Abstract
This paper describes the capabilities of DISCO, an extensible approach that supports integrative Web-based information dissemination. DISCO is a component of the Neuroscience Information Framework (NIF), an NIH Neuroscience Blueprint initiative that facilitates integrated access to diverse neuroscience resources via the Internet. DISCO facilitates the automated maintenance of several distinct capabilities using a collection of files 1) that are maintained locally by the developers of participating neuroscience resources and 2) that are “harvested” on a regular basis by a central DISCO server. This approach allows central NIF capabilities to be updated as each resource’s content changes over time. DISCO currently supports the following capabilities: 1) resource descriptions, 2) “LinkOut” to a resource’s data items from NCBI Entrez resources such as PubMed, 3) Web-based interoperation with a resource, 4) sharing a resource’s lexicon and ontology, 5) sharing a resource’s database schema, and 6) participation by the resource in neuroscience-related RSS news dissemination. The developers of a resource are free to choose which DISCO capabilities their resource will participate in. Although DISCO is used by NIF to facilitate neuroscience data integration, its capabilities have general applicability to other areas of research.
Keywords: Data integration, database federation, database interoperation, neuroinformatics
Introduction
DISCO is a Web-based discovery and data integration framework that is a component of the Neuroscience Information Framework (NIF), an NIH Neuroscience Blueprint initiative. The goal of the NIF as a whole is to provide a broad range of capabilities to facilitate the integrated access to diverse neuroscience resources (databases, Web sites, and other online resources) via the Internet (http://neuinfo.org, Gardner et al., 2008; Gupta et al., 2008). DISCO provides an extensible framework to facilitate the automated maintenance of several distinct integrative capabilities. The development of these DISCO capabilities was initially guided by the Interoperability Subcommittee of the Society for Neuroscience’s Neuroinformatics Committee (http://www.sfn.org/index.aspx?pagename=committee_NIC), and is currently driven by the needs of the NIF.
There is a rapidly growing set of neuroscience resources available via the Web. These undergo continual changes as new resources appear, as old resources are phased out, and as the content of existing resources evolve over time. DISCO is designed to assist in the automated updating of a spectrum of Web-based capabilities to help deal with these continual changes. For example, when a resource changes the scope of its contents, the resource developers can make corresponding changes to a local DISCO file describing their resource. The information in this file is then “harvested” by a central DISCO server on a regular basis and incorporated into the NIF Registry entry describing the resource.
The term DISCO (DISCOvery) is inspired by the UDDI (Universal Description, Discovery and Integration) concept. The current NIF DISCO implementation is oriented towards facilitating resource services discovery and integration via the NIF by automating the updating of the information that drives the NIF registration, information retrieval, and information sharing processes.
The broad goals of DISCO are: 1) to support a range of integrative capabilities designed to enhance the utility of the evolving set of Web-based resources to NIF users, 2) to assist resource developers in maintaining those capabilities as the various resources evolve over time, and 3) to help add power and robustness to broad integrative efforts such as the NIF.
The paper describes DISCO capabilities currently in use by the NIF, and plans to incorporate other capabilities in the future.
Background
Neuroscience research data are characterized by a high level of complexity and heterogeneity. These data are generated by research in many domains (e.g., genetics and genomics, physiology, pharmacology, synaptic, neuronal, circuit, and brain pathway function, 3D and 4D imaging of whole brains and of cells, and behavior). Many different types of data are generated. These data are increasingly accessible via the Internet. A number of approaches have been explored to facilitate the integrated discovery of, and access to, this diverse information.
Powerful search engines, such as Google, are used as primary tools for researchers to find information on the Internet. These systems have significant limitations, however, in their ability to find certain types of information, to interrelate concepts in found in different resources, and to integrate the results of searching multiple resources. A major limitation is the fact that much data is buried in the “hidden Web,” stored in databases whose contents are not accessible to Web crawlers.
A different approach involves the use of catalogs, as seen in the Neuroscience Database Gateway (NDG), developed for the Society for Neuroscience (NDG, 2008). The NDG provides a database of information about neuroscience resources that allows the user to search for resources on a given neuroscience topic, to read summary information about each resource, and to link directly to each resource for further information.
The NIF expands upon this general approach and includes 1) a Resource Registry (similar to the NDG), 2) a Database Mediator to allow integrated retrieval directly from within selected resources, 3) a Text Archive for retrieval of neuroscience publications, 4) terminology services to assist the integration of information from different resources, and 5) other related capabilities.
