SUMMARY
Streptococcus mutans is generally considered to be the principal etiological agent for dental caries. Many of the proteins necessary for its colonization of the oral cavity and pathogenesis are exported to the cell surface or the extracellular matrix, a process that requires the assistance of the export machineries. Bioinformatic analysis revealed that the S. mutans genome contains a prsA gene, whose counterparts in other gram positive bacteria, including Bacillus and Lactococcus encode functions involved in protein post-export. In this study, we constructed a PrsA-deficient derivative of S. mutans and demonstrated that the prsA mutant displayed an altered cell wall/ membrane protein profile as well as cell surface related phenotypes, including auto-aggregation, increased surface hydrophobicity, and abnormal biofilm formation. Further analysis revealed that the disruption of the prsA gene resulted in reduced insoluble glucan production by cell surface localized glucosyltransferases, and mutacin as well as cell surface-display of a heterologous expressed GFP fusion to the cell surface protein SpaP. Our study suggested that PrsA in S. mutans encodes functions similar to the ones identified in Bacillus, and thus is likely involved in protein post-export.
Keywords: foldase protein PrsA, protein secretion, Streptococcus mutans
INTRODUCTION
As the primary etiological agent of dental caries in humans (Hamada & Slade, 1980), Streptococcus mutans relies on the activities of secreted or cell surface localized proteins to interact with other oral bacteria, colonize the oral cavity and exert its pathogenesis. Previous studies have shown that S. mutans employs various mechanisms to deliver proteins across the cell membrane. The general secretory (Sec) translocation channel is the major secretion apparatus in proteins translocation across the cytoplasmic membrane (Fekkes & Driessen, 1999; Muller et al., 2001). In addition, S. mutans also contains certain specific secretion systems. For example, specific ATP-binding cassette transporters were found to be responsible for direct translocation of the bacteriocins across the cytoplasmic membrane (van Belkum et al., 1997). Most proteins translocated across the cell membrane via above pathways are considered to be delivered in an unfolded conformation across the cytoplasmic membrane into the interface between cytoplasmic membrane and cell wall peptidoglycan (Matias & Beveridge, 2006; Matias & Beveridge, 2008; Sarvas et al., 2004). Due to the high cation concentration, as well as the low pH and high density of negative charge within the cytoplasmic membrane- cell wall peptidoglycan interface (Sarvas et al., 2004), protein chaperones as well as foldases are required to ensure the proper folding of these proteins after export. However, the components and their roles in the folding and stabilize proteins to their active form are poorly characterized in S. mutans.
The foldase protein PrsA is found ubiquitously in the genomes of all Gram-positive bacteria including S. mutans. In the Group A streptococcus including S. pyogenes, PrsA was found to be required for the final maturation steps of SpeB, a pluripotent cysteine protease and an important virulence factor (Ma et al., 2006). The role of PrsA in assisting the folding and stability of exported proteins has been extensively studied in Bacillus and Lactococcus. In B. subtilis, the extracytoplasmically located PrsA has been shown to be critically important in vivo for the proper conformation of various exoproteins (Jacobs et al., 1993), and considered as an essential rate-limiting component of the secretion machinery (Kontinen & Sarvas, 1993). It influences neither the expression nor the translocation of exoproteins but is required for their correct conformation and stability in the post-translocational phase of secretion (Hyyryläinen et al., 2001; Vitikainen et al., 2001). In L. lactis, the PrsA- like protein triggers the folding of the translocated lipase (Drouault et al., 2002). Over-expression of B. subtilis PrsA also resulted in increased heterologous protein expression in L. lactis, presumably by allowing more efficient protein folding (Lindholm et al., 2006).
In S. mutans, the predicted PrsA protein contains 333 amino acids with a molecular mass of ~36 kDa. It includes a predicted signal sequence at the N-terminus. However, no cellular function has been assigned to this protein. In this study, by constructing a prsA-deficient strain and performing an array of phenotypic analyses, we sought to investigate the biological functions of PrsA in S. mutans.
