Abstract
Enveloped viruses escape infected cells by budding through limiting membranes. In the decade since the discovery that the Human Immunodeficiency Virus (HIV) recruits cellular ESCRT (endosomal sorting complexes required for transport) machinery to facilitate viral budding, this pathway has emerged as the major escape route for enveloped viruses. In cells, the ESCRT pathway catalyzes the analogous membrane fission events required for the abscission stage of cytokinesis and for a series of “reverse topology” vesiculation events. Studies of enveloped virus budding are therefore providing insights into the complex cellular mechanisms of cell division and membrane protein trafficking (and vice versa). Here, we review how viruses mimic cellular recruiting signals to usurp the ESCRT pathway, discuss mechanistic models for ESCRT pathway functions, and highlight important research frontiers.
Keywords: Enveloped viruses, Virus-host interactions, Cytokinesis, Exosomes, Shedding microvesicles
1 Enveloped Virus Budding
Cell membranes present significant dissemination barriers and viruses have therefore developed sophisticated mechanisms for entering and exiting cells. Here, we review how enveloped viruses bud through membranes and thereby acquire their lipid bilayers, with a particular focus on viruses that employ the ubiquitous strategy of usurping the cellular ESCRT (endosomal sorting complexes required for transport) pathway. Although ESCRT-dependent budding is best studied for retroviruses, particularly HIV-1, a remarkable variety of enveloped viruses use this pathway to escape cells (Supplemental Table 1). Comparative virology is providing important insights not only into virology, but also into the cellular functions and mechanisms of the ESCRT machinery (reviewed in Agromayor and Martin-Serrano, 2013; Hanson and Cashikar, 2012; Henne et al., 2011; Hurley and Hanson, 2010; McCullough et al., 2013; Weissenhorn et al., 2013). This comparative approach is particularly powerful because ESCRT pathways are conserved across Eukarya and are even found in Archaea; for example, in crenarchaeal hyperthermophiles such as Sulfolobus solfataricus the ESCRT pathway is used for egress of the enveloped Sulfolobus Turreted Icosahedral Virus (STIV) (Snyder et al., 2013).
Enveloped viruses commonly assemble and bud at the plasma membrane, although some bud into internal compartments (Lorizate and Krausslich, 2011). In the latter cases, the internal compartment must ultimately fuse with the plasma membrane to release the virus from the cell. The topology of virus budding differs from classical cellular vesiculation processes such as endocytosis, where the membrane constricts away from the cytoplasm and membrane fission is catalyzed by cytoplasmic dynamin, which acts from the outside of the bud neck. In contrast, the membranes of budding viruses must be constricted toward the cytoplasm, and cytoplasmic host factors that catalyze membrane fission must work from within the bud neck. The ESCRT pathway remains the only well characterized cellular pathway known to perform such “reverse topology” membrane fission events, and this capability seems to explain why so many different viruses have evolved to usurp this machinery.
Conceptually, virus budding can be divided into two stages: 1) membrane deformation, when the membrane is “wrapped” around the assembling virion; and 2) membrane fission, when the bud neck is severed. The structural proteins of enveloped viruses typically bind membranes and form spherical or helical assemblies. Thus, assembly and budding are often inextricably linked processes. It has generally been assumed that the energy provided by protein-protein and protein-membrane interactions is sufficient to drive membrane envelopment, although there are intriguing hints that cellular factors may sometimes be recruited to help with membrane bending, much as membrane bending proteins cooperate with clathrin scaffolds to create endosomal vesicles (McMahon and Boucrot, 2011). The ESCRT machinery then typically draws the opposing membranes together and mediates the final membrane fission step required for virus release.
The different stages of virus budding are nicely illustrated by the process of HIV-1 assembly (Sundquist and Krausslich, 2012). As in other retroviruses, the HIV-1 Gag polyprotein functions as the major viral structural protein. Gag is targeted to the inner leaflet of the plasma membrane by a bipartite signal comprising an N-myristoyl fatty acid modification and a binding site for the plasma membrane-specific phosphatidyl inositol, PI(4,5)P2. Gag molecules capture the viral RNA genome and assemble into a spherical virion that is organized on a semi-regular hexagonal net (Bharat et al., 2012). Recombinant HIV-1 Gag molecules can form spherical particles in vitro, and Gag assembly can therefore contribute to the energy required for membrane deformation (Campbell and Rein, 1999). Under some conditions, however, retroviral assembly can arrest before fully spherical virions are formed, suggesting that host factors may also participate in membrane deformation and Gag assembly (Dooher et al., 2007; Gottwein et al., 2003; Le Blanc et al., 2002; Zhang et al., 2011). Assembling HIV-1 Gag molecules also recruit early-acting factors of the ESCRT pathway required to complete the membrane fission step. In the absence of ESCRT factor recruitment, virus assembly typically arrests at a late stage in which the fully assembled viral Gag shell remains connected to the plasma membrane through a thin membrane “stalk” (Gottlinger et al., 1991). Thus, the ESCRT pathway functions primarily to mediate the membrane fission step required for virion release. In the following, we discuss components and functions of the ESCRT pathway, describe key insights gathered from virology, and outline important next steps for future research.