Evolution of the DISCO Approach
The DISCO approach evolved from early work in the Yale SenseLab project (Shepherd et al., 1997, Miller et al., 2001). In an attempt to facilitate external interoperation from SenseLab databases, researchers at Yale created the EAV/CR Dataset Protocol (EDSP), an XML metadata-driven mechanism used to share database information at various integrative levels, including database description, schema, data, metadata, and terminology (Marenco et al., 2003). The goal was to allow external applications to interact with SenseLab databases in as flexible a fashion as possible, and to explore different approaches to achieving robust integration. Once these capabilities were developed, there was a clear need for a broadcasting mechanism to help disseminate and maintain them. Although a variety of standard formats existed to implement this general type of mechanism, none provided the flexibility we felt was needed to support these capabilities robustly. As a result, the DISCO approach was created.
Once the DISCO capabilities were piloted in the context of SenseLab, they were first deployed broadly in the NDG and subsequently in the NIF. All the DISCO capabilities described in this paper are operational in one or more of these contexts (SenseLab, NDG, and/or the NIF).
Biositemaps
Biositemaps is an initiative under development that has certain long-term goals that overlap with those of DISCO. Biositemaps is being developed by the National Centers for Biomedical Computing (http://biositemaps.ncbcs.org) and aims to help in locating, querying, integrating, and mining biomedical resources on the Internet. Its current implementation allows the population of a registry of resources using a limited number of resource descriptors. As the DISCO and Biositemaps projects evolve over time, we plan to harmonize the two approaches to the extent possible. For example, DISCO is currently able to accept resource descriptions expressed in either the DISCO or Biositemaps formats.
The DISCO Approach
Figure 1 provides a schematic overview of the DISCO approach. DISCO involves a collection of files that reside on each participating resource. The files are maintained locally by the resource developers and “harvested” by a central DISCO server on a regular basis. In this way, central NIF capabilities can be updated in a timely fashion as the various local resources evolve/change over time. DISCO supports several distinct capabilities and the developers of a resource are free to participate in whichever DISCO capabilities they chose. In the remainder of this section we describe each of the capabilities currently provided by DISCO for the NIF, as well as certain capabilities that might be included in the future. Additional technical details can be found at http://disco.neuinfo.org.
Figure 1.
A schematic overview of the DISCO operational framework. (1) The DISCO server updates its registry of participating Web resources from the NIF Registry. (2) All resources are checked for their DISCO files. (3) When found, each file is processed by its corresponding DISCO capability engine, and its content is synchronized with the associated NIF component. Regular users of NIF and the Entrez systems use the information provided by the DISCO server to find the information from the collection of participating Web resources. (In addition to supporting this operational framework, DISCO also provides tools to allow resource staff to create and update their DISCO files, and to help NIF staff coordinate the various DISCO-enabled activities.)
The DISCO Main File
Each participating resource must maintain a file named “disco.xml” in the root directory of that resource. For example, Figure 2 shows the main DISCO file from the NeuronDB database (http://senselab.med.yale.edu/NeuronDB). The file is written in a simple XML format, providing one section for basic technical information about the resource (e.g., location URL, technical contact email address), and another section describing the various DISCO capabilities in use by the resource. Each capability is described using XML to indicate its service type, URL, and format. DISCO also allows using more than one type of format to describe any capability. Thus a resource’s DISCO main file might point to two resource descriptions, one in NIF format and the other in Biositemaps format. The functionality of the main DISCO file is similar to that of the Robots exclusion standard or “robots.txt protocol” file (http://en.wikipedia.org/wiki/Robots_exclusion_standard) used by search engines to determine what content should be indexed in a Web site.
Figure 2.

The DISCO main file for NeuronDB (http://senselab.med.yale.edu/neurondb/disco.xml) contains several “service” nodes, corresponding to the different DISCO capabilities in which NeuronDB is participating. Each node has a type describing the capability, a format and a URL pointer to the DISCO XML file corresponding to that specific capability. A single type of DISCO capability can be described in more than one format (as shown in this figure for the “Resource Description” and “Interoperation” service nodes).