METHODS
Bacterial strains and growth conditions
E. coli strain DH5α was used for cloning as well as plasmid amplifications and grown in Luria-Bertani (LB) medium aerobically at 37 °C. S. mutans strain UA140 (wild type), UA140 prsA-deficient strain, as well as UA140 pHluorin-SpaP fusion strain and its corresponding prsA-deficient strain were cultured in Todd-Hewitt (TH) media (Difco) at 37° Cin the presence of 5% CO2. For selection of antibiotic-resistant colonies after genetic transformation, spectinomycin (100 μg/mL for E. coli or 800 μg/mL for S. mutans) or kanamycin (150 μg/mL for E. coli or 800 μg/ml for S. mutans) was added to the medium.
Strain construction
The open reading frame for the predicted prsA gene (GenBank accession no. AAN58382) was annotated in the S. mutans UA159 genome database (http://www.genome.ou.edu/smutans.html). BLASTn and BLASTp sequence homology analyses were performed by using the BLAST network service of the National Center of Biotechnology Information (NCBI), Bethesda, MD. The pFW5 vector (Podbielski et al., 1996) was employed for generating a mutant derivative of S. mutans wild type strain UA140 (Qi et al., 2001) carrying a deletion in the prsA gene. S. mutans UA140 genomic DNA served as a template to amplify the prsA upstream region with the primer pair upF (5′-CCGCTCGAGCGCAAACCACATCCACAGGG) which contains a XhoI site incorporated at its 5′ end and upR (5′-CCCAAGCTTCACAAGTCCTGTAGCAATCG) which has a HindIII site incorporated at its 5′ end, while the corresponding downstream region was obtained using downF (5′-ACATGCATGCCAGCAGCAAGCGGAAGTGGC) which carries a SphI site incorporated at its 5′ end and downR (5′-TCCCCCCGGGAGCATCATCACGGAAGTAAT) with a XmaI site incorporated at its 5′ end (the restriction enzyme recognition sites are underlined). The fragments were generated using Pfu polymerase (Stratagene) and subsequently inserted into the two multiple cloning sites of pFW5 vector respectively. The resulting recombinant plasmid pFW5-prsA was confirmed by restriction analysis, PCR amplification and DNA sequencing. Plasmid pFW5-prsA was then linearized using a unique NheI site in the vector backbone and transformed into S. mutans UA140 via competence-stimulating peptide (CSP)-induced natural transformation (Kreth et al., 2005). CSP was a gift from C3 Jian Inc. (Los Angeles, CA). Transformants were selected on TH agar containing 800 μg/mL of spectinomycin. The resulting prsA deletion mutant was confirmed by PCR and sequencing.
The GFP coding sequence was in-frame inserted via overlapping PCR between the second and third amino acids after the identified signal-peptide cleavage site of SpaP (Kelly et al., 1989), a surface protein antigen-encoding gene of S. mutans. The resulting fragment was then ligated downstream of the lactate dehydrogenase gene (ldh) promoter (Merritt et al., 2005) and cloned into pFW5 vector to generate pFW5Φ(ldhp-leading sequence-gfp-spaP). The recombinant construct was transformed into S. mutans strain UA140 as well as the prsA-deficient mutant to generate the cell surface displayed GFP-SpaP fusion derivative (Guo et al., unpublished data), respectively. The integration of gfp-spaP into the chromosome of S. mutans via single crossover homologous recombination was confirmed by PCR, and cell surface localization of the GFP-SpaP fusion was revealed by Western blot analysis using an anti-GFP antibody (Guo et al., unpublished data).
General phenotypic characterization assays
Growth kinetics were measured for S. mutans UA140, the prsA-deficient strain and the strain containing the cell surface displayed GFP. Autolysis assay was also performed as previously described (Wen & Burne, 2002).