2 The ESCRT Pathway
2.1 ESCRT Pathway Functions
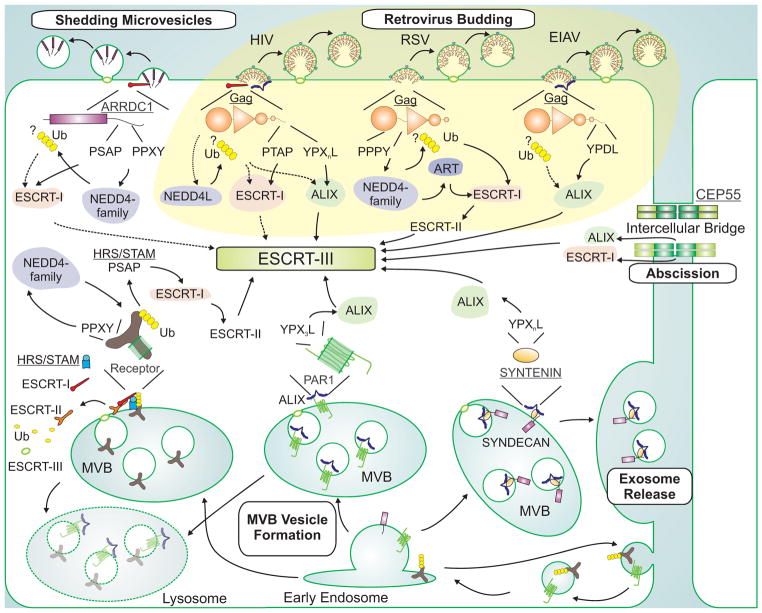
The ESCRT pathway was initially identified through genetic analyses in yeast that defined the factors required to target membrane proteins for degradation within vacuoles (or lysosomes in mammalian cells) (Hanson and Cashikar, 2012; Henne et al., 2011). Such ubiquitylated membrane proteins are sorted into vesicles that bud inward into the lumen of maturing endosomes or multivesicular bodies (MVB), and are then degraded when the MVB fuses with the vacuole. As illustrated in Figure 1, the MVB vesiculation/trafficking pathway is expanded in mammalian cells because MVBs can either fuse with lysosomes to release vesicle contents for degradation, or fuse with the plasma membrane to release extracellular exosomes (Akers et al., 2013; Raposo and Stoorvogel, 2013).
Figure 1. ESCRT pathway recruitment.
Cellular and viral adaptor proteins and complexes that recruit early-acting ESCRT factors and NEDD4 family ubiquitin E3 ligases to different sites of ESCRT-dependent membrane fission are depicted schematically. The figure emphasizes how different retroviruses hijack the ESCRT pathway using late assembly domains within their Gag polyproteins (yellow background, Gag proteins in orange, with their constituent domains shown schematically), and how these interactions mimic analogous interactions between cellular adaptors (underlined) and ESCRT factors (white background). Polyubiquitin chains are depicted as connected yellow hexagons, solid arrows denote known protein-protein interactions, dashed arrows denote inferred or indirect protein-protein interactions, and question marks indicate that the site(s) of ubiquitin attachment are uncertain (see text). Abbreviations: HIV, Human Immunodeficiency Virus; RSV, Rous sarcoma virus; EIAV, Equine Infectious Anemia Virus; MVB, multivesicular body; ART, arrestin-related trafficking adaptor. For illustration purposes, we selected retroviruses that bud primarily through P(T/S)AP (HIV-1), PPXY (RSV) and YPXL (EIAV) late assembly domains, but note that these viruses all also use auxiliary late assembly domains, including an YPXL motif in RSV (not shown) (Dilley et al., 2010).
Since the initial discovery that HIV-1 usurps the ESCRT pathway to bud from the plasma membrane (Demirov et al., 2002; Garrus et al., 2001; Martin-Serrano et al., 2001; Patnaik et al., 2000; VerPlank et al., 2001), many other enveloped viruses have been shown to utilize this pathway (Supplemental Table 1). Moreover, even in uninfected cells, vesicles can bud directly from the plasma membrane (termed shedding microvesicles or ectosomes) (Akers et al., 2013; Nabhan et al., 2012). These different ESCRT-dependent processes share the same reverse topology and produce small vesicles of similar sizes (30–120 nm), but differ in the target membrane from which vesiculation occurs (endosome vs. plasma membrane), the site of vesicle release (lysosome vs. extracellular space), and/or the cargo (membrane proteins vs. exosomal cargoes vs. viral genomes).
In 2007, Martin-Serrano and colleagues demonstrated that the ESCRT pathway also severs the thin intercellular bridges that connect daughter cells during the final step of cell division (termed abscission) (Agromayor and Martin-Serrano, 2013; Carlton and Martin-Serrano, 2007; Fededa and Gerlich, 2012). This unexpected discovery emerged from their work on HIV budding, and beautifully illustrates how analyses of microbe-host interactions can illuminate important cell biology (and vice versa). Abscission appears to have been the primordial ESCRT pathway function because crenarchaeal organisms that lack internal membranes nevertheless use a primitive ESCRT pathway to divide (Samson and Bell, 2009). During abscission, the ESCRT pathway again mediates membrane fission from the cytoplasmic face of the intercellular bridge, but the membrane must be constricted over a much larger distance, starting from a diameter of ~1 μm in mammalian cells. Moreover, ESCRT pathway functions must be integrated with other complex mitotic processes, including cleavage furrow ingression, microtubule severing, chromosome segregation, and the abscission checkpoint (Agromayor and Martin- Serrano, 2013; Fededa and Gerlich, 2012; McCullough et al., 2013). The complexity of cytokinesis, together with the variety of different ESCRT pathway functions, seem to explain why the mammalian ESCRT pathway has more than 30 components, including multiple isoforms of nearly all core ESCRT factors.
2.2 ESCRT Pathway Components
Although the ESCRT pathway functions as an integrated membrane fission machinery, three broad classes of factors are recruited sequentially to perform distinct functions: 1) adaptor proteins define sites of ESCRT action at specific membranes (Figure 1); 2) early-acting factors initiate ESCRT assembly and stabilize membrane curvature; and 3) late-acting factors mediate membrane constriction and fission. Examples of the growing list of ESCRT adaptors include HRS/STAM (MVB vesicles), viral structural proteins such as retroviral Gag proteins, arenaviral Z proteins, and filoviral VP40 proteins (viruses), CEP55 (abscission), ARRDC1 (shedding microvesicles) and Syntenin/Syndecan (exosomes) (Figure 1). These adaptors localize to different membranes and membrane domains – often by recognizing specific phospholipids – where they concentrate vesicle cargoes and recruit the early-acting ESCRT factors. Two different classes of early-acting ESCRT factors have been identified: Bro1 domain proteins and ESCRT-I/-II complexes. Although distinct in architecture, these factors share several key mechanistic activities, including the ability to bind ubiquitin and recruit late-acting ESCRT-III subunits. ESCRT-III subunits then form filaments and recruit VPS4 ATPases, which together constrict membranes and mediate fission. The following sections summarize the structures and functions of core early- and late-acting ESCRT factors. We refer readers to other recent reviews for more comprehensive descriptions of the ESCRT machinery and full primary references (Agromayor and Martin-Serrano, 2013; Hanson and Cashikar, 2012; Henne et al., 2011; Hurley and Hanson, 2010; McCullough et al., 2013; Weissenhorn et al., 2013).
