Abstract
The CCR5 chemokine receptor acts as a co-receptor for HIV-1 viral entry. Here we report the 2.7 Å resolution crystal structure of human CCR5 bound to the marketed HIV drug Maraviroc. The structure reveals a ligand binding site that is distinct from the proposed major recognition sites for chemokines and the viral glycoprotein gp120, providing insights into the mechanism of allosteric inhibition of chemokine signaling and viral entry. A comparison between CCR5 and CXCR4 crystal structures, along with models of co-receptor/gp120-V3 complexes, suggests that different charge distributions and steric hindrances caused by residue substitutions may be major determinants of HIV-1 co-receptor selectivity. These high-resolution insights into CCR5 can enable structure-based drug discovery for the treatment of HIV-1 infection.
Chemokine receptors and their peptidic ligands, chemokines, are the main organizers of leukocyte trafficking, and are validated therapeutic targets due to their involvement in many physiopathological disorders (1, 2). The chemokine receptor CCR5 binds and responds to four endogenous chemokine agonists, RANTES, MIP-1α, MIP-1β, and MCP-2 (3). CCR5 and another chemokine receptor CXCR4 are required for human immunodeficiency virus type 1 (HIV-1) infectivity, acting as co-receptors of the viral envelope glycoprotein gp120 (4). The structure of CXCR4 has been determined (5), but the details of chemokine recognition and viral infectivity remain poorly understood. HIV can infect a variety of CD4-expressing immune cells, and evolves inside the infected organism to encompass a wider range of susceptible cells by changing its co-receptor specificity, a phenomenon related to HIV tropism. HIV-1 strains using the co-receptor CCR5 are termed R5, while the strains using CXCR4 are termed X4, and those using either co-receptor are R5X4 (6). Intense research into the development of inhibitors capable of blocking HIV entry by targeting the co-receptor CCR5 has led to approval of the allosteric CCR5 inhibitor Maraviroc for treatment of HIV-1 infection (7–11).
Structural studies were carried out using an engineered human CCR5 construct, purified and crystallized in complex with Maraviroc (figure S1 and figure S2) (12). The crystal structure of the CCR5/Maraviroc complex was determined at 2.7 Å resolution with two complexes per asymmetric unit (ASU) (Fig. 1, figure S3, figure S4 and table S1). Molecule A will be used for discussion purposes.
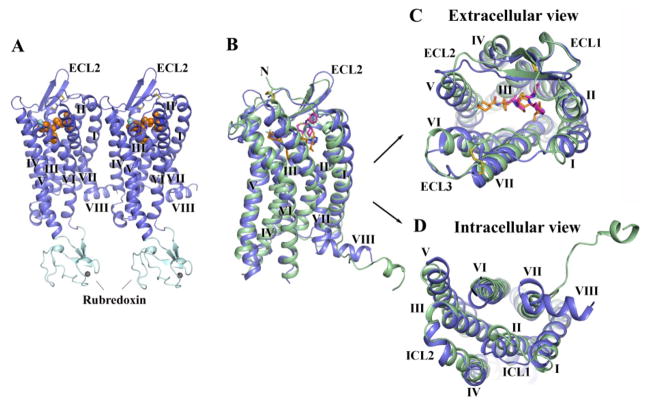
Fig. 1.
Overall fold of the CCR5/Maraviroc complex and comparison with CXCR4. (A) Overall structure of the two CCR5-rubredoxin molecules related by pseudo-translational symmetry in one ASU. The receptor is colored blue, and the rubredoxin is light-cyan. The ligand Maraviroc is shown in orange sphere representation. The disulfide bonds are shown as yellow sticks. Zinc ions are shown as grey spheres. (B–D) Structure comparison between CCR5 (blue) and CXCR4 (PDB ID: 3ODU, green). The ligands are shown in stick representation. Maraviroc in CCR5 and IT1t in CXCR4 have orange and magenta carbons, respectively. C, top view of the extracellular side of CCR5 and CXCR4; D, bottom view of the intracellular side of CCR5 and CXCR4.
