene products in intricate spatial and temporal patterns that determine cellular differentiation and morphogenesis. A rigorous understanding of these developmental processes requires automated methods that quantitatively record and analyze complex morphologies and their associated patterns of gene expression at cellular resolution. Here we summarize light microscopy based approaches to establish permanent, quantitative datasets—atlases—that record this information. We focus on experiments that capture data for whole embryos or large areas of tissue in three dimensions, often at multiple time points. We compare and contrast the advantages and limitations of different methods and highlight some of the discoveries made. We emphasize the need for interdisciplinary collaborations and integrated experimental pipelines that link sample preparation, image acquisition, image analysis, database design, visualization and quantitative analysis.
Introduction
Although quantitative measurements of morphology and gene expression have long been a component of developmental research1, 2, qualitative descriptions have predominated, especially in molecular studies. Qualitative statements describe in a yes/no manner for example, which tissues a gene is expressed in or if two groups of cells move relative to one another. This basic information is insufficient, though, to address many fundamental questions in developmental biology.
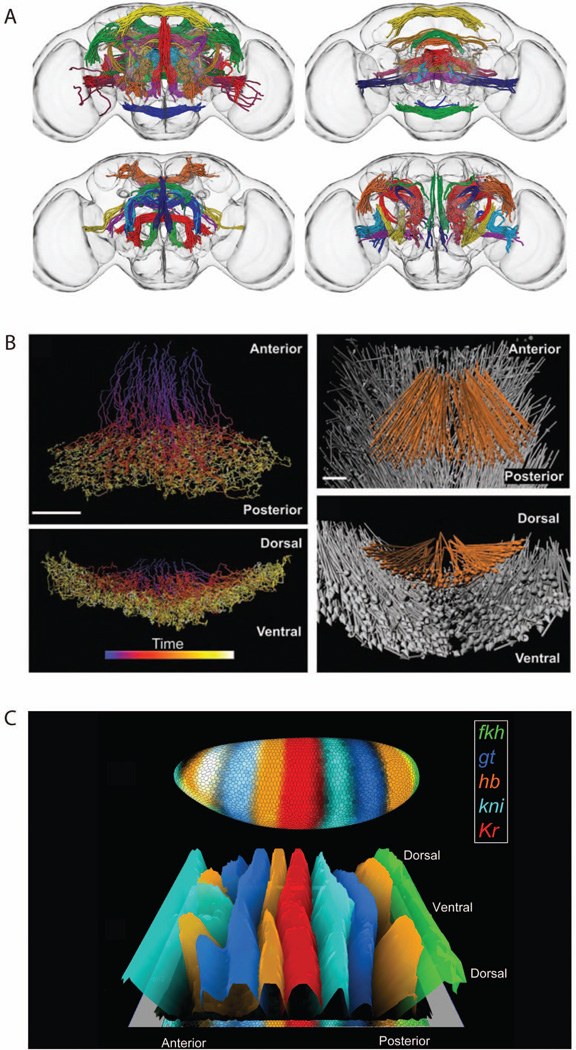
Advances in labeling, imaging and computational image analysis, especially over the last 12 years, are allowing quantitative measurements to be made more readily and in much greater detail than in the past in a range of organisms including Arabidopsis, Ciona, Drosophila, C. elegans, mice, Platynereis, and zebra fish3–16. For example, a cellular resolution, three dimensional atlas has been constructed that records cell type, time of developmental origin, and connections for each of tens of thousands of neurons8 (Fig. 1A). The movements of thousands of cells have been tracked in real time relative to one another3–7 (Fig 1B). Quantitative maps of gene expression in each cell of an embryo have been produced9–13 (Fig. 1C)). Changes in the shapes of cells over time have been measured14, 15.
Figure 1. Examples of three dimensional atlases.
A. The FlyCircuit atlas of neuronal connectivity in Drosophila brains8. Each color groups members of a neuronal tract. Each panel displays different subsets of tracts. B. Cell migration during gastrulation in Drosophila embryos3. The two left panels show cell movements over time. The two right panels show the net displacement vectors, with mesoderm cells shown in orange and ectoderm cells show in grey. C. Patterns of mRNA expression of transcription factors in Drosophila blastoderm embryos10. The upper view shows a three dimensional representation of the embryo. The lower view shows a cylindrical projection in which height indicates the level of expression of each transcription factor in each cell. Both views were generated using the visualization tool PointCloudXplore75, 78
These large scale, quantitative data provide new insights that could not have been gained through qualitative analyses. For instance, sets of individual neurons that form local processing units were discovered that form a basic substructure of the brain8; a subset of gastrulation movements in Drosophila were shown not to require Fibroblast growth factor (FGF), whereas previous qualitative analyses had suggested that FGF was an essential signal3; and regulators of dorsal/ventral cell fates were found to weakly affect the expression of anterior/posterior regulators in Drosophila, which previous non-quantitative studies had failed to detect17.
Just as comprehensive datasets of genomic sequence have revolutionalized biological discovery, large scale quantitative measurements of gene expression and morphology will certainly be of great assistance in enabling computational embryology in the future. Such datasets will form the essential basis for systems level, computational models of molecular pathways and how gene expression concentrations and interactions alter to drive changes in cell shape, movement, connection and differentiation. In this review, we discuss the strategies and methods used to generate such datasets.
The initial inputs for deriving quantitative information of gene expression and embryonic morphology are raw image data, either of fluorescent proteins expressed in live embryos or of stained fluorescent markers in fixed material. These raw images are then analyzed by computational algorithms that extract features such as cell location, cell shape, and gene product concentration. Ideally, the extracted features are then recorded in a searchable database, an atlas, that researchers from many groups can access. Building a database with quantitative graphical and visualization tools has the advantage of allowing developmental biologists who lack specialized skills in imaging and image analysis to use their knowledge to interrogate and explore the information it contains.