The Resource Description Capability
The DISCO resource description capability is used to help keep the NIF Registry up-to-date as the resources described change in their scope and contents over time. This capability allows resource developers to create and maintain information describing their resource in the format required by the NIF Registry. During the evolution of DISCO, several Resource Description formats have been defined or adopted: one for the Neuroscience Database Gateway, one based on BrainML (http://brainml.org) for the first version of NIF, and one for the current version of NIF.
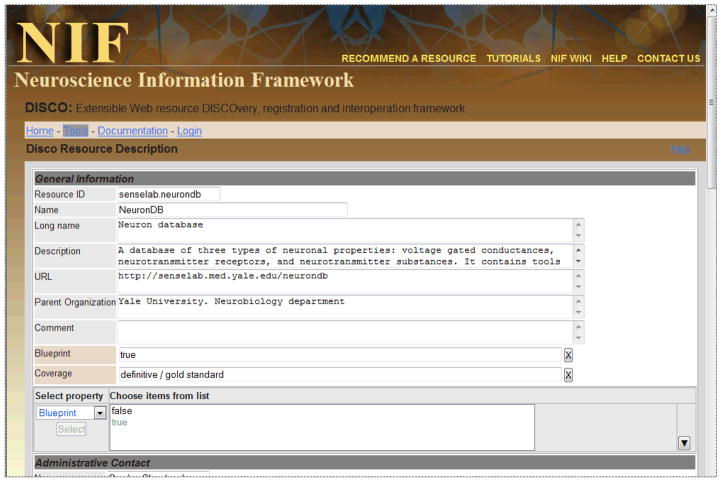
Figure 3 shows the Web-based DISCO resource description authoring tool currently operational on the NIF DISCO server. This tool can be used by a resource developer to create and edit the DISCO description of a resource. Once the information has been entered, the tool creates a resource description file expressed in XML. This file is placed locally on the resource (in a URL pointed to by the resource’s DISCO main file) to allow it to be “harvested” and used to populate the NIF Registry, where it is integrated with similar information gathered from other resources. In addition to the DISCO resource description format, the NIF DISCO server can also accept Biositemap files. These files are authored using the Biositemap editor and placed in an URL that is listed in the resource’s main DISCO file.
Figure 3.
The resource description authoring tool on the DISCO server that allows a resource developer to create and edit the DISCO resource description for a resource.
The LinkOut Capability
The National Center for Biotechnology Information (NCBI) has implemented a capability called “LinkOut” that allows users of NCBI Entrez (who might for example be looking at an article in PubMed) to link to related information in resources external to the NCBI (NCBI, 2008). To allow this to happen, the NCBI needs to maintain an up-to-date listing of such links.
The DISCO LinkOut capability, initially described in (Marenco et al., 2008), is designed to allow NIF resources to participate in this NCBI LinkOut process. It provides a mechanism to collect resource’s data links related to elements in the NCBI databases, and forward those links to NCBI for incorporation into its Entrez LinkOut system. NCBI database users will then find these links when using the NCBI’s LinkOut feature. For example, Figure 4 shows an NCBI page implementing the ability to “LinkOut” from a selected Entrez entity (in this case, a PubMed article) to related information in external databases. Seven NIF databases currently use this functionality, providing items to which an NCBI user can link out to. To participate in this DISCO capability, a resource needs to identify the URLs of items in its database (or Web site) that relate to NCBI entities, along with related information, and place this information into a file using the XML-based DISCO LinkOut format (Marenco et al., 2008). Figure 5 shows a portion of the Cell Centered Database’s (CCDB; http://ccdb.ucsd.edu) DISCO LinkOut file. Entries in this file specify Entrez database IDs (e.g.: PubMed IDs) and related URLs of entries in CCDB’s database.
Figure 4.
This figure shows how the NCBI Entrez system uses LinkOut information provided by three neuroscience resources via the NIF DISCO LinkOut capability. The left screen shows Entrez’s “LinkOut - more resources” information (expanded) for a PubMed paper. Down at the bottom of that page, under “Miscellaneous”, are three links provided by the NIF. To the right are three data-pages (from ModelDB, NeuronDB, and Neuromorpho.org respectively) generated by clicking on those URLs, all of which relate to the PubMed paper.
Figure 5.