The overall CCR5 fold shares a similar architecture with previously solved class A G protein-coupled receptor (GPCR) structures, containing seven transmembrane (7TM) α-helices (I to VII) connected by three extracellular loops (ECL1–3) and three intracellular loops (ICL1–3) (Fig. 1A). CCR5 is structurally similar to the chemokine receptor CXCR4 [Cα RMSD within the 7TM bundle between CCR5/Maraviroc and CXCR4/IT1t is 1.8 Å (sequence identity = 34%)]. The largest loop in CCR5, ECL2, forms a β-hairpin structure; the conformations of the N-terminal segment (residues 19–26) and ECLs are constrained by two disulfide bonds, one linking Cys1013.25 (13) with Cys178 of ECL2, and another one connecting Cys20 at the N-terminus with Cys2697.25 (Fig. 1, B and C). Despite the overall similarity, CCR5 and CXCR4 structures differ substantially in a number of regions. In the CXCR4 crystal structures, the C-terminus after helix VII adopted an extended disordered conformation (5), while in CCR5, a short α-helix VIII is observed. This difference could be explained by the presence of an α-helical sequence motif F(RK)xx(FL)xxx(LF) in CCR5’s helix VIII, while in CXCR4 this motif is partially modified (FKxxAxxxL) (5, 14). However, it cannot be excluded that this difference might also be due to different crystal packing interactions. Helix IV in CCR5 is tilted by about 15° with respect to the corresponding helix in CXCR4; its intracellular portion is 1.5 turns shorter than in CXCR4 and forms a classical α-helix in contrast with a distorted π-helix in CXCR4. ICL2, which is unstructured in CXCR4, contains a two-turn α-helix in CCR5 running parallel to the membrane (Fig. 1D). Phe135 and Ala136 in the α-helix of ICL2 form a hydrophobic cluster with Leu1283.53 and Ala1293.54 at the intracellular end of helix III to stabilize the conformation of ICL2.
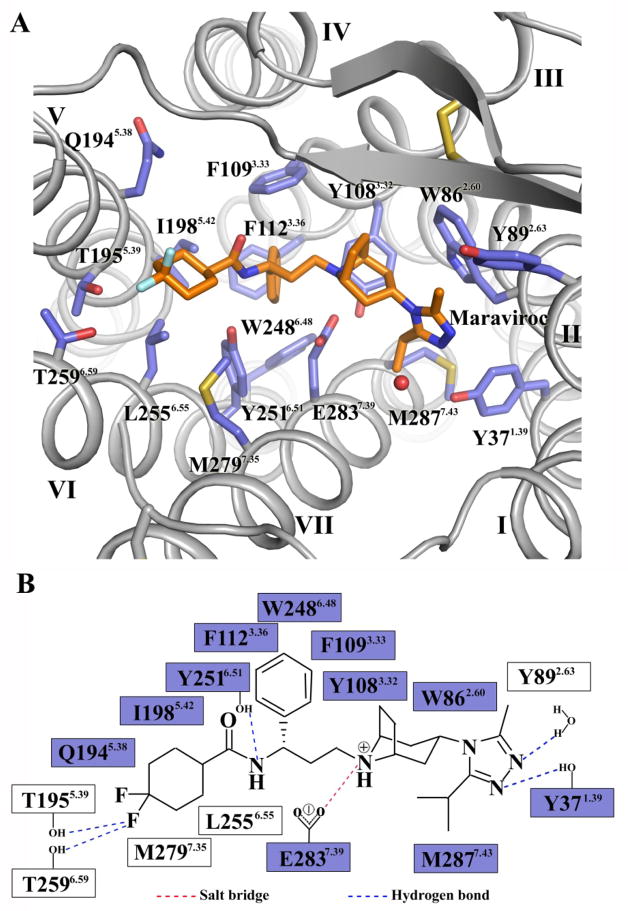
In the CCR5/Maraviroc structure, the ligand occupies the bottom of a pocket defined by residues from helices I, II, III, V, VI and VII (Fig. 2A and figure S5). The nitrogen of the tropane group is likely protonated and engaged in a salt-bridge interaction with Glu2837.39. The carboxamide nitrogen forms a hydrogen bond with Tyr2516.51. The length of the carbon chain between the above-mentioned two nitrogens was reported to be critical for the anti-HIV infection activity of the inhibitors (15), which correlates with the spatial locations of Glu2837.39 and Tyr2516.51. The amine moiety of the triazole group hydrogen bonds with Tyr371.39 and with a water molecule. Another two hydrogen bonds are formed by one of the fluorines in the cyclohexane ring with Thr1955.39 and Thr2596.59. In addition, the phenyl group reaches deep into the pocket to form hydrophobic interactions with five aromatic residues, Tyr1083.32, Phe1093.33, Phe1123.36, Trp2486.48 and Tyr2516.51. The triazole, tropane and cyclohexane groups also fit into small subpockets and make hydrophobic contacts with CCR5. The above interactions are supported by previous mutagenesis studies (Fig. 2B) (11, 16).