We focus on approaches that capture information with cellular resolution in three dimensions because the cell is the basic building block for all animals and morphology is almost invariably three dimensional. Lower resolution studies or two dimensional image analyses have proven useful for addressing some important questions18–26, but space does not permit discussion of these approaches here.
Creating three dimensional atlases: overview
Creating an atlas is more encompassing than image acquisition and analysis. It requires a clear understanding of the biological questions to be addressed. Then appropriate labeling, sample preparation, imaging, image analysis, visualization, and data management methods must be selected (Fig. 2). An interdisciplinary team is required that collectively possess the needed expertise. Generating useful atlases is still in its infancy. Which methods to use at each step along the pipeline will depend greatly on what analysis is required. There is currently no "magic toolbox" that scientists can use to apply to their specific task. Each step has to be tailored to suit the experiment.
Figure 2. A pipeline for building and using a three dimensional atlas.
Imaging three dimensional specimens is particularly challenging. Optical lenses with high magnificiations and resolving powers produce high quality images from thin, two dimensional samples. However, because of their short depth-of-fields such lenses project blurry, mostly out-of-focus images from thick, three dimensional samples. Also, three dimensional samples like embryos, tissues and other multicellular systems, are partially opaque. This limits the depth into a three dimensional sample that can be imaged. These hurdles are continuously being addressed by the development of new fluorescent probes, contrast agents, and image acquisition and image analysis techniques.
Three dimensional atlas projects such as those in Figure 1 generate large amounts of raw data, involving many embryos or tissue samples. Assembly of these data into an atlas, from which desired biological information can be extracted, requires a fully automated analysis pipeline. Fortunately, advances in computer hardware, data storage, image analysis and computer vision have kept pace with improvements in biolabeling and three dimensional bioimaging methods.
Labeling and mounting
The first step in building an atlas is deciding, based on the biological questions to be addressed, what macromolecules need to be labeled in the system being studied, and if live cell or fixed material should be used. It is not practical to build a universal atlas that contains all the information needed by a wide range of developmental biologists. Instead, different atlases will be required to address each question. It is currently not possible to label a single embryo with tens of probes for different biomarkers. Typically, only two to four different labels can be efficiently incorporated and distinguished in a single image, certainly in high throughput studies where sample preparation must be robust. Consequently, atlases require the amalgamation of data from many images.
If cell shape measurements are required, a cell membrane stain would be useful14. Nuclear stains, such as DNA binding dyes or a histone-Green Fluorescent Protein fusion, are ideal for identifying the locations of cells17, 27. If cell migration is to be studied, a live cell approach is called for3, 11, 14, 27. If gene expression measurements must be made in opaque tissue, then fixed material that has been made translucent by soaking it in an optically clear mountant is the practical approach17. If the expression levels of many mRNAs is to be measured, then it is more practical to use nucleic acid in situ hybridization to label fixed material17 than it is to fluorescently tag mRNAs in live embryos, as the latter requires the construction of complex transgenic lines28.
Where data for many specific biomolecules are to be incorporated into the atlas, images from multiple differently labeled embryos or tissue samples must be registered into a common coordinate system. This requires that in addition to each sample being labeled for one of the specific biomolecules, they must also bear a common reference label. The reference could be a spatially patterned protein or mRNA10. Alternatively, if the morphology and number of cells is sufficiently constant between samples, as it is between nematode embryos of the same developmental stage, then a general stain for nuclei or other biomarker of cell location can be used27. In some cases, the overall morphology of the sample has been used successfully for registration8. Since the reference label is to be used throughout the atlas building project, particular care should be taken in its selection.
For three dimensional datasets, especially ones that make quantitative measurements of gene expression, the labels used are almost always fluorescent. In live embryo experiments, transgenic lines expressing fluorescent proteins with different emission spectra are employed29, 30. Here the difficulty of creating transgenic animals that target the specific genes of interest is commonly the rate limiting step. With fixed material, a wider range of fluorophores can be deployed, including DNA binding dyes and antibody conjugated Alexa dyes or quantum dots17, 31. When several different fluorescent probes are used to stain the same sample, the emission spectrum of the probes must be optimized to give the greatest spectral separation. It should also be borne in mind that many probes are bulky and can potentially interfere with ligand-receptor interactions.
Proper mounting of labeled samples prior to image acquisition is paramount. Proper mounting maximizes the optical clarity of the sample, reduces the damaging effects of free-radicals and minimizes the blurring created by optical aberrations. The overall goal of mounting is to minimize changes in the refractive index between the lens, the mountant and through the tissue sample. Fixed material can be cleared, but care must be taken that the solvents used do not disrupt the biology of interest, the fluorophores or the morphology. Live biology has to be mounted so that it can freely exchange oxygen and carbon dioxide and has room to grow, again so its morphology is not disrupted.
Imaging in Three Dimensions
The next step is to select the appropriate imaging method and the associated image acquisition parameters. A number of technologies are available for capturing three dimensional images. These use different ways to remove the out-of-focus information that is collected when a three dimensional image is projected onto a two dimensional image capture device32, 33. Some basic measures can be used to compare different imaging systems. Optical efficiency is the ratio of the number of photons that are collected to create the image to the number of photons used to excite the fluorescence. High optical efficiency lowers the amount of damaging excitation light required and increases the potential acquisition speed. Signal to noise ratio is the fraction of image signal intensity divided by the noise intensity. Image noise results from ubiquitous fluorescence from the sample, such as autofluorescence, and from random thermal noise generated by the detector. It is important to maintain the signal to noise ratio above a critical minimum otherwise the image noise dominates, making subsquent image analysis challenging. Signal to noise is an important consideration when fast acquisition speeds are required or when imaging low numbers of fluorophores.