Hydrophobicity assay by the bacterial adherence to hydrocarbons (BATH)
The bacterial hydrophobicity was measured using the BATH method as described previously (Dillon et al., 1986). Briefly, the bacterial cultures which were dispersed by drawing up and expelling the bacterial suspension 10 times through a 26 gauge, 5/8 inches (15.9 mm) long needle (Dunning et al., 2008) were suspended in PUM buffer (K2HPO4, 16.87 g/L; KH2PO4, 7.26 g/L; MgSO4·7H2O, 0.2 g/L; urea, 1.8 g/L; pH 7.1) to a final OD600 of 0.6. A 1.2 mL volume of the bacterial suspension was dispensed into each one of 12 round bottom test tubes with 10 mm in diameter. Four tubes with four different volumes (0.2, 0.15, 0.1, and 0.05 mL) of N-hexadecane were kept at room temperature for 10 min, vortex-mixed for 2 min and followed by incubation at room temperature for 15 min to allow hydrocarbon separation. The absorbance at 400 nm of the aqueous phase was measured before and after treatment (Spectronic Genesys 5 UV-Visible Spectrophotometers) and results were recorded as the percentage absorbance of the aqueous phase after treatment relative to the initial absorbance of the bacterial suspension.
Atomic force microscopy (AFM) force spectroscopy
AFM force spectroscopy was performed to measure cell hydrophobicity. AFM force-distance curves were obtained in deionized water using a combined inverted optical (Bruker, Santa Barbara, CA) system. Oxide-sharpened microfabricated Si3N4 cantilevers with a spring constant of 0.01 N/m (MLCT, Bruker, Santa Barbara, CA) were coated by electron beam thermal evaporation with a 5-nm-thick Cr layer followed by a 30-nm-thick Au layer (Sharma et al., 2009). Gold-coated cantilevers were immersed for 14 h in 1 mM solutions of HS (CH2)11CH3 in ethanol and then rinsed with ethanol. To probe S. mutans cell surface hydrophobicity, cells were grown on glass cover-slips for 3 h in TH medium supplemented with 0.5% sucrose. Surface immobilization of the bacterial cells was tested by gently imaging them at low forces (~200pN) prior to force curve measurements. Force-distance curves over a 400 × 400 nm area were obtained using hydrophobic tips with z ramp size of 10μms, 1024 × 1024 samples/line and 0.5Hz. The adhesion strength was calculated for each force curve using SPIP™ software (Image Metrology, Horsholm, Denmark.).
Sonication-resistance analysis
The resistance of S. mutans UA140 and its prsA-deficient derivative to sonication was assayed by monitoring the bacterial viability after sonication treatment. Bacterial suspensions with a density of 109 CFU/ml were prepared from overnight cultures and subjected to sonication at a constant frequency of 22 kHz and output power of 10 watts for different periods of time ranging from 1 to 9 min. Cell viability after sonication treatment was determined by CFU counting on TH agar plates.
Early biofilm formation assay
In order to compare the sucrose-independent and sucrose-dependent adhesion abilities between S. mutans UA140 wild-type and the prsA-deficient mutant, overnight cultures of both strains were dispersed by needle and syringe, and resuspended in glucose or sucrose-supplemented (20 mM) minimal medium (Loo et al., 2000) to a final OD600 of 0.1. Four hundred μL of bacterial suspension was added to the well of a 24-well flat-bottomed polystyrene microtiter plate (Corning, New York, NY). After 3 h incubation in the presence of 5% CO2, the plates were rinsed with PBS for three times to remove planktonic and loosely bound cells. The biofilms were detached using cell scrapers (Thermo, Rochester, NY) and clumps were broken up and dispersed by needle and syringe. The biomass of early biofilm was then calculated as CFU/mL by viability counting on agar plates.
Scanning electron microscopy (SEM)
Overnight cultures of S. mutans UA140 and its prsA-deficient derivative were harvested and resuspended in fresh TH medium to an OD600 of 0.6. A 100-fold dilution of the bacterial suspension into defined minimal medium supplemented with 0.5% (wt/vol) sucrose was then added to each well of six-well polystyrene microtiter plates in which sterile coverglasses had been placed. After 16 h incubation, the medium containing the remaining planktonic cells was aspirated and the cover glasses were carefully rinsed twice with 1 mL PBS without disturbing the attached biofilms. The biofilms were fixed with 1% glutaraldehyde. After another wash with phosphate buffer, the samples were mounted on a stub with silver adhesive (Electron Microscopy Sciences, Hatfield, PA, U.S.A.), sputter coated with a 40-nm layer of platinum and examined with an SEM operating at 5 kV in the secondary electron mode (XL 30 S, FEG, FEI Company, Hillsboro, OR, U.S.A.).