Fig. 2.
CCR5 ligand binding pocket for Maraviroc. (A) Key residues in CCR5 for Maraviroc binding. Maraviroc (orange carbons) and receptor residues (blue carbons) involved in ligand binding are shown in stick representation. Other elements are colored as follows: oxygen, red; nitrogen, dark blue; sulfur, yellow; fluorine, light-cyan. The water molecule involved in interacting with Maraviroc is shown as a red sphere. (B) Schematic representation of interactions between CCR5 and Maraviroc. Mutations reported to be critical for Maraviroc binding are indicated with blue squares (11, 16).
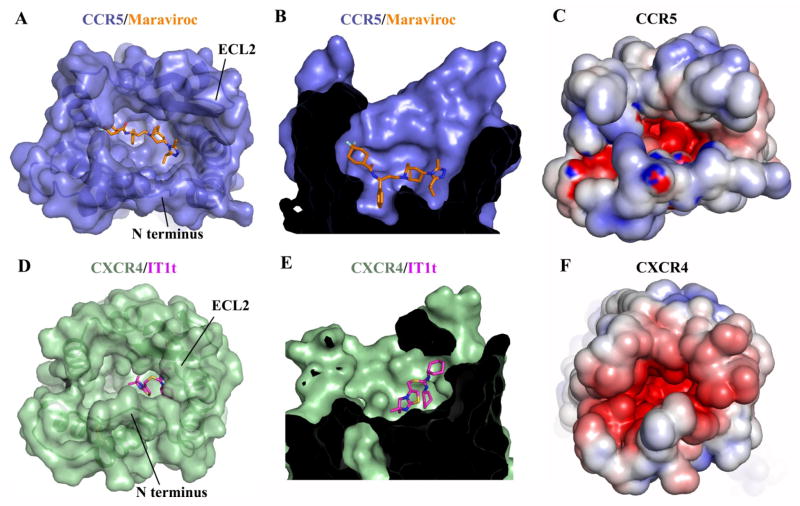
Compared to the CXCR4/IT1t structure, the binding site for Maraviroc in CCR5 is deeper, and occupies a larger area of the pocket, making no contacts with the ECLs (Fig. 3, A, B, D, and E). In CCR5, the extracellular end of helix VII shifts away from the central axis of the receptor by ~3 Å compared with CXCR4, leading to a corresponding shift of CCR5’s N-terminal fragment connected to helix VII by the disulfide bond Cys20-Cys2697.25. Also in CXCR4, Asp972.63 and Arg183 in ECL2 form a salt bridge, which is absent in CCR5 due to substitutions to Tyr892.63 and His175, resulting in a 6 Å shift at the β-hairpin tip of ECL2 of CXCR4 toward the ligand binding pocket compared with CCR5. As a consequence, the entrance to the CXCR4 ligand binding pocket is partially covered by its N-terminus and ECL2, while the CCR5 ligand binding pocket is more open.
Fig. 3.
Comparison of the ligand binding pockets between CCR5/Maraviroc and CXCR4/IT1t. (A, D) Top views of the ligand binding pockets in CCR5 (A, blue) and CXCR4 (D, green), showing a more open ligand binding pocket in CCR5. The receptors are shown in both cartoon and molecular surface representations. The ligands are shown in stick representation. Maraviroc in CCR5 and IT1t in CXCR4 have orange and magenta carbons, respectively. (B, E) Side views of the ligand binding pockets in CCR5 (B) and CXCR4 (E), showing that Maraviroc binds deeper in CCR5 than IT1t in CXCR4. (C, F) Top views of the ligand binding pockets in CCR5 (C) and CXCR4 (F). Both CCR5 and CXCR4 surfaces are colored according to their electrostatic potential from red (negative) to blue (positive), showing different charge distribution within the ligand binding pockets of these two receptors.
Maraviroc has been characterized as an inverse agonist of CCR5 (17), suggesting that Maraviroc stabilizes CCR5 in an inactive conformation. The major evidence for an inactive state of the CCR5/Maraviroc structure is the conformation of the highly conserved class A GPCR residues Trp2486.48 and Tyr2446.44, which are involved in relaying ligand-stabilized conformational changes in the binding pocket into conformational changes in the cytoplasmic domain. In the CCR5/Maraviroc structure, these residues are in similar conformations to those observed in other inactive structures and distinct from their active state conformations (18, 19). Moreover, the phenyl group of Maraviroc forms a hydrophobic interaction with Trp2486.48, preventing its activation-related motion. The inactive state of the CCR5/Maraviroc structure is also manifested by the close packing of helix VI with other helices of the 7TM bundle at the intracellular side of receptor that precludes G-protein binding. The inactive conformation of helix VI can be stabilized by an “ionic lock” formed by a salt bridge interaction between the conserved Arg3.50 in helix III and Asp/Glu6.30 at the intracellular end of helix VI (20, 21). Although wild-type CCR5 lacks any acidic residue at position 6.30, the thermostabilizing mutation Ala2336.33 Asp makes a salt-bridge contact with Arg3.50, potentially locking the receptor in an inactive conformation (figure S2).