Choosing the imaging method depends on many factors. For live cell experiments it is critical that the total light exposure be kept to the minimum because excited fluorescent molecules in the presence of oxygen create charged free-radicals that disrupt biochemical pathways and cross-link macromolecular cellular components. Speed of image acquisition is also important in live cell imaging to properly capture living dynamics. The choice of objective lens is important. Objective lenses are defined by many properties but essentially by their magnification, numerical aperture, and working distance. Magnification is the ratio of lengths in the image to corresponding lengths in the sample. Numerical aperture (NA) is a measure of the ability of a lens to collect light. It is defined by the largest angle that light emitted from a point on the sample will be captured by the lens and in practice by the refractive index of the fluid that couples the objective to the sample. The NA of objective lenses vary from low values between 0.3 and 0.5, through medium values between 0.5 and 1.0, to high values between 1.0 and 1.5. NA is a measure of and increases with the resolving power of the imaging system. In choosing the NA for the objective lens, it should be borne in mind that its axial (z-axis) resolving power will be generally less than its lateral (x-y axis) resolving power. Working distance is the distance between the front of the lens and the point or plane in the sample being imaged. Working distance is important in three dimensional imaging because it is the maximum depth into the sample that can be imaged by that lens. Objective lenses with low and medium NAs are usually air-coupled to the sample, have low to medium magnifications and allow the greatest working distances. Objective lenses with high NA have the shortest working distances and are coupled to the sample with fluids that are more dense than air, like water, glycerol or oil. While these criteria complicate image acquisition, high NA lenses have the highest magnifications and resolving powers, producing images of exceptional quality. Still, high NA lenses may not be practical for some applications, like imaging live embryo that have been mounted in configurations requiring longer working distances. Each imaging method has its strengths and weakness and ultimately the choice depends on the scientific question driving the atlas construction. Choosing the imaging method that is most appropriate for a given application requires an understanding of key technical details.
Wilson-grating structured illumination microscopy34 capitalizes on the shallow depth-of-field of objective lenses by using an opaque grating in the illumination path. An image of the grating is projected and illuminates part of the sample in a striped pattern. Multiple images are acquired with the grating shifted into different positions until the entire sample is illuminated. The resulting images are combined in such a way that the out-of-focus component, common to them all, is removed. What remains is a single optical slice of the sample at the focal plane of the objective. Three dimensional images are constructed by repeatedly imaging the sample at different optical planes along the optical axis. The technique is moderately optically efficient in that a large percent of fluorescently emitted photons are captured. In this method high NA lenses give the thinest optical slices and thus the best axial resolution. It uses two dimensional image capture, so the image acquisition is fast, and as a result this method is useful for imaging live as well has fixed cell biology. More recently, other forms of structured "standing-wave" illumination have been used to double the resolving power of wide-field fluorescence microscopy35 and create super resolution optical microscopes, with resolving powers an order of magnitude beyond the theoretical diffraction limit36–39. These will be ideally suited for building atlases of subcellular structures within single cells, rather than cellular resolution atlases of entire embryos.
Deconvolution microscopy40 records a series of images at different planes of focus through a biological sample. Each image can be thought of as the sum of an in-focus optical section and many out-of-focus sections. With knowledge of the sample, which is gained from the image, and the optical response of the imaging system, termed the point spread function, a three dimensional in-focus image can be reconstructed mathematically. Various computational methods have been developed to remove the out-of-focus components from the individual images, which are then combined to create a single three dimensional image. This method is optically efficient because light is not thrown away - it is deconvolved and reassigned. The technique uses a two dimensional image capture device, so image acquisition is fast and thus well suited to live and fixed cell biology. Axial resolving power is highest with high NA lenses. The technique works well in fluorescence but can also be applied to other types of microscopy.
Confocal laser scanning microscopy uses galvanometer mirrors to raster-scan a focused laser beam across the sample41. Because the beam illuminates one point on the sample at a time, a three dimensional image is built sequentially, point by point. However, although objective lenses focus a laser to pinpoint accuracy in the focal plane at the sample, the focused point is still too large along the optical-axis and illuminates many out-of-focus planes. To remove the light collected from the out-of-focus planes, the image is filtered through a pinhole at the image plane. This allows light to pass from the conjugate sample point, blocking the light collected from the out-of-focus planes. The method is optically inefficient because most of the excited photons are excluded from the image by the pinhole. Further, because the image is collected one point at a time, the image acquisition times are long, and this limits the usefulness of this method for live cell imaging. Never-the-less, confocal laser scanning microscopy has become very popular because of its simplicity of implementation.
Spinning disk technology42 is an adaptation of confocal laser scanning microscopy. A mechanical circular array of pinholes, a Nipkow disk, allows thousands of focused laser beams to scan the object at the same time. In the implementation by Yokogawa Electric Corporation, two disks are used - one an array of microlenses which focus the laser beams onto the sample and the other an array of pinholes that confocalize the image. The advantage of this approach is that it is fast, uses two dimensional image capture, and thus it has been widely used in live cell imaging. However, it has low optical sensitivity because of the use of pinholes and because of its rapid acquisition speed, care must be taken that the signal to noise ratio of the images is maintained.