2.2.1 Bro1 Domain Proteins
ALIX is the founding member of a family of related Bro1 domain-containing mammalian proteins that also include HD-PTP, BROX, RHPN1 and RHPN2. ALIX is recruited by many viral structural proteins ((Strack et al., 2003) and see Supplemental Table 1), and also by CEP55 during cytokinesis (Carlton and Martin-Serrano, 2007) and by Syndecan/Syntenin during exosome formation (Baietti et al., 2012). These observations imply that ALIX can initiate ESCRT assembly. Recently, the yeast Bro1p homolog was shown to act early in the yeast MVB pathway, indicating that Bro1p is likely a true ALIX homolog (Pashkova et al., 2013). Bro1 domains bind and recruit downstream ESCRT-III subunits of the CHMP4/Snf7p and CHMP5 families. ALIX Bro1 also can bind membranes, particularly lysobisphosphatidic acid (LBPA) (Bissig et al., 2013), and may help stabilize (or drive) membrane curvature at bud necks owing to its crescent shape (Kim et al., 2005; Pires et al., 2009). The activities of different Bro1 family members can be modulated by a series of auxiliary domains and inputs, including: 1) calcium and calcium-responsive binding partners (ALIX) (Bissig et al., 2013; Okumura et al., 2013); 2) downstream V domains (in ALIX/Bro1p and HD-PTP) that can bind both ubiquitin (Dowlatshahi et al., 2012; Keren-Kaplan et al., 2013; Pashkova et al., 2013) and YPXL motifs (see below); 3) autoinhibitory elements and their activating partners (ALIX) (Zhai et al., 2011a; Zhou et al., 2010); and 4) post-translational modifications such as ubiquitylation (ALIX) and farnesylation (BROX).
2.2.2 ESCRT-I/-II
Mammals express a variety of heterotetrameric ESCRT-I complexes, each of which contains a single copy of the unique TSG101 subunit and single copies of one of the different isoforms of VPS28, VPS37 and MVB12/UBAP1 (McCullough et al., 2013). ESCRT-I, in turn, can bind the heterotetrameric ESCRT-II complex, which contains single copies of EAP45 and EAP30, and two copies of EAP20. The large, crescent-shaped ESCRT-I/-II supercomplex concentrates within the necks of budding vesicles (Boura et al., 2012; Hurley and Hanson, 2010; Wollert and Hurley, 2010) and contains a series of accessory domains and motifs that can bind adaptors, ubiquitin, membranes, and specific phosphatidyl inositides (Henne et al., 2011; Hurley and Hanson, 2010; McCullough et al., 2013). The two EAP20 subunits of ESCRT-II bind the ESCRT-III protein, CHMP6 (Im et al., 2009), which in turn binds CHMP4 (ESCRT-III) (Carlson and Hurley, 2012; Henne et al., 2012). Thus, both ALIX and ESCRT-I/-II ultimately function to recruit CHMP4 and promote ESCRT-III filament formation within the bud neck.
2.2.3 ESCRT-III Proteins
Mammals have twelve different ESCRT-III subunits that comprise eight subfamilies; designated CHMP1-7 and IST1, with three isoforms of CHMP4 (designated A-C) and two isoforms each of CHMP1 and CHMP2. The ESCRT-III core is an unusual four helix bundle in which the first two helices form a long antiparallel hairpin and the two shorter helices pack against the open end of the hairpin (Henne et al., 2011; Hurley and Hanson, 2010; McCullough et al., 2013). C-terminal tails of variable length and sequence fold back on the ESCRT-III core and function as autoinhibitory elements. When autoinhibition is relieved, ESCRT-III proteins can form homo- and heterooligomeric filaments that can further assemble into helices, both in vitro and in vivo. In several case, ESCRT-III helices have been shown to associate tightly with membranes and induce extrusion of membrane tubules from the plasma membrane (Bodon et al., 2011; Effantin et al., 2013; Hanson et al., 2008), indicating that ESCRT-III proteins likely also form helices or spirals within bud necks. The different ESCRT-III subunits engage in a hierarchy of protein-protein interactions in which their different core domains make preferential pairwise interactions (Babst et al., 2002; Carlson and Hurley, 2012), while their C-terminal tails bind VPS4 ATPases and other binding partners, particularly during abscission. The structures of ESCRT-III filaments are not well understood, but 3–4 nm filaments are common and paired CHMP4 filaments can form in vitro, consistent with the observation that each nucleating ESCRT-II complex contains two copies of the EAP20 ESCRT-III binding subunit (Henne et al., 2012). Similarly, ALIX can also dimerize and crosslink CHMP4 filaments (Pires et al., 2009), although the functional purpose (if any) of paired filament formation is not known.
2.2.4 VPS4 ATPases
As the only known enzymes in the ESCRT pathway, VPS4 ATPases power the pathway by converting the energy of ATP hydrolysis into mechanical work. Mammals express two closely related VPS4 proteins (A and B), whose functions are interchangeable, at least in some contexts. Like other AAA ATPases, VPS4 proteins function as hexameric rings whose assembly and ATPase activities are regulated by other ESCRT factors, particularly the stimulatory LIP5 cofactor (Hill and Babst, 2012). VPS4 enzymes use an unusual N-terminal three helix bundle, termed the MIT domain, to bind several different types of motifs within the exposed C-terminal tails of polymerized ESCRT-III substrates (Hill and Babst, 2012; McCullough et al., 2013). ESCRT-III subunits are then apparently “pulled” up into the central pore of the hexameric ring in an ATP-dependent manner (Hill and Babst, 2012), and this activity can depolymerize ESCRT-III filaments in vitro (Lata et al., 2008). ESCRT-III subunits are thereby released for multiple rounds of MVB vesicle formation, and there is some evidence that VPS4 remodeling of ESCRT-III filaments also may be required for membrane fission (Baumgartel et al., 2011; Elia et al., 2011; Jouvenet et al., 2011).