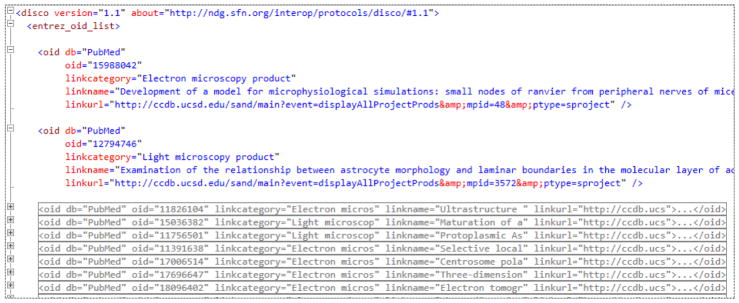
This figure shows a portion of CCDB’s DISCO LinkOut XML file. The first “oid” node shows how a specific PubMed ID (15988042) is linked to a specific CCDB URL (http://ccdb.ucsd.edu/sand/main?event=displayAllProjectProds&mpid=48&ptype=sproject).
Web Interoperation Capability
A major goal of the NIF is to allow flexible interoperation and integrated querying of the data stored in NIF resources. When a resource is a database with a direct API interface, this can be accomplished by SQL querying of that database. Not all resources, however, have an API interface.
The Web Interoperation capability is designed to provide a data integration capability for resources that 1) do not have a direct API interface for their back-end database, or 2) do not have a back-end database at all. (For example, a resource might just be implemented as a collection of Web pages.). Using the DISCO Web Interoperation capability, such resources can still be integrated with the NIF federated integration system which allows a NIF user to specify a single query that is automatically transformed to allow retrieval of information from multiple relevant databases.
The DISCO Web Interoperation capability can only be implemented for resources that provide a mechanism to query data that is presented in a consistently structured format. The information can be gathered by following URLs to static or dynamic pages, or by posting queries to Web interfaces to retrieve data. The goal is to allow data from these resources to be integrated with the NIF Mediator system as if the Mediator was directly connected to a database containing that information.
The data gathering process can be quite simple: for example, traversing well-structured Web pages displaying organized sets of data. The process can also be quite complex, for example, using a list of the values needed to fill-in the parameters in a Web form, launching a query with one or more of those values, and then parsing the results retrieved to obtain the desired data element(s). The broad process of providing such Web interoperation is a complex challenge, and we have identified a number of distinct scenarios that have been successfully implemented and others that are currently being explored. Those scenarios will be described later in this section.
One by-product of this work may be the development of guidelines as to how a Web interface might best be designed to facilitate this type of integration. Figure 6 shows the content of the DISCO interoperability file created to extract Disorders information from the National Institute of Neurological Disorders and Stroke (NINDS) Web site. This file uses the “disco.interop” format, an XML format that has been implemented to provide the logic necessary to perform this type of capability. This file contains information about a resource’s Web documents or interfaces and allows scripted navigation, parsing, querying and extraction of content using XML or regular expression queries. Figure 7 shows the DISCO Web Interoperability Control Panel with the NINDS script executed and the data retrieved. Figure 8 shows how that information is presented by the NIF integrated search interface when searching for “Ataxia”.
Figure 6.
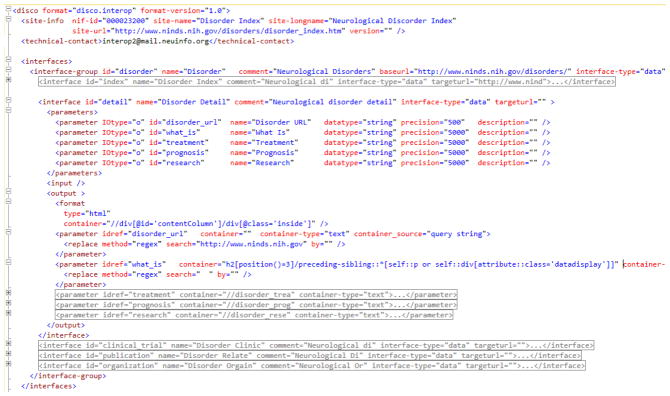
This screen shows a portion of the DISCO Web Interoperation file created to extract neurological disorder information from the National Institute of Neurological Disorders and Stroke Web site. The format is XML-based and includes information about interfaces and parameters (analogous to database tables and fields) among other elements to be passed, and the necessary logic to navigate the Web site to locate, extract, and transform the information into relational tables.
Figure 7.