Two-photon laser scanning microscopy is another adaptation of laser scanning microscopy43, 44. Instead of exciting fluorescence by the absorption of single photons at a fluorophore's absorption energy, fluorescence is excited by simultaneous absorption of two half-energy photons. This non-linear absorption significantly reduces the volume within the biological sample in which fluorescence occurs. The density of photons is only high enough for simultaneous absorption of photons within a tiny volume of the focused laser beam. As a result, out-of-focus planes are not fluorescently excited, and this eliminates the need for a confocal pinhole. Two-photon microscopy has higher optical efficiency than confocal microscopy. In addition, because half-energy photons have longer wavelengths, they scatter less and penetrate further into biological samples (Fig 2B). Because fluorophores outside of the excitation volume are not excited, phototoxicity and photobleaching during scanning are also significantly reduced.
Light-sheet, selective plane microscopy38, 45, does away with the traditional Köhler light source that illuminates the out-of-focus planes in the first place. Rather than illuminating along the optical axis, selective plane microscopy creates a transverse sheet of light that excites a single optical plane through the sample. No scanning in the X-Y direction is involved, and the illuminated optical section is imaged onto a two dimensional capture device. In the latest versions of selective plane microscopy, multiple objective lenses can be used to image the sample from different angles, and the sample stage is designed to allow rotational symmetry about the light sheet45–48. Depending on the orientation of the objective lenses, the optical penetration through the sample can be doubled (objectives at 180° orientation), or the axial resolving power of one lens can be increased by the second lens (objectives at 90° orientation). Further, by rotating the stage the sample can be imaged at multiple orientations. Selective plane microscopy is optically efficient, image acquisition is fast and it allows both single-photon and two-photon excitation49. This techniques is versatile and ideally suited to live cell dynamics. Some tricks are needed to correct images for the transverse shadowing effects created by illuminating with a light sheet. This can be done computationally after the images are acquired or by oscilating the angle of the light sheet. Multiple lens imaging also requires post acquisition analysis to construct the final image50. Mounting the biological samples is complicated by the rotational symmetry required about the light sheet. Although iSPIM51 uses a cleaver optical adaptation which allows selective plane microscopy on a regular inverted microscope. Longer working-distance objective lenses with medium NA are needed, and this reduces the resolving power of the imaging from what is possible. Thus for example, the quality of images acquired from live cell biology will never be as high as that possible from fixed-cell methods which allow the sample to be optically cleared and imaged with the highest optical resolving powers currently possible (Fig. 3, compare panels A and B.).
Figure 3. Comparing image quality of SPIM and laser scanning multi photon microscopy.
Multiphoton optical sections are shown for stage-16 Drosophila embryos stained to label nuclei. A) The top two images are of a live embryo expressing GFP-histone. The images were taken using the two-photon SPIM technique of "simultaneous multiview imaging" (SiMView) and were kindly provided by Philipp J. Keller48. B) The bottom two images are of a fixed emrbyo stained with SYTOX Green and were acquired using standard two-photon laser scanning microscopy. The embryo's dorsal / ventral direction is shown from top to bottom in each image. Optical sections were selected through the midplane of each three dimensional embryo image to show, from left to right, its anterior / posterior (left) and it's left / right (right) directions.
Segmentation, Feature Extraction and Registration
Once a set of high quality images have been obtained, the next step is to use these data to build a quantitative atlas. Three dimensional atlas projects such as those in Figure 1 generate large amounts of raw image data that needs to be combined into a computationally analyzeable atlas. As we define it, an atlas is essentially a large spread sheet, a table with rows and columns of numbers and other descriptions. These may give the x,y,z coordinates in space of cells at successive time points, the concentrations and locations of gene products in each cell, the histological cell type of each cell, and/or the indices of neighboring or connecting cells. Building an atlas from raw image data involves broadly three types of image analysis: segmentation, feature extraction and registration52–55 and these need to be done in an automated way.
Segmentation
Segmentation is the subdivision of an image into regions belonging to—and not belonging to—objects of interest, such as nuclei, cell membranes, and tissues. Each voxel is assigned in a yes/no manner into one the categories being defined. Many segmentation techniques are available and rely on different properties of an image, such as brightness, color or texture. Examples of segmentation techniques include Thresholding, Template Matching, Watershed, Region Growing, Laplacian of Gaussian, Difference of Gaussians, Level Set and Fast Marching methods56, 57. In the world of image analysis, the problem of segmentation has been solved for many applications. However, and particularly for fluorescence-based bioimaging, the complexity of the images means that existing methods do not work “out of the box” and need specific tailoring for specific application. For example, total DNA-staining combined with relatively simply segmentation algorithms can be used to detect the position and number of cells. However, more sophisticated segmentation approaches will be needed to delineate cells if their packing density is too high. If accurate determination of cellular or subcellular volumes is required, then segmentation techniques that detect edges as well as blobs in an image may be needed in combination with the staining of other cellular components58. Often the segmentation analysis may need to be supplied with a priori information, such as the number, size, shape, and packing density of the objects in the image to be segmented..For multicellular systems, hierarchical segmentation maybe required so that biological components can be segmented on a subcellular, cellular, tissue and organ level.
The results of a segmentation analysis are labeled segmentation masks that delineate individual objects in the image. However, before these can be used to direct subsequent quantitative evaluation, their accuracy must be determined. The biggest difficulty is to obtain an accurate ground truth to compare the segmentation to. One approach is to compare the results of segmentation to the raw image data using a visualization tool59. In high throughput studies, however, such labor intensive “by eye” scoring can only be performed on a small sample, usually only small portions from a few images. Alternatively, automatic approaches can be devised. For example, the correlation between the number of segmented nuclei versus the overall volume of the embryo measures the relative accuracy of different nuclear segmentation methods, though this approach cannot determine the absolute accuracy10. Determining the accuracy of large scale segmentation analyses remains a challenge for the field.