SDS-PAGE
Cell wall/membrane was prepared from S. mutans as described by Yamashita et al. (1998) with some modifications. Briefly, bacterial cells from the overnight cultures were collected by centrifugation. The cell pellets were resuspended in 50mM Tris·HCl (pH 8.0) containing 1 mM PMSF, transferred to a chilled 2 ml microcentrifuge tube containing 425–600μm diameter glass beads (Sigma) and disrupted with a Mini-Bead Beater homogenizer (Biospec Products) for 10 min. The glass beads were removed and undisrupted cells were separated by centrifugation at 2,000 × g for 10 min. The supernatants were further centrifuged at 150,000 × g for 2 h to collect the crude cell wall fraction. The pellets were washed twice with warm distilled water and resuspended in 50mM Tris•HCl (pH 8.0) containing 1 mM EDTA, 1 mM PMSF, 10 mg/L RNase and 10 mg/L DNase and incubated at 37°C for 1 h. The samples were boiled in SDS-PAGE loading buffer for 10 min before being loaded onto 10% SDS-PAGE gel. The protein concentration was determined by the Bradford method with the Pierce BCA protein assay kit (Thermo, Rockford, IL, USA).
Glucan Analysis
Overnight cultures of S. mutans strains were collected by centrifugation at 6,000 × g for 5 min. The pellets were washed twice with PBS and resuspended in minimal defined medium to a cell density of 108 cells/ml. Sucrose was added to the cell suspension to a final concentration of 100 mM. The cells were incubated at 37°C for 16 h in the presence of 5% CO2, and collected by centrifugation at 10,000 × g for 10 min. Pellet and supernatant were used to assess the production of insoluble and soluble glucan, respectively. For soluble glucan analysis, the supernatant was precipitated by chilled ethanol. As for insoluble glucan analysis, the pellet was washed with distilled water three times to remove the remaining soluble glucan. NaOH (1.0 N) was added and the alkali-soluble polysaccharides were precipitated with chilled ethanol. The amounts of glucan were measured by the phenol sulphuric acid method (Dubois et al., 1956). A derivative of S. mutans UA140 lacking gtfBC (kindly provided by H. Kuramitsu, University of Buffalo, NY, USA) was used as a negative control.
Mutacin IV production assay
Mutacin IV production was measured by deferred antagonism according to the protocol by Tsang et al. (2006). Five microlitres of S. mutans overnight culture was spotted onto TH agar plates and the plates were incubated at 37°C in the presence of 5% CO2. After 16 h incubation, 5 mL of a soft agar overlay containing S. sanguinis ATCC 10556 or S. gordonii DL1 at an OD600 of 0.1 as indicator strain was poured on top of the plates spotted with S. mutans. The growth inhibition zone of S. sanguinis or S. gordonii in the overlay agar was inspected after overnight incubation. The distance from the colony edge to the edge of the clearance zone was measured to calculate the inhibition area. Results are expressed as percentage of inhibition area induced by the prsA mutant relative to its parent strain.
Confocal laser scanning microscope (CLSM)
The 3-h biofilms of S. mutans GFP-SpaP fusion derivatives of wild type and the prsA mutant were formed according to aforementioned procedure in the section of early biofilm formation assay except a final OD600 of 0.3 was used for the mutant in order to normalize the number of bacteria attached to the well. Biofilms were grown in medium supplemented with sucrose in each well of a sterile 8-well Lab-Tek™ Chambered Coverglass (Nalge Nunc International; Naperville, IL). The biomass of wild type and prsA mutant strains was normalized according to their respective CFU counts. All biofilm images were collected with a Zeiss LSM 5 PASCAL confocal laser scanning microscope (CLSM) using LSM 5 PASCAL software (Zeiss, Jena, Germany). Excitation at 488 nm with an argon laser in combination with a 505–530 nm bandpass emission filter was used for GFP fluorescence imaging. The scanning module of the system was mounted onto an inverted microscope (Axiovert 200M). A 40 oil-immersion objective (numerical aperture 1.3) was used for imaging. Image stacks (1024 by 1024-pixel tagged image file format) of eight randomly chosen spots were collected for each biofilm and quantified using the image analysis software COMSTAT. The fluorescence intensities in the biofilms of S. mutans GFP-SpaP strain and its prsA-deficient derivative were normalized to the number (CFU counts) of bacteria present in the well.