2.2.5 Models for Membrane Fission
A key unanswered question is how ESCRT-III filaments can drive membrane constriction and fission (Hanson and Cashikar, 2012; Henne et al., 2011; McCullough et al., 2013). Although a consensus has not been reached, mechanistic models envision that ESCRT-III filaments could form tapering spirals or whorls that pull the opposing membranes toward a central fission point (e.g., the “dome” (Fabrikant et al., 2009), “whorl” (Boura et al., 2012), and “hourglass” (Dobro et al., 2013) models) and/or that spiraling filaments could constrict membranes by sliding past themselves, perhaps with the assistance of VPS4 (e.g., the “break and slide” (Elia et al., 2011) and “purse string” (Saksena et al., 2009) models).
3 Lessons from Virology
Our understanding of ESCRT pathway functions and mechanisms has benefited greatly from the interplay between studies of cell biology and virology. Viral systems offer a series of useful experimental features, including: 1) ease of genetic manipulation and powerful experimental readouts, such as virion release and infectivity; and 2) plasma membrane assembly, which is particularly amenable for imaging studies. Furthermore, viruses may highlight essential core ESCRT activities because they appear to utilize a streamlined machinery that bypasses the elaborate regulatory controls required by cellular processes.
3.1 Viral Recruitment of the ESCRT Pathway
Virus budding is usually coupled tightly to virion assembly, and most viruses therefore use their structural proteins to recruit the ESCRT pathway. The first indication that virus budding might be mediated by host factors came from the work of Götlinger and colleagues, who showed that the p6 region of the HIV-1 Gag polyprotein, though not required for Gag polymerization, is required to detach the nascent virions from the plasma membrane (Gottlinger et al., 1991). Subsequent studies identified two different short peptide motifs within p6Gag that contributed to the efficiency of HIV-1 budding (Gottlinger et al., 1991; Huang et al., 1995; Strack et al., 2003). In parallel, others identified distinct short peptide motifs within the structural proteins of other viruses, and called these motifs “late assembly domains” because their mutation typically arrested virus assembly at a very late stage (Parent et al., 1995; Puffer et al., 1997). We now understand that different enveloped viruses use at least five distinct classes of late assembly domains, which can often function interchangeably. In several instances, the discovery of new viral late assembly domains has subsequently led to the identification of analogous motifs within cellular proteins that bind and recruit ESCRT factors, implying that the late assembly domains act by mimicking comparable cellular interactions (Figure 1 and Supplemental Table 1).
3.1.1 P(T/S)AP Late Assembly Domains Bind the TSG101 Subunit of ESCRT-I
The tetrapeptide late assembly domain motif P(T/S)AP (where the second amino acid can either be a Thr or a Ser) was first identified within HIV-1 p6Gag (Gottlinger et al., 1991; Huang et al., 1995), and was subsequently found within the structural proteins of filoviruses, arenaviruses, rhabdoviruses, and reoviruses (see Supplemental Table 1 for a full, referenced list). The P(T/S)AP late assembly domain functions by binding directly to the UEV domain of TSG101, thereby recruiting ESCRT-I to virus assembly sites (Demirov et al., 2002; Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001). UEV domains are structurally related to ubiquitin E2 conjugating enzymes, but lack the active site cysteine required for ubiquitin transfer. The P(T/S)AP peptides bind in a groove of the TSG101 UEV domain which, in functional E2 enzymes, is normally occluded by two terminal helices. Each of the four P(T/S)AP residues makes specific contacts within the UEV groove, which explains the high conservation of this late assembly domain sequence (Im et al., 2010).
P(T/S)AP motifs were subsequently identified in a series of cellular proteins that act as adaptors that recruit ESCRT pathway components, including the endosomal adaptors HRS/STAM, GGA3 and TOM1L1 (Hanson and Cashikar, 2012; McCullough et al., 2013) and the plasma membrane shedding microvesicle adaptor, ARRDC1 (Nabhan et al., 2012). P(T/S)AP motifs are also present in some membrane proteins, such as Gap-junction proteins, that appear to be downregulated through the ESCRT pathway (Gaietta et al., 2002).
3.1.2 YPXL Late Assembly Domains Bind ALIX
The YPXL late assembly domain was initially identified when it was shown that the p9Gag YPDL tetrapepide is required for budding of the Equine Infectious Anemia Virus (EIAV) (Figure 1) (Puffer et al., 1997). YPXL late assembly domains function by binding directly to the central V-domain of ALIX (Fisher et al., 2007; Lee et al., 2007; Strack et al., 2003; Usami et al., 2007; Zhai et al., 2008; Zhai et al., 2011b). YPXL late assembly domains have since been identified in other retroviruses, including HIV-1 and Murine Leukemia Virus (MLV), and ALIX now is known to facilitate release of paramyxoviruses, arenaviruses, flaviviruses, hepadnaviruses, herpesviruses, and tombusviruses (Supplemental Table 1).
Structural studies revealed that the ALIX binding motif is best described by the complex consensus sequence: ΦYX0/2(P/Φ)X0/3(L/I), (where Φ denotes a hydrophobic residue and X denotes amino acid spacers of different lengths) (Zhai et al., 2008; Zhai et al., 2011b). The anchoring tyrosine residue binds in a pocket in the second arm of the ALIX V domain, and the flanking hydrophobic residues bind in shallower pockets on the surface of arm 2. Remarkably, the variability in spacing between the conserved hydrophobic residues is accommodated by different intervening secondary structures, with extended, strand-like conformations used to span short spacers and α-helices used to span longer spacers.
The discoveries of ALIX-YPXL interactions in viruses and in PalA, which is a component of the ESCRT-dependent pH sensing system in Aspergillus (Vincent et al., 2003), contributed to the subsequent identification of analogous ALIX-recruiting motifs in other cellular proteins, which include the MVB/exosome adaptor, Syndecan/Syntenin (Baietti et al., 2012), as well as substrates of the MVB pathway, such as PAR1, a mammalian G protein-coupled receptor that is downregulated through the MVB pathway following activation (Dores et al., 2012).