Feature Extraction
Once the accuracy of the segmentation has been confirmed, the segmentation masks can be used to direct quantitative evaluation of the features required for the atlas. Many image features can be measured. These are broadly divisible into hard-features, such as positions, dimensions, rates of motion and the brightness of cells, and soft-features, like the statistical analysis of texture, pattern recognition, context matching, clustering and classification53, 60–62.
Registration
Image registration methods have been developed in the field of vision research and applied to remote sensing and medical imaging for years63. Many of these techniques are applicable to multicellular biological systems. To create an atlas, information from many images must be placed onto a common morphological framework. This is essential because biological samples, like embryos, are rarely identical. Biological variability means that samples of the same biological system may have different numbers of cells or an equivalent number in different relative positions, and this biological variability varies with developmental time. Each image from a series of different samples or from the same sample at different times will have unique information, such as the expression pattern of a gene or the neighborhood connections between cells. Information from multiple images must be combined in such a way that the resulting comprehensive atlas accurately represents the biology. To do this, registration methods first define sets of biologically equivalent locations in each image: for example, corresponding cells. There are many ways of doing this that use the expression of specific genes or the inherent morphological complexity within the sample to define such equivalences3, 9–11, 16. Registration is performed either on the raw image data prior to segmentation or on sets of segmented features. When raw image data are registered, a single representative image can be chosen as the reference coordinate system onto which other images are mapped8. When segmented features are registered, a statistical average model can be created as the reference coordinate system onto which extracted data are placed10. In either case, registration involves the determination of sets of equivalences in space and time that allow points or segmented objects in one image to be registered with the equivalent points or objects in another.
As with segmentation analysis, the accuracy of the registration must be determined. For example, coarsely registering multiple embryo images may accurately align the principal body axes of a sample, but it will blur information from non equivalent, neighboring cells, due to the biological variability. The only way to create atlases that correctly represent the biology is to register images at cellular resolution. In this way one will be able to demonstrate that quantitative features derived from the atlas replicate results derived from the analysis of multiple individual embryos10.
Image Analysis Packages
To support automated high-throughput image-based investigations of multicellular systems, many groups are creating image analysis tool-boxes specifically for bioimaging informatics64. These toolboxes bring the latest developments in segmentation, feature extraction, and registration to a broad audience from fields that span biology to computer vision. In doing so, these toolboxes are helping to form a new community with multidisciplinary expertise.
Particularly useful are open source toolboxes that are compatible with multiple operating systems. The National Institutes of Health’s NIH Image and ImageJ were some of the earliest open source initiatives65, and have more recently undergone a further round of development with the introduction of Fiji66. ICY is another bioinformatics and image analysis platform. It leverages the open-source Visualization Toolkit (VTK, http://www.vtk.org)67. BioImageXD68 is yet another collaborative effort providing image analysis, processing and visualizing for multi-dimensional microscopy images. It is also based on the Visualization Toolkit VTK, and the National Library of Medicine's Insight Segmentation and Registration Toolkit (ITK, http://www.itk.org). DIPimage (http://www.diplib.org) is a scientific image processing and analysis toolbox written specifically for MATLAB (http://www.mathworks.com). DIPimage harnesses the power of MATLAB, while allowing programmer flexibility in creating image analysis pipelines. Other groups have created toolboxes for specific application. For example, CellProfiler allows quantitative evaluation for cultured cell phenotype69. Considerable effort has also gone into development of database and image analysis environments for bioimage informatics. Examples of these are Bisque70 and Open Microscopy Environment (OME) and more recently OME Remote Objects (OMERO)71.
Databases
Projects that produce large amounts of raw and processed data require a database. Many atlases are based on images from hundreds or thousands of biological samples. For each image, sample preparation and imaging involves multiple steps, often with several associated variables such as the biomolecule labeled, the developmental stage, image quality, the date of experiment etc. Thus, in addition to data files, extensive metadata describing the experiments associated with each file must also be recorded in the database. This requires the construction of a relational database to allow rapid search and retrieval of files based on a variety of criteria. In addition to allowing ready access to data for subsequent analysis, such a database greatly aids quality control during atlas construction. For example, it allows a user to work backwards along the pipeline to locate the cause of any data analysis failures, determining what variables are associated with a given failure or artifact. The database should have an associated web site for access by internal researchers working on atlas construction and quite likely a separate web site for public access to published datasets. For examples, see: (http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=summary); (http://www.flycircuit.tw); (http://caltech.wormbase.org/virtualworm/).
Discovery using atlases
Once a searchable atlas has been constructed there are fundamentally two approaches that can be used to analyze the data: one visual, the other mathematical. The challenge is that while biologists best understand the questions that can be addressed using the atlas, they may not always possess the computational and mathematical skills needed to conduct sophisticated analyses of such data files. For this reason, biologists generally collaborate with computational scientists. It is not always clear, though, what is the best way to frame the analysis. Here, visualization tools can provide important guidance. These tools provide a point and click environment in which biologists can explore various features of the data on their own, for example looking for interesting correlations. This exploration may itself lead to novel discoveries, but will also help the biologist better understand the quality and nature of the dataset, improving his or her ability to suggest analyses to computational colleagues. The results of the subsequent mathematical analysis can often be exported as additional rows and columns into an updated version of the atlas and then explored by the biologist using the visualization tool.