Mutagenesis and biochemical studies suggest that Maraviroc and some other small-molecule CCR5 inhibitors are allosteric modulators (9–11, 16, 17). The N-terminal region of CCR5, together with ECL2, have been identified as the major binding determinants for its chemokine ligands (22). In the so-called “two-site” model, this region is viewed as a chemokine recognition site (Site 1), which interacts with the chemokine core (16). Several residues in the 7TM region were found to be important for CCR5 activation upon binding to the chemokine ligand, such as Tyr371.39 and Trp2486.48 (11), which are involved in Maraviroc binding. This region is considered as an activation site (Site 2), which interacts with the flexible N-terminus of the chemokine (23). In the CCR5 structure, Maraviroc is buried in a cavity within the 7TM domain, which is distinct from Site 1 of chemokine recognition, but potentially overlaps with Site 2. Thus, the CCR5/Maraviroc structure indicates that Maraviroc most likely inhibits chemokine function by blocking receptor activation through interactions with Site 2, which explains the allosteric inhibition of chemokine signaling by Maraviroc. It has also been reported that CCR5’s N-terminus and ECL2 play an important role in gp120 binding and HIV-1 infection (11, 24, 25), suggesting that Maraviroc interferes with the effects of gp120 binding in a similar allosteric fashion. It should be noted that the above conclusion does not rule out the possibility that Maraviroc may reduce chemokine and gp120 binding in an allosteric inverse agonism manner by stabilizing CCR5 in an inactive conformation. Thus, Maraviroc may alter chemokine signaling and gp120 binding by allosterically blocking agonist recognition and/or inhibiting receptor activation.
The third variable region, V3 loop, of gp120 forms a β-hairpin structure, and has been identified as the major determinant of cellular tropism and co-receptor specificity (24, 26, 27). The stem region of the V3 loop has been reported to be responsible for gp120 binding to the co-receptor’s N-terminus, while the V3 crown interacts with the co-receptor’s ECL2 and residues inside the ligand binding pocket (24, 25, 28). Sequence analysis of the V3 region shows that it is more positively charged in the X4-tropic viruses than the R5-tropic viruses (29, 30). Several acidic residues in CXCR4, Asp972.63, Asp1714.60, Asp187 (ECL2), Asp1935.32 and Asp2626.58, are key for ligand binding in the CXCR4 structures (5), and have been reported to be critical for HIV-1 infectivity (28). In CCR5, these acidic residues are substituted by uncharged residues, Tyr892.63, Gly1634.60, Ser179, Gln1885.32 and Asn2586.58, respectively. Additionally, the N-terminus of CXCR4 contains nine acidic residues, while CCR5 only has three (Fig. 3, C, F and figure S6A). These differences may correlate with the different charge distribution in the V3 loops of X4- and R5-tropic viruses. It was reported that the binding of gp120 to CCR5 was sensitive to mutations of some uncharged residues of CCR5, such as Trp862.60, Trp94, Tyr1083.32, Trp2486.48 and Tyr2516.51 etc. (11), providing additional evidence for the importance of the net charge in the V3 loop for co-receptor selectivity. These residues form a cluster within the CCR5 ligand binding pocket, which composes a potential binding site for gp120 (figure S7).
To further understand the mechanism of co-receptor selectivity, we have built models of CCR5/R5-V3 and CXCR4/X4-V3 complexes based on the CCR5 structure and previous studies (5, 25, 30) (figure S6B). The models suggest that the different charge distributions in the co-receptor ligand binding pockets and steric hindrances caused by residue substitutions may be major determinants of HIV-1 co-receptor selectivity (figure S8) (12). These models initiate our understanding of HIV-1 tropism in a structural perspective; however, additional structures of the co-receptors in an apo state or complemented with the same or similar allosteric or orthosteric ligands, and complexes between the co-receptors and gp120-CD4, are needed to fully understand the mechanisms of HIV-1 tropism.