3.1.3 PPXY Late Assembly Domains Bind NEDD4 Family Proteins
The PPXY late assembly domain motif (where X can be any amino acid but is most commonly proline) was first identified within the p2bGag region of Rous Sarcoma Virus (Figure 1) (Parent et al., 1995). PPXY late assembly domains also are present in a wide range of other retroviruses, as well as in rhabdoviruses, filoviruses, arenaviruses, reoviruses, and hepadnaviruses (Supplemental Table 1). These motifs function by binding the WW domains present in members of the NEDD4 family of HECT E3 ubiquitin ligases (Garnier et al., 1996; Macias et al., 2002). WW domains are three-stranded, antiparallel β-sheets, and the PPXY motif binds in a shallow groove on one side of the sheet. The first two prolines dock between conserved aromatic residues of the WW domain, and the tyrosine contacts residues from the second and third strands (Macias et al., 2002).
PPXY motifs were initially identified in cellular proteins that are binding partners and often ubiquitylation substrates of NEDD4 family members that are sorted through the MVB pathway (Rotin and Kumar, 2009). Ubiquitin transfer is also important for NEDD4 family-dependent virus budding (Chung et al., 2008; Martin-Serrano et al., 2005; Spidel et al., 2004; Usami et al., 2008; Weiss et al., 2010; Zhadina et al., 2007). The relevant substrates, ubiquitin sensor(s) and protein-protein interactions that connect NEDD4 family members and their substrates to the ESCRT pathway are not well understood, but there is evidence that arrestin-related trafficking adaptors (ARTs, especially ARRDC1-4) participate (Rauch and Martin-Serrano, 2011). This observation is consistent with known connections between ARTs, ESCRT factors, and the sole yeast NEDD4 homolog, Rsp5p (Lin et al., 2008), and with the observation that ARRDC1 also recruits NEDD4 family E3 ligases and ESCRT factors to release shedding vesicles from the plasma membrane (Figure 1) (Nabhan et al., 2012).
3.1.4 Ubiquitin-Dependent Recruitment of the ESCRT Pathway
Retroviral virions concentrate ubiquitin (Putterman et al., 1990), and ubiquitin appears to function at the budding step because: 1) ubiquitin depletion inhibits virus budding (Patnaik et al., 2000; Schubert et al., 2000; Strack et al., 2000); 2) covalent ubiquitin can sometimes function as a late assembly domain when fused directly to retroviral Gag proteins (Joshi et al., 2008; Patnaik et al., 2000); 3) the known early-acting mammalian ESCRT factors ALIX, ESCRT-I and ESCRT-II, all contain ubiquitin-binding domains (UBDs); and 4) genetic analyses indicate that ALIX binding to K63-linked ubiquitin chains enhances EIAV and HIV-1 budding (Dowlatshahi et al., 2012; Keren-Kaplan et al., 2013). In most cases, however, the key ubiquitin sensor(s) have not been identified, perhaps because ESCRT pathway UBDs function redundantly. Similarly, the functional target(s) of ubiquitylation have not been defined unambiguously, although the viral structural proteins themselves are leading candidates (Sette et al., 2013; Weiss et al., 2010), and budding efficiency correlates with formation of K63-linked poly-Ub chains on HIV-1 Gag (Weiss et al., 2010). Thus, ESCRT recruitment by viral structural proteins may be analogous to ESCRT recruitment by mono- and K63-linked poly-ubiquitylated membrane proteins that are being sorted into MVB vesicles (Shields and Piper, 2011). However, ubiquitin-dependent budding of the retrovirus Prototypic Foamy Virus does not require Gag ubiquitylation (Zhadina et al., 2007). In this and perhaps other cases, ubiquitylation of the ESCRT machinery itself may play a key functional role.
3.1.5 Additional Late Assembly Domains and Partners
A series of observations indicate that additional late assembly domains and/or their ESCRT binding partners remain to be identified and characterized: 1) “FPIV” late assembly domains (consensus sequence, ØPxV, where Ø is an aromatic residue) within the M proteins of the paramyxoviruses human Parainfluenzavirus Type 5 (hPIV-5) and Mumps stimulate ESCRT-dependent virus budding (Li et al., 2009; Schmitt et al., 2005); 2) AMOTL1 can also bind hPIV-5 M and facilitate virus release (Pei et al., 2010); 3) α-taxilin is required for efficient release of Hepatitis B (a hepadnavirus) and can associate with TSG101 (Hoffmann et al., 2013); 4) isolated Bro1 domains and Bro1 family members that lack YPXL binding activities can interact with retroviral NCGag domains (Bello et al., 2012; Dussupt et al., 2009; Popov et al., 2008, 2009); 5) in several cases, NEDD4 family members can stimulate release of retroviral Gag proteins that lack PPXY late assembly domains, implying novel interaction modes (Calistri et al., 2009; Chung et al., 2008; Jadwin et al., 2010; Usami et al., 2008; Weiss et al., 2010). This phenomenon is best characterized for HIV-1 where overexpression of NEDD4L can “rescue” the release and infectivity of viral constructs that lack TSG101 and ALIX binding sites (Chung et al., 2008; Usami et al., 2008; Weiss et al., 2010). This activity requires the NEDD4L ubiquitin E3 ligase activity and is not observed for other NEDD4 family members, implying specific connections between NEDD4L and the assembling virion; and 6) The actin remodeling protein IQGAP is required for efficient release of Ebola virus-like particles (Lu et al., 2013). IQGAP also can bind the structural proteins of Ebola and MLV and can bind TSG101 (Leung et al., 2006; Lu et al., 2013; Morita et al., 2007), although the functional relevance of these interactions remains to be established.