Visualization tools
Developing visualization tools for an atlas of three dimensional morphology and expression is challenging59, 72–74. The complexity of the data quickly become uninterpretable to the human eye. Many thousands of cells are layered on top of one another, each with multiple quantitative attributes assigned to them. The challenge is to find ways to view only defined parts of the data to reduce the complexity and thus allow visualization of, say, correlations between one attribute and another. It is important that the tool allow the user flexibility in choosing which attributes to compare, ideally with different graphing and display options.
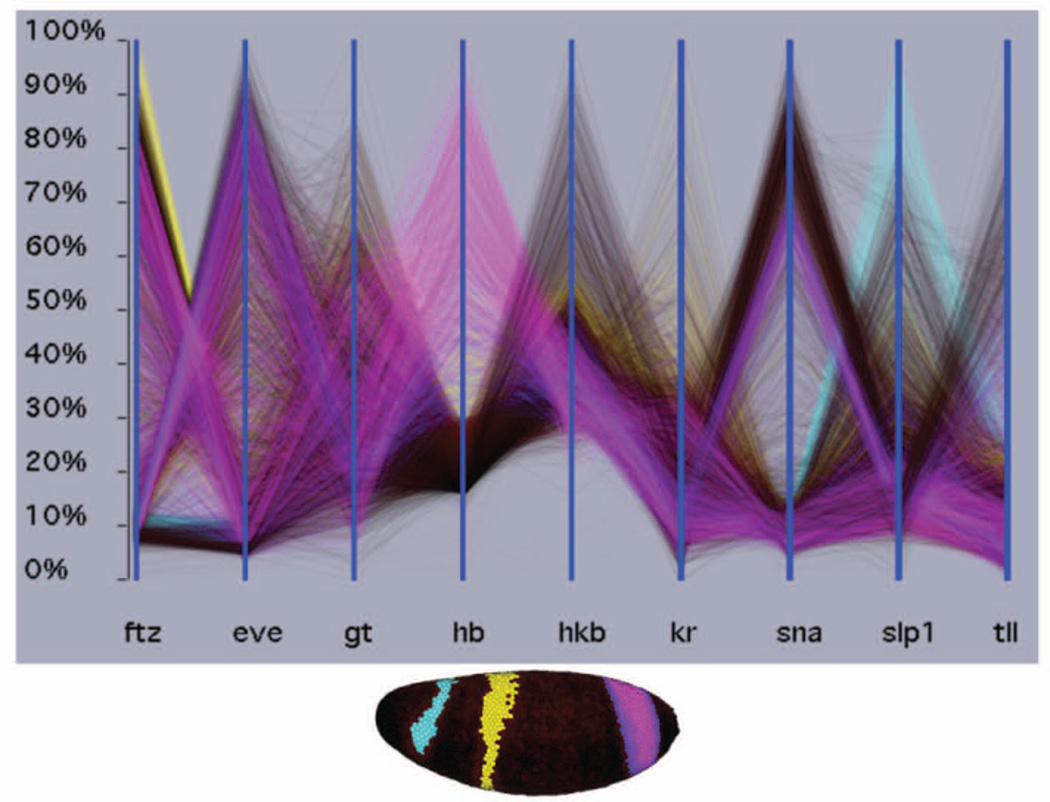
Figure 1 provides some examples. In A, defined subsets of neurons belonging to specific tracts are displayed in each panel. In B, two ways to visualize how cells move during gastrulation are shown, one showing continuous change over time, the other showing mean vectors over a defined time interval. In C, the differences in mRNA expression between cells are shown in two views, a physical three dimensional view (upper) and a cylindrical projection (lower) in which height is used to better illustrate differences in expression levels. Visualization tools, however, can go beyond these relatively straightforward ways of displaying data. For example, only a relatively few gene expression patterns can be visualized at once in a physical view such as that in Figure 1C. The expression of tens of genes can be compared at once, however, if the levels of expressions each gene in all cells is represented along one of a series of one dimensional, parallel coordinates (Fig. 4)75. At the same time, results for a subset of genes can be projected back into a three dimensional physical view, once they are identified as being of interest (Fig. 4).
Figure 4. Visualizing 3D gene expression by parallel coordinates.
Each vertical axis shows the mRNA expression levels for one gene in each of the 6,000 cells in the Drosophila blastoderm embryo. The lines connect data for the same cells. Blue lines connect cells expressing the anterior most stripe of hunchback (hb), yellow lines the central hb stripe and pink lines the posterior stripe. The locations of these cells are shown in the physical 3D view below. In the parallel coordinates, it can be readily seen that the anterior stripe on hb coincides with high slp1 expression, the central hb stripe with high ftz expression, and 50% of the posterior hb stripe with high eve expression. These views were generated using PointCloudXplore, an interactive visualization tool (http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=pcx)75.
Mathematical analysis
Ultimately, though, the most powerful way to analyze a three dimensional atlas is by sophisticated mathematical approaches. Only in this way can the combination of multiple quantitative features within the data be rigorously compared. A wide array of analyses have been made using three dimensional atlases. For instance, a model of mesodermal cell movement during gastrulation showed, among other things, that neither ectodermal cell movements or the orientations of cell divisions correlate with the direction of mesoderm cell movement3. An analysis of the correlation between the relative concentrations of transcription factor protein molecules and temporal changes in target gene mRNA expression established putative regulatory relationships within a transcription network76. Quantification of changes in plant stem cell volumes and divisions showed that both play key roles in shaping specific morphologies14. Quantifying interspecies divergence showed that even small changes in regulatory networks result in significient differences in the placement and number of equivalent cells77.
There is every reason to believe that in future a wider array of developmental processes will be studied by mathematical analysis of three dimensional atlases - created using optical imaging and image analysis techniques. As other data classes—such as molecular interaction data—are folded in, more complex systems models can be expected that seek to link the biochemistry of regulatory networks to morphological dynamics. The creation and exploitation of large scale quantitative atlases will lead to a more precise understanding of development.