The DISCO Web Interoperability Control Panel. This tool executes “disco.interop” scripts. The figure shows the URL of the National Institute of Neurological Disorders and Stroke (NINDS) DISCO interoperability script that collects disorder information from the site’s Web pages. The NINDS disorder information is scattered among several pages. During the extraction process, this tool first extracts disorder URLs from index pages. Later it queries each of those URLs to extract the disorder information. This process is recursive into linked pages containing related information about the disorder to create other tables. All information collected by this script is passed to NIF servers where it is stored along with similar information from other NIF resources.
Figure 8.

Data Federation results in the NIF’s user search interface after searching for “ataxia”. On the left, the “Disease (24)” category was expanded, and the “NINDS: ninds (24)” source was clicked. The first disease “ataxia” is presented in a list of a total of 24 diseases. Notice the relation between the columns shown in this figure (Name, What_is, Treatment and Prognosis) with “Disorder Detail” fields shown in figure 7.
In this example, all the desired data from the resource is extracted in a batch run (on a regular scheduled basis) and placed into a relational table on a NIF server. This approach has the advantage that NIF user queries can run much more efficiently (against the extracted data rather than against the resource itself). In addition, a NIF user query will still work even if the resource is “down” or even if its Web interface has been changed. (If the resource’s Web interface has changed, the DISCO Web Interoperation file needs to be changed before that resource’s data extract can be refreshed, but in the meantime NIF queries can run against the most recently extracted data set.) DISCO’s Web Interoperability capability has currently been developed for three different types of Web interfaces, with certain limitations to one case.
Web sites with unstructured text may require additional HTML tagging (e.g., sites like crcns.org). These sites have content that while structured, cannot be consistently extracted and organized in structured datasets needed for robust data integration. These sites require annotation of their content with easily-parsed lexical markup. This annotation can then be parsed and stored in a database along with pointers to each annotated Web page. That database can then be searched using the lexical terms used for annotation, thereby allowing the Web-based content of the resource to be retrieved in a fashion that is integrated with retrieval from other NIF resources.
Many Web sites (e.g.: http://www.ninds.nih.gov/disorders) have structured content described in HTML, XML, delimited text, or some other type of well-structured text that can be extracted using XML Query or Regular expressions algorithms.
A third class of web site allows data to be retrieved (e.g., from a database) using query parameters. These sites account for most of the hidden Web sites that most search engines cannot index. Such sites provide data by user interaction via Web forms or Web services. Examples include the WebQTL/GeneNetwork database (http://www.genenetwork.org/) and the NCBI Entrez system. DISCO Web interoperation retrieval from this type of web site has been implemented but there are some limitations related to the number of parameters that can be used to query the interface. When numerous, as in the NCBI Entrez case, the list of parameters may result in a large number of queries. As described above, these can be performed in batch modes with the extracted data stored on a NIF server.
The Lexicon/Ontology Capability
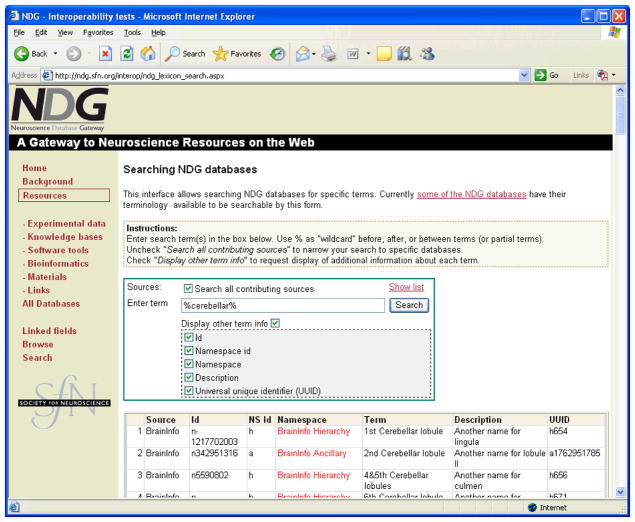
This capability was created to allow a resource to share its lexical terms, including an indication as to how are they used in the database. These terms come with associated information, including mappings to standard terminologies, such as the NIFSTD (Bug et al., 2008), the ontology of neuroscience used by the NIF. These mappings are intended to facilitate semantic data integration from multiple resources. Figure 9 shows a portion of the Olfactory Receptors Database (ORDB) resource’s DISCO Lexicon data. Figure 10 shows the Web interface built into NDG that allows users to search the lexicon data imported from participating NDG databases. In this example, the user has requested all terms containing the string “cerebellar”. For each term returned, the results include the database in which a matching term was found, the exact name of the term in that database, and other related information describing that term. This capability is operational on the NDG Web site for several databases using the “disco.lexicon” format. The Lexicon/Ontology capability in NIF has been designed to expose ontologies in OWL and other formats, but no code has yet been generated on NIF to process this type of information.