Supplementary Material
Acknowledgments
The authors thank I. Wilson, V. Katritch and T. Handel for careful review and scientific feedback on the manuscript, A. Walker for assistance with manuscript preparation, C. Wang and D. Wacker for help on collection of X-ray diffraction data, and E. Kellenberger for providing CCR5 model. Atomic coordinates and structure facors have been deposited in the Protein Data Bank with identification code 4MBS. This work was supported by “National Basic Research Program of China” grants 2012CB518000 and 2012CB910400, National Institutes of Health R01 AI100604, National Science Foundation of China grants 31270766, 31170683 and 81025017, and Shanghai Science and Technology Committee grants 11JC1414800 and 12PJ1410500. Additionally, VC and RCS acknowledge support from National Institutes of Health U54 GM094618 (Target GPCR-28); IK acknowledges support from U01 GM094612, U54 GM094618 and R01 GM071872.
Footnotes
Materials and Methods
The data presented in this paper are tabulated in the main paper and in the supplementary materials.
References and Notes
- 1.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 2.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, et al. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899. [PubMed] [Google Scholar]
- 4.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 5.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV. Curr Opin Infect Dis. 2005;18:9. doi: 10.1097/00001432-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Palani A, Tagat JR. Discovery and development of small-molecule chemokine coreceptor CCR5 antagonists. J Med Chem. 2006;49:2851. doi: 10.1021/jm060009x. [DOI] [PubMed] [Google Scholar]
- 8.FDA notifications. Maraviroc approved as a CCR5 co-receptor antagonist. AIDS Alert. 2007;22:103. [PubMed] [Google Scholar]
- 9.Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharmacol. 2005;67:1268. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- 10.Muniz-Medina VM, et al. The relative activity of “function sparing” HIV-1 entry inhibitors on viral entry and CCR5 internalization: is allosteric functional selectivity a valuable therapeutic property? Mol Pharmacol. 2009;75:490. doi: 10.1124/mol.108.052555. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Perez J, et al. Allosteric model of maraviroc binding to CC chemokine receptor 5 (CCR5) J Biol Chem. 2011;286:33409. doi: 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Materials, methods and discussion are available as supplementary materials on Science Online.
- 13.In Ballesteros-Weinstein numbering, a single most-conserved residue among the class A GPCRs is designated x.50, where x is the transmembrane helix number. All other residues on the helix are numbered relative to this conserved position.
- 14.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr; and x, any amino acid.
- 15.Imamura S, et al. Discovery of a piperidine-4-carboxamide CCR5 antagonist (TAK-220) with highly potent Anti-HIV-1 activity. J Med Chem. 2006;49:2784. doi: 10.1021/jm051034q. [DOI] [PubMed] [Google Scholar]
- 16.Scholten DJ, et al. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2012;165:1617. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Perez J, et al. New insights into the mechanisms whereby low molecular weight CCR5 ligands inhibit HIV-1 infection. J Biol Chem. 2011;286:4978. doi: 10.1074/jbc.M110.168955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2012;477:549. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 22.Duma L, Haussinger D, Rogowski M, Lusso P, Grzesiek S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J Mol Biol. 2007;365:1063. doi: 10.1016/j.jmb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 23.Blanpain C, et al. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem. 2003;278:5179. doi: 10.1074/jbc.M205684200. [DOI] [PubMed] [Google Scholar]
- 24.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biscone MJ, et al. Functional impact of HIV coreceptor-binding site mutations. Virology. 2006;351:226. doi: 10.1016/j.virol.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 26.Speck RF, et al. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80:6093. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 29.Cardozo T, et al. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23:415. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- 30.Xiang SH, Pacheco B, Bowder D, Yuan W, Sodroski J. Characterization of a dual-tropic human immunodeficiency virus (HIV-1) strain derived from the prototypical X4 isolate HXBc2. Virology. 2013;438:5. doi: 10.1016/j.virol.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haycock-Lewandowski SJ, Wilder A, Ahman J. Development of a Bulk Enabling Route to Maraviroc (UK-427,857), a CCR-5 Receptor Antagonist. Org Process Res Dev. 2008;12:1094. [Google Scholar]
- 33.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.C. BUSTER v. 2.8.0. Global Phasing Ltd; U. K: 2009. [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 40.Basmaciogullari S, Babcock GJ, Van Ryk D, Wojtowicz W, Sodroski J. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J Virol. 2002;76:10791. doi: 10.1128/JVI.76.21.10791-10800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.