Acknowledgements
The image of the live, late-stage Drosophila embryo was kindly provided by Philipp J. Keller. Ann-Shyn Chiang kindly provided the brain wiring figure. Pablo Arbelaez, Damir Sudar, Anil Aswani, and Soile V.E. Keränen gave helpful suggestions on the manuscript. It's writting was funded by U.S. National Institutes of Health (NIH) grant 1R01GM085298-01A1 (to DWK). Work at Lawrence Berkeley National Laboratory is conducted under Department of Energy contract DE-AC02-05CH11231.
References
- 1.Bookstein FL. Morphometric tools for landmark data. Geometry and biology. Cambridge University Press; 1997. [Google Scholar]
- 2.Thompson DAW. On growth and form. Cambridge, New York: University Press; 1942. [Google Scholar]
- 3.McMahon A, Supatto W, Fraser SE, Stathopoulos A. Dynamic analyses of Drosophila gastrulation provide insights into collective cell migration. Science. 2008;322:1546–1550. doi: 10.1126/science.1167094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Dev Biol. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDole K, Xiong Y, Iglesias PA, Zheng Y. Lineage mapping the pre-implantation mouse embryo by two-photon microscopy, new insights into the segregation of cell fates. Dev Biol. 2011;355:239–249. doi: 10.1016/j.ydbio.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivier N, Luengo-Oroz MA, Duloquin L, Faure E, Savy T, Veilleux I, Solinas X, Debarre D, Bourgine P, Santos A, et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science. 2010;329:967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- 7.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 9.Bao Z, Murray JI, Boyle T, Ooi SL, Sandel MJ, Waterston RH. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowlkes CC, Luengo Hendriks CL, Keranen SVE, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray JI, Boyle TJ, Preston E, Vafeados D, Mericle B, Weisdepp P, Zhao Z, Bao Z, Boeck M, Waterston RH. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 2012;22:1282–1294. doi: 10.1101/gr.131920.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles DW. Three-dimensional morphology and gene expression mapping for the Drosophila blastoderm. Cold Spring Harb Protoc. 2012;2012:150–161. doi: 10.1101/pdb.top067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods. 2010;7:547–553. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- 15.Tassy O, Daian F, Hudson C, Bertrand V, Lemaire P. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr Biol. 2006;16:345–358. doi: 10.1016/j.cub.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 16.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 17.Luengo Hendriks CL, Keranen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biology. 2006;7 doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 19.Surkova SY, Myasnikova EM, Kozlov KN, Samsonova AA, Reinitz J, Samsonova MG. Methods for Acquisition of Quantitative Data from Confocal Images of Gene Expression in situ. Cell and tissue biol. 2008;2:200–215. doi: 10.1134/S1990519X08020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- 21.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen EH, Overly CC, Jones AR. The Allen Human Brain Atlas: Comprehensive gene expression mapping of the human brain. Trends Neurosci. 2012 doi: 10.1016/j.tins.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawrylycz M, Baldock RA, Burger A, Hashikawa T, Johnson GA, Martone M, Ng L, Lau C, Larson SD, Nissanov J, et al. Digital atlasing and standardization in the mouse brain. PLoS Comput Biol. 2011;7:e1001065. doi: 10.1371/journal.pcbi.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frise E, Hammonds AS, Celniker SE. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol. 2010;6:345. doi: 10.1038/msb.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 30.Mavrakis M, Pourquie O, Lecuit T. Lighting up developmental mechanisms: how fluorescence imaging heralded a new era. Development. 2010;137:373–387. doi: 10.1242/dev.031690. [DOI] [PubMed] [Google Scholar]
- 31.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 32.Fischer RS, Wu Y, Kanchanawong P, Shroff H, Waterman CM. Microscopy in 3D: a biologist's toolbox. Trends Cell Biol. 2011;21:682–691. doi: 10.1016/j.tcb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lidke DS, Lidke KA. Advances in high-resolution imaging--techniques for threedimensional imaging of cellular structures. J Cell Sci. 2012;125:2571–2580. doi: 10.1242/jcs.090027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neil MA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Lett. 1997;22:1905–1907. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- 35.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung BO, Chou KC. Review of super-resolution fluorescence microscopy for biology. Appl Spectrosc. 2011;65:967–980. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 38.Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat Methods. 2011;8:811–819. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- 39.Kanchanawong P, Waterman CM. Advances in light-based imaging of three-dimensional cellular ultrastructure. Curr Opin Cell Biol. 2012;24:125–133. doi: 10.1016/j.ceb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally JG, Karpova T, Cooper J, Conchello JA. Three-dimensional imaging by deconvolution microscopy. Methods. 1999;19:373–385. doi: 10.1006/meth.1999.0873. [DOI] [PubMed] [Google Scholar]
- 41.Amos WB, White JG. How the confocal laser scanning microscope entered biological research. Biol Cell. 2003;95:335–342. doi: 10.1016/s0248-4900(03)00078-9. [DOI] [PubMed] [Google Scholar]
- 42.Wilson T. Spinning-disk microscopy systems. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.top88. 2010:pdb top88. [DOI] [PubMed] [Google Scholar]