3.1.6 Viruses Frequently Use Multiple Late Assembly Domains
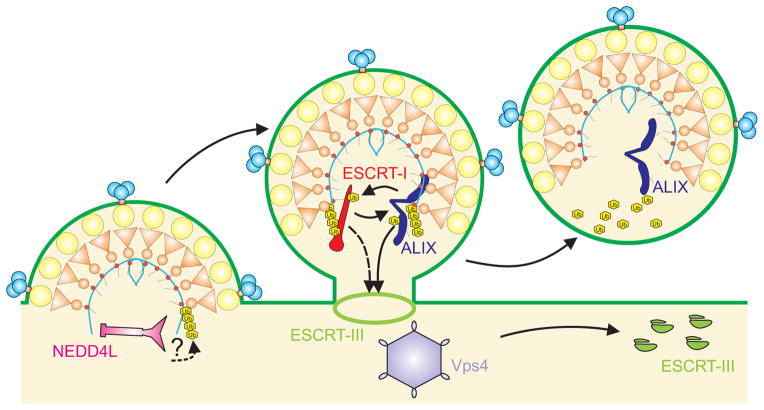
Most ESCRT-dependent viruses appear to enter the ESCRT pathway using multiple late assembly domains and binding partners (Supplemental Table 1). For example, HIV-1 recruits three different ESCRT-associated factors: TSG101/ESCRT-I, ALIX, and NEDD4L. The use of multiple late assembly domains may help extend viral tropism. For example, HIV-1 p6Gag P(T/S)AP-TSG101 is the dominant interaction for viral budding from transformed epithelial cell lines, but the p6Gag YPXL-ALIX interaction apparently can play a more important role in some T cells (Fujii et al., 2009). Different late assembly domain partners may also work together to provide temporal control or complementary budding activities. As illustrated in Figure 2, one possibility is that HIV-1 could initially engage NEDD4L to induce Gag ubiquitylation, which in turn could allow high affinity binding of TSG101 and ALIX because both of these early-acting factors can bind both HIV-1 Gag and ubiquitin, and can also bind one another. Thus, although late assembly domains and their ESCRT binding partners are often studied in isolation, they actually may form dynamic supercomplexes whose multiple components work together to orchestrate the stepwise transformations necessary to terminate virus assembly and initiate ESCRT factor recruitment and activation.
Figure 2. Multiple late assembly domains of HIV-1 Gag recruit different ESCRT-associated factors that may work together to facilitate virus budding.
The model suggests how three different early-acting ESCRT-associated factors recruited by HIV-1 Gag -- NEDD4L (pink), ESCRT-I (red) and dimeric ALIX (dark blue) -- could work together in a stepwise fashion to facilitate virus budding. The three regions of HIV-1 Gag are depicted in yellow (MA), orange (CA) and red (NC, bound to blue RNA), the subunits of the trimeric viral Env protein are depicted in blue (SU/gp120) and pink (TM/gp41), and ESCRT-III proteins (light green) are depicted schematically as either polymeric filaments (central panel, light green ring) or soluble, autoinhibited subunits (right panel, discrete subunits). ESCRT-I is missing from the final panel because the ultimate fate of this complex is not yet clear.
3.2 Late-Acting ESCRT Factors and Virus Budding
The late-acting ESCRT-III and VPS4 factors are recruited to viral budding sites immediately prior to virion release (Baumgartel et al., 2011; Jouvenet et al., 2011) and appear to catalyze membrane constriction and fission. Although the roles of the many different ESCRT-III proteins remain to be characterized in detail, siRNA depletion experiments indicate that of the eight different human ESCRT-III protein families, only CHMP4 and CHMP2 family members are absolutely critical for HIV and EIAV budding (Morita et al., 2011; Sandrin and Sundquist, 2013). Modest effects on HIV-1 budding were also seen upon depletion of CHMP1 and CHMP3 proteins, and there are other indications that both of these ESCRT-III families may participate in HIV-1 release (Carlson and Hurley, 2012; Effantin et al., 2013; Jouvenet et al., 2011). EIAV is the best understood and most streamlined model system for ESCRT-mediated membrane fission because unlike HIV-1, EIAV virus does not use ESCRT-I. Instead, the sole YPDL late assembly domain in Gag recruits ESCRT factors through a set of ordered, direct protein-protein interactions: Gag:ALIX:CHMP4:CHMP2:VPS4 (Figure 1) (Sandrin and Sundquist, 2013). Structural models are available for each of these protein-protein interactions, with the exception of the CHMP4:CHMP2 interaction which is not well understood.
In addition to their uniquely critical roles in HIV-1 and EIAV budding, there are several other indications that the CHMP4 and CHMP2 families play especially fundamental roles in membrane deformation and scission. For example, several simple eukaryotes like P. falciparum and E. histolytica have apparent CHMP4 and CHMP2 homologs, but lack other core ESCRT-III homologs (Leung et al., 2008). Furthermore, both CHMP2 and CHMP4 proteins can extrude helical tubes from the plasma membrane when overexpressed (Bodon et al., 2011; Hanson et al., 2008), whereas this activity has not been reported for other ESCRT-III proteins. CHMP4 subunits appear to constitute the major building block of ESCRT-III filaments (Teis et al., 2008) that may be capped (or copolymerize) with CHMP2, which in turn recruits VPS4. CHMP3 may help to stabilize or bridge CHMP2-CHMP4 interactions, particularly for the CHMP2A isoform (Carlson and Hurley, 2012; Effantin et al., 2013), and CHMP1 may arrive late and contribute to VPS4 recruitment (Carlson and Hurley, 2012).
4 Frontiers
4.1 ESCRT Networks and Other Host Factors
Although viruses have contributed substantially to our understanding of ESCRT pathway recruitment and function, important gaps in our knowledge remain to be filled. As discussed above, several late assembly domains and partners remain to be identified, the complete sequence of ESCRT protein-protein interactions required for viral budding through the P(T/S)AP, PPXY and FPIV late assembly domains remains to be determined, the role(s) of ubiquitin remains to be defined, and models for ESCRT-III filament structure and function need to be tested. Furthermore, although the membrane lipid compositions of several enveloped viruses are known, the role(s) of the lipid in virus budding is not well understood (Lorizate and Krausslich, 2011). Finally, it also will be important to determine the functions and ESCRT connections (if any) of the growing list of host factors implicated in enveloped virus trafficking, assembly, and polarized release, and to learn how the cell monitors and controls viral use of the ESCRT pathway (Grover et al., 2013; Kuang et al., 2011).