Figure 9.

This figure shows a portion of the DISCO Terminology data (in XML format) describing the terms used in the Olfactory Receptors Database (ORDB), http://senselab.med.yale.edu/ordb.
Figure 10.
This figure shows a tool designed to search the DISCO Terminology files of multiple NDG resources. In this example, the search string “%cerebellar%” (the percent sign is used as a wildcard) has been entered and all matching terms are returned from multiple resources, together with information describing each term
The Database Schema Capability
This capability is designed for resources that store their information in a database. Sharing a resource’s database schema helps technically knowledgeable personnel better understand the information in that database. Schema information is necessary to allow others to construct views into the database and to formulate queries. At present no specific format is recommended for use in DISCO. Any format, including database schema dumps, reverse engineered formats, and XML proprietary formats are allowed. This capability is currently being used by SenseLab databases using the EDSP format.
The RSS News Feed Capability
RSS is a standard approach used by Web sites to communicate to their users information about changes to a Web site’s content, for example when new information has been added. Each participating resource may publicize its “news” using this DISCO approach. The DISCO server can then gather this information from multiple resources, and communicate it to interested users in an integrated fashion. In this way, for example, users can receive one message that contains integrated news rather than multiple messages from many different sites. In addition, in the future this capability might be extended so that users could indicate specific topics of interest (e.g., selected from a list of SfN Annual Meeting topics) and resources could “tag” their news items to indicate which topic(s) each piece of news related to. This approach would allow users to receive integrated RSS news tailored to their specific interests. RSS news feeds have been added to DISCO to three SenseLab databases (NeuronDB, ModelDB, and ORDB). This capability may be included in a future version of NIF.
DISCO’s Implementation
The DISCO approach has been in use for several years by the NDG, and by early versions of the NIF. DISCO is currently implemented in production mode on the NIF DISCO server at http://disco.neuinfo.org. The DISCO server is an open source Web application written in Java, using Tomcat, and hosted on a Linux server. It uses two PostgreSQL databases for backend storage. For communication with other servers, it uses Web services and a variety of Web formats including text, HTML, XML, and others (see Figure 1).
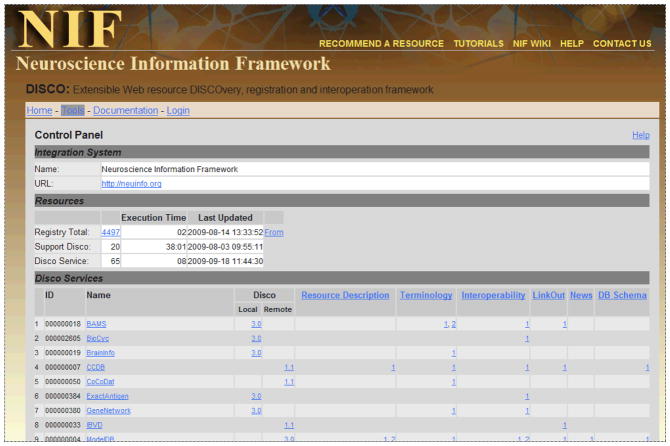
The DISCO server also provides authoring and administrative tools with technical documentation to assist resource developers and DISCO administrators perform the various tasks needed to successfully integrate their information, and to keep that integration up-to-date. Authoring tools are provided as Web applications to help the resource developer in building DISCO content (e.g., disco main file, resource description, and LinkOut) and in the form of documentation. Management and control tools are provided via the DISCO dashboard panel (see Figure 11).
Figure 11.
This figure shows the DISCO Dashboard Control Panel. For each participating resource, this panel indicates which DISCO capabilities are being utilized. In addition the Dashboard provides controls allowing NIF staff who are logged in to import DISCO files provided by the participating resources, as well as tools to export and synchronize that information with other NIF components.