- 43.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 44.So PT, Dong CY, Masters BR, Berland KM. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 45.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 46.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 47.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomer R, Khairy K, Amat F, Keller PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- 49.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 50.Preibisch S, Saalfeld S, Schindelin J, Tomancak P. Software for bead-based registration of selective plane illumination microscopy data. Nat Methods. 2010;7:418–419. doi: 10.1038/nmeth0610-418. [DOI] [PubMed] [Google Scholar]
- 51.Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Bao Z, Colon-Ramos D, Shroff H. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:17708–17713. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wollman R, Stuurman N. High throughput microscopy: from raw images to discoveries. J Cell Sci. 2007;120:3715–3722. doi: 10.1242/jcs.013623. [DOI] [PubMed] [Google Scholar]
- 53.Hamilton N. Quantification and its applications in fluorescent microscopy imaging. Traffic. 2009;10:951–961. doi: 10.1111/j.1600-0854.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- 54.Truong TV, Supatto W. Toward high-content/high-throughput imaging and analysis of embryonic morphogenesis. Genesis. 2011;49:555–569. doi: 10.1002/dvg.20760. [DOI] [PubMed] [Google Scholar]
- 55.Long F, Zhou J, Peng H. Visualization and analysis of 3D microscopic images. PLoS Comput Biol. 2012;8:e1002519. doi: 10.1371/journal.pcbi.1002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marr H. Vision. A computational investigation into the human representation and processing of visual information. W. H. Freeman & Co; 1982. [Google Scholar]
- 57.Li G, Liu T, Tarokh A, Nie J, Guo L, Mara A, Holley S, Wong ST. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 2007;8:40. doi: 10.1186/1471-2121-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khairy K, Keller PJ. Reconstructing embryonic development. Genesis. 2011;49:488–513. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- 59.Rueden CT, Eliceiri KW. Visualization approaches for multidimensional biological image data. Biotechniques. 2007;43:31, 33–36. doi: 10.2144/000112511. [DOI] [PubMed] [Google Scholar]
- 60.Belongie S. Shape Matching and Object Recognition Using Shape Contexts. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24:509–522. doi: 10.1109/TPAMI.2005.220. [DOI] [PubMed] [Google Scholar]
- 61.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Shamir L, Delaney JD, Orlov N, Eckley DM, Goldberg IG. Pattern recognition software and techniques for biological image analysis. PLoS Comput Biol. 2010;6:e1000974. doi: 10.1371/journal.pcbi.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zitova B, Flusser J. Image registration methods: a survey. Image and Vision Computing. 2003;21:977–1000. [Google Scholar]
- 64.Eliceiri KW, Berthold MR, Goldberg IG, Ibanez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, et al. Biological imaging software tools. Nat Methods. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 68.Kankaanpaa P, Paavolainen L, Tiitta S, Karjalainen M, Paivarinne J, Nieminen J, Marjomaki V, Heino J, White DJ. BioImageXD: an open, general-purpose and highthroughput image-processing platform. Nat Methods. 2012;9:683–689. doi: 10.1038/nmeth.2047. [DOI] [PubMed] [Google Scholar]
- 69.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: a platform for bioimage analysis and management. Bioinformatics. 2010;26:544–552. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- 71.Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, Macdonald D, Moore WJ, Neves C, Patterson A, et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weber GH, Rubel O, Huang MY, DePace AH, Fowlkes CC, Keranen SVE, Hendriks CLL, Hagen H, Knowles DW, Malik J, et al. Visual Exploration of Three-Dimensional Gene Expression Using Physical Views and Linked Abstract Views. Ieee-Acm Transactions on Computational Biology and Bioinformatics. 2009;6:296–309. doi: 10.1109/TCBB.2007.70249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubel O, Weber GH, Huang MY, Bethel EW, Biggin MD, Fowlkes CC, Hendriks CLL, Keranen SVE, Eisen MB, Knowles DW, et al. Integrating Data Clustering and Visualization for the Analysis of 3D Gene Expression Data. Ieee-Acm Transactions on Computational Biology and Bioinformatics. 2010;7:64–79. doi: 10.1109/TCBB.2008.49. [DOI] [PubMed] [Google Scholar]
- 74.Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S, et al. Visualization of image data from cells to organisms. Nat Methods. 2010;7:S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rübel O, Weber GH, Keränen SVE, Fowlkes CC, Luengo Hendriks CL, Shah NY, Biggin MD, Hagen H, Knowles DW, Malik J, Sudar D, Hamann B. PointCloudXplore: Visual analysis of 3D gene expression data using physical views and parallel coordinates. In: Santos BC, Ertl T, Joy K, editors. Data Visualization 2006 (Proceedings of the Eurographics/IEEEVGTC Symposium on Visualization, Lisbon, Portugal, May 2006) 2006. [Google Scholar]
- 76.Aswani A, Keranen SV, Brown J, Fowlkes CC, Knowles DW, Biggin MD, Bickel P, Tomlin CJ. Nonparametric identification of regulatory interactions from spatial and temporal gene expression data. BMC Bioinformatics. 2010;11:413. doi: 10.1186/1471-2105-11-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fowlkes CC, Eckenrode KB, Bragdon MD, Meyer M, Wunderlich Z, Simirenko L, Hendriks CLL, Keranen SVE, Henriquez C, Knowles DW, et al. A Conserved Developmental Patterning Network Produces Quantitatively Different Output in Multiple Species of Drosophila. Plos Genetics. 2011;7 doi: 10.1371/journal.pgen.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber GH, Rubel O, Huang MY, DePace AH, Fowlkes CC, Keranen SV, Luengo Hendriks CL, Hagen H, Knowles DW, Malik J, et al. Visual exploration of threedimensional gene expression using physical views and linked abstract views. IEEE/ACM Trans Comput Biol Bioinform. 2009;6:296–309. doi: 10.1109/TCBB.2007.70249. [DOI] [PMC free article] [PubMed] [Google Scholar]