4.2 ESCRT-Independent Virus Budding
Not all enveloped viruses utilize the ESCRT pathway for egress, and the exceptions are interesting and informative (Weissenhorn et al., 2013). ESCRT independence is typically inferred from insensitivity to overexpression of a dominantly inhibitory VPS4 mutant that inhibits the ESCRT pathway. This is indeed strong evidence for a lack of ESCRT pathway involvement, although of course this conclusion will only be certain once the alternative budding mechanism is fully understood. One important ESCRT-independent class includes Semliki Forest Virus (a togavirus) (Taylor et al., 2007). Here, ESCRT independence is likely explained by the robust assembly of an external membrane-associated glycoprotein shell, which is apparently sufficient to drive membrane deformation and fission without the assistance of host factors (Cockburn et al., 2004; Jose et al., 2009; Taylor et al., 2007; Weissenhorn et al., 2013).
Another intriguing example is Influenza A Virus (IAV, an orthomyxovirus), which can bind ESCRT-I but appears to be ESCRT-independent as judged by a lack of susceptibility to VPS28 depletion or VPS4 inhibition (Bruce et al., 2009). Instead, IAV appears to encode its own membrane fission machinery within the viral M2 transmembrane protein (Rossman et al., 2010). Remarkably, helical amphipathic peptides corresponding to the M2 tails exhibit membrane fission activity in vitro, and it will be important to determine how this system works and why other viruses instead utilize the much more elaborate ESCRT machinery.
Respiratory Syncytial Virus (a paramyxovirus) is also insensitive to VPS4 inhibition (Utley et al., 2008), and Respiratory Syncytial Virus, IAV (Bruce et al., 2010) and Andes Virus (a bunyavirus) (Rowe et al., 2008) all require a functional Rab11 pathway for release. Rab11 depletion causes extruded Respiratory Syncytial Virus and IAV virions to accumulate at the plasma membrane, implying a budding defect. However, Rab11 typically functions in vesicle targeting and fusion at recycling endosomes, so it remains unclear whether Rab11 is required for proper trafficking of viral or cellular factors, or participates directly in viral membrane budding/fission reactions (and if so, how).
4.3 Non-Enveloped Viruses and Exosome Cargoes
Unlike enveloped viruses, non-enveloped viruses are typically released by cell lysis. However, “non-enveloped” viruses such as Bluetongue (a reovirus) and Hepatitis A (a picornavirus) can also recruit the ESCRT pathway and be released within membranes (Feng et al., 2013; Wirblich et al., 2006). Moreover, the viral RNAs from cells infected with Hepatitis C (a flavivirus) can be released within exosomes and subsequently trigger immune responses within nonpermissive plasmacytoid dendritic cells and establish infection (Dreux et al., 2012; Ramakrishnaiah et al., 2013). These phenomena have important implications for immune recognition and evasion, and blur the distinctions between enveloped and non-enveloped viruses, and exosomes (Gould et al., 2003).
4.4 ESCRT Requirements for Virus Entry
In addition to their importance in viral egress, ESCRT factors are also required for the entry of some rhabdoviruses, arenaviruses, flaviviruses, and baculoviruses (see Supplemental Table 1). These viruses enter cells via receptor-mediated endocytosis and are then released into the cytoplasm when they cross the limiting membrane of the late endosome. Their ESCRT dependence remains to be understood in mechanistic detail, but it is difficult to envision how the canonical membrane fission activity of the ESCRT pathway could contribute directly to membrane fusion. One intriguing alternative is that these viruses may fuse with intraluminal MVB vesicles (whose formation is ESCRT-dependent), and are then released into cytoplasm when the intraluminal vesicles backfuse with the limiting endosomal membrane (Le Blanc et al., 2005). It will therefore be critical to document and characterize this unusual backfusion reaction, which also has important implications for intercellular sharing of exosomal cargoes.
5. Summary
To spread infection, viruses must cross cellular membranes efficiently, and both entry and budding require complex interactions between host and viral factors. A key difference, however, is that while most enveloped viruses encode their own membrane fusion proteins to mediate entry, many viruses instead harness the cellular ESCRT machinery to effect membrane fission during egress. Relentless selective pressure has led enveloped viruses to mimic the remarkable variety of different cellular ESCRT recruitment mechanisms, and studies of virus budding will continue to inform ESCRT pathway functions in cellular vesiculation and abscission, and shed light on the continuum between biogenesis of viruses and extracellular vesicles.
Supplementary Material
Acknowledgments
We thank Leremy Colf, John McCullough and Matt Lalonde for helpful comments on the manuscript. This research was supported by NIH grants AI051174 and GM082545 (WIS) and the Deutsche Forschungsgemeinschaft (DFG) fellowship VO 1836/1-1 (JV). We apologize to our many colleagues whose primary literature references we were unable to cite owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.04.006. Epublication ahead of print. [DOI] [PubMed] [Google Scholar]
- Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Journal of neuro-oncology. 2013 doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann D, Estepa-Sabal E, Meerloo T, Emr S. Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- Bello NF, Dussupt V, Sette P, Rudd V, Nagashima K, Bibollet-Ruche F, Chen C, Montelaro RC, Hahn BH, Bouamr F. Budding of retroviruses utilizing divergent L domains requires nucleocapsid. J Virol. 2012;86:4182–4193. doi: 10.1128/JVI.07105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- Bissig C, Lenoir M, Velluz MC, Kufareva I, Abagyan R, Overduin M, Gruenberg J. Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev Cell. 2013;25:364–373. doi: 10.1016/j.devcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, Eaton WA, Hummer G, Hurley JH. Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure. 2012;20:874–886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Calistri A, Del Vecchio C, Salata C, Celestino M, Celegato M, Gottlinger H, Palu G, Parolin C. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J Cell Physiol. 2009;218:175–182. doi: 10.1002/jcp.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci USA. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn JJ, Abrescia NG, Grimes JM, Sutton GC, Diprose JM, Benevides JM, Thomas GJ, Jr, Bamford JK, Bamford DH, Stuart DI. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432:122–125. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley KA, Gregory D, Johnson MC, Vogt VM. An LYPSL late domain in the gag protein contributes to the efficient release and replication of Rous sarcoma virus. J Virol. 2010;84:6276–6287. doi: 10.1128/JVI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL, Bell SD, Jensen GJ. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–2327. doi: 10.1091/mbc.E12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic. 2007;8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol. 2012;197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR. ALIX Is a Lys63-Specific Polyubiquitin Binding Protein that Functions in Retrovirus Budding. Developmental cell. 2012;23:1247–1254. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS pathogens. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, Schoehn G, Weissenhorn W. ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding. Cellular microbiology. 2013;15:213–226. doi: 10.1111/cmi.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–447. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Garnier L, Wills JW, Verderame MF, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc Natl Acad Sci USA. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover JR, Llewellyn GN, Soheilian F, Nagashima K, Veatch SL, Ono A. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J Virol. 2013;87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. Multivesicular Body Morphogenesis. Annu Rev Cell Dev Biol. 2012;28:1–26. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Hill CP, Babst M. Structure and function of the membrane deformation AAA ATPase Vps4. Biochimica et Biophysica Acta. 2012;1823:172–181. doi: 10.1016/j.bbamcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J, Boehm C, Himmelsbach K, Donnerhak C, Roettger H, TSW, Ploen D, Hildt E. Identification of alpha-taxilin as an essential factor for the life cycle of Hepatitis B virus. Journal of hepatology. 2013 doi: 10.1016/j.jhep.2013.06.020. Epublication ahead of print. [DOI] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature reviews Molecular cell biology. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, Liu F, Burke TR, Jr, Bonifacino JS, Freed EO, Hurley JH. Crystallographic and functional analysis of the ESCRT-I /HIV-1 Gag PTAP interaction. Structure. 2010;18:1536–1547. doi: 10.1016/j.str.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, Hurley JH. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Developmental cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J Virol. 2010;84:704–715. doi: 10.1128/JVI.01319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009;4:837–856. doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008;9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Kaplan T, Attali I, Estrin M, Kuo LS, Farkash E, Jerabek-Willemsen M, Blutraich N, Artzi S, Peri A, Freed EO, et al. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. The EMBO journal. 2013;32:538–551. doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Seo EJ, Leis J. The Mechanism of Inhibition of Retrovirus Release from Cells by Interferon Induced Gene ISG15. Journal of Virology. 2011;85:7153–7161. doi: 10.1128/JVI.02610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I, Luyet PP, Pons V, Ferguson C, Emans N, Petiot A, Mayran N, Demaurex N, Faure J, Sadoul R, et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7:653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I, Prevost MC, Dokhelar MC, Rosenberg AR. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J Virol. 2002;76:10024–10029. doi: 10.1128/JVI.76.19.10024-10029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Yueh A, Appah FS, Jr, Yuan B, de los Santos K, Goff SP. Interaction of Moloney murine leukemia virus matrix protein with IQGAP. Embo J. 2006;25:2155–2166. doi: 10.1038/sj.emboj.7601097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol. 2009;83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Lorizate M, Krausslich HG. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011;3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Qu Y, Liu Y, Jambusaria R, Han Z, Ruthel G, Freedman BD, Harty RN. Host IQGAP1 and Ebola Virus VP40 Interactions Facilitate Virus-Like Particle Egress. J Virol. 2013;87:7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III Protein Requirements for HIV-1 Budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Katsuyama AM, Shibata H, Maki M. VPS37 Isoforms Differentially Modulate the Ternary Complex Formation of ALIX, ALG-2, and ESCRT-I. Bioscience, biotechnology, and biochemistry. 2013 doi: 10.1271/bbb.130280. [DOI] [PubMed] [Google Scholar]
- Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Sunshine AB, Rich M, Dunham MJ, Yu L, Piper RC. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev Cell. 2013;25:520–533. doi: 10.1016/j.devcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A, Chau V, Wills JW. Ubiquitin is part of the retrovirus budding machinery. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Bai Y, Schmitt AP. PIV5 M protein interaction with host protein angiomotin-like 1. Virology. 2010;397:155–166. doi: 10.1016/j.virol.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A Crescent-Shaped ALIX Dimer Targets ESCRT-III CHMP4 Filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83:7185–7193. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterman D, Pepinsky RB, Vogt VM. Ubiquitin in avian leukosis virus particles. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. Journal of virology. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Rowe RK, Suszko JW, Pekosz A. Roles for the recycling endosome, Rab8, and Rab11 in hantavirus release from epithelial cells. Virology. 2008;382:239–249. doi: 10.1016/j.virol.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17:507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Sandrin V, Sundquist WI. ESCRT requirements for EIAV budding. 2013. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Sette P, Nagashima K, Piper RC, Bouamr F. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology. 2013;10:79. doi: 10.1186/1742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Samson RY, Brumfield SK, Bell SD, Young MJ. Functional interplay between a virus and the ESCRT machinery in Archaea. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1301605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spidel JL, Craven RC, Wilson CB, Patnaik A, Wang H, Mansky LM, Wills JW. Lysines close to the Rous sarcoma virus late domain critical for budding. J Virol. 2004;78:10606–10616. doi: 10.1128/JVI.78.19.10606-10616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX Is a Binding Partner for HIV-1 p6 and EIAV p9 Functioning in Virus Budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb Perspect Med. 2012:2. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. Journal of virology. 2007;81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE., Jr Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci USA. 2008;105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Rainbow L, Tilburn J, Arst HN, Jr, Penalva MA. YPXL/I Is a Protein Interaction Motif Recognized by Aspergillus PalA and Its Human Homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Gottlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS pathogens. 2010:6. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Poudevigne E, Effantin G, Bassereau P. How to get out: ssRNA enveloped viruses and membrane fission. Curr Opin Virol. 2013;3:159–167. doi: 10.1016/j.coviro.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirblich C, Bhattacharya B, Roy P. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J Virol. 2006;80:460–473. doi: 10.1128/JVI.80.1.460-473.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci USA. 2007;104:20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. Activation of the retroviral budding factor ALIX. Journal of virology. 2011a;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol. 2011b;85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zang T, Wilson SJ, Johnson MC, Bieniasz PD. Clathrin facilitates the morphogenesis of retrovirus particles. PLoS Pathog. 2011;7:e1002119. doi: 10.1371/journal.ppat.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.