DISCO Information Workflow
To operate, the DISCO server needs a list of participating resources. The NIF provides this information via the following URL: http://neuinfo.org/nif_registry/disco/resourceDescList_1_0. This list is kept up-to-date by regular synchronization of the DISCO server with the NIF Registry. Using this list, the DISCO server searches each resource for DISCO files which are parsed to identify the capabilities being used by that resource. A DISCO administrator then evaluates the submitted information and coordinates its incorporation into NIF. Many of the capabilities are scheduled for automated update. When specified, the system also automatically sends confirmation emails of success or failure of each of the steps described above to DISCO server administrators and DISCO capability developers.
Summary
In summary, the DISCO approach is designed to play a critical role in facilitating a range of diverse information integration capabilities on the Web. Most of these capabilities are now implemented in the current Neuroscience Information Framework (NIF) system acting as a portal to data and information across the field of neuroscience. The flexibility of this approach is designed to enable users to keep pace with this rapidly developing and extraordinarily broad multidisciplinary field of research.
Acknowledgments
This research was supported in part by NIH grants P01 DC04732 and R01 DA021253, by NIH Neuroscience Blueprint Contracts HHSN271200577531C and NIDA-SSSAB-2008-1 via NIDA, and by the Society for Neuroscience. In addition we appreciate the collaboration on implementing the DISCO approach by Mihail Bota at UCSD (Brain Architecture Management System), Willy WaiHo Wong and Maryann Martone (Cell Centered Database), Rolf Kotter (CocoDat), David Kennedy (Internet Brain Volume Database), Maryam Halavi and Giorgio Ascoli (Neuromorpho.org), and Daniel Gardner (Neurodatabase.org).
Footnotes
Information Sharing Statement
All information and program code related to DISCO protocols are publicly available through the Web sites mentioned in this publication.
Contributor Information
Luis Marenco, Center for Medical Informatics, Department of Anesthesiology, Yale University School of Medicine, New Haven, CT 06520, USA.
Rixin Wang, Center for Medical Informatics, Department of Anesthesiology, Yale University School of Medicine, New Haven, CT 06520, USA.
Gordon M. Shepherd, Department of Neurobiology, Yale University School of Medicine, New Haven, CT 06520, USA
Perry L. Miller, Center for Medical Informatics, Department of Anesthesiology, Yale University School of Medicine, New Haven, CT 06520, USA. Department of Molecular, Cellular, and Developmental Biology, Yale University School of Medicine, New Haven, CT 06520, USA
References
- Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6(3):175–94. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, Goldberg DH, Grafstein B, Grethe JS, Gupta A, Halavi M, Kennedy DN, Marenco L, Martone ME, Miller PL, Müller HM, Robert A, Shepherd GM, Sternberg PW, Van Essen DC. The Neuroscience Information Framework: A data and knowledge environment for neuroscience. Neuroinformatics. 2008;6(3):149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bug W, Marenco L, Qian X, Condit C, Rangarajan A, Müller HM, Miller PL, Sanders B, Grethe JS, Astakhov V, Shepherd G, Sternberg PW, Martone ME. Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF) Neuroinformatics. 2008;6(3):205–217. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenco L, Tosches N, Crasto C, Shepherd G, Miller PL, Nadkarni PM. Achieving evolvable Web-database bioscience applications using the EAV/CR framework: Recent advances. J Am Med Inform Assoc. 2003;10(5):444–53. doi: 10.1197/jamia.M1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenco L, Giorgio A, Ascoli GA, Martone ME, Shepherd GM, Miller PL. The NIF LinkOut Broker: A web resource to facilitate federated data integration using NCBI identifiers. Neuroinformatics. 2008;6(3):219–227. doi: 10.1007/s12021-008-9025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PL, Nadkarni P, Singer M, Marenco L, Hines M, Shepherd G. Integration of Multidisciplinary Sensory Data: A Pilot Model of the Human Brain Project Approach. J Am Med Inform Assoc. 2001;8:34–48. doi: 10.1136/jamia.2001.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. Entrez LinkOut Service. 2008 http://www.ncbi.nlm.nih.gov/projects/linkout.
- NDG. Neuroscience Database Gateway. 2008 http://www.sfn.org/index.cfm?pagename=NDG_main.
- Shepherd GM, Mirsky JS, Healy MD, Singer MS, Skoufos E, Hines MS, Nadkarni PM, Miller PL. The Human Brain Project: Neuroinformatics tools for integrating, searching, and modeling multidisciplinary neuroscience data. Trends in Neuroscience. 1998;21:460–468. doi: 10.1016/s0166-2236(98)01300-9. [DOI] [PubMed] [Google Scholar]