RESULTS
Bioinformatic analysis revealed the presence of a prsA gene in the S. mutans genome. Sequence homology analysis showed that the prsA gene of S. mutans exhibited 69% homology to the one found in Streptococcus agalactiae, 67% homology to Streptococcus pyogenes, and 66% homology to Streptococcus dysgalactiae at the nucleotide level. The deduced PrsA amino acid sequences exhibited a homology of ~57–62% to other PrsA proteins among these streptococci, suggesting that PrsA from these streptococci are not well conserved. And the PrsA from S. mutans displayed only 32% identity to the PrsA of Bacillus subtilis. Since there are two types of PrsA proteins differing in the presence or absence of peptidyl-prolyl cis/trans-isomerases (PPIase) signature motif, the PrsA protein sequence of S. mutans was further aligned with both the B. subtilis and Listeria monocytogenes PrsA sequence using the ClustalW tool (www.ebi.ac.uk/clustalw). However, the signature motif for PPIase is absent in S. mutans PrsA.
prsA deletion mutant auto-aggregates when grown as planktonic culture
The expression of the downstream gene of prsA is not affected by the prsA deletion mutant construction method (data not shown). The disruption of prsA did not affect cell viability either, and no significant difference in cell autolysis was observed between the prsA mutant and its parent strain (data not shown). However, the prsA-deficient strain displayed a striking auto-aggregation phenotype and the cells tended to clump and precipitate at the bottom of the glass tubes, while the wild-type strain showed a uniformly turbid appearance in TH medium after overnight growth (Fig 1A). Light microscopy observations showed that the mutant strain formed about 12 clumps per field of view, in contrast to the parent strain for which no clumps were observed (Fig 1B and C).
Figure 1.
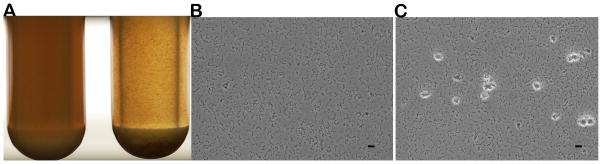
Cell morphological characteristics in TH medium. 24-h cultures of S. mutans UA140 (left) and the prsA-deficient strain (right) (A). Light microscopic observation of a 24-h culture of S. mutans UA140 (magnification, ×400) (B) and the prsA-deficient strain (C). The scale bar equals to 10 μm.
prsA-deficient strain has altered cell surface characteristics
Enhanced auto-aggregation could result from altered cell surface properties, thus we examined the cell surface hydrophobicity of the prsA mutant. Using the BATH assay, we demonstrated that the percentage absorbance of the aqueous phase in the prsA mutant after treatment with hydrocarbon relative to the initial absorbance was significantly less than that of the parent strain (37% vs. 61%) (Fig 2A), suggesting an increased bacterial surface hydrophobicity in prsA-deficient strain. Cell surface hydrophobicity was also quantitatively measured using chemically modified AFM. Fig. 2B shows the adhesion histograms and representative force curves obtained for the prsA-deficient and wild type strains. “Saw tooth-like” force rupture events were observed in the retract regions of the force curves with hydrophobic tips (shown in inset images in 2B). Adhesion forces measurements showed higher adhesion forces over prsA-deficient cell surfaces compared to wild-type cell surfaces with mean values of 370±17pN and 120±12pN respectively. These results confirm the qualitative findings obtained from the BATH assay at the single bacterial level and indicate that the prsA-deficient strain is about three-fold more hydrophobic compared to the wild-type (P<0.05).
Figure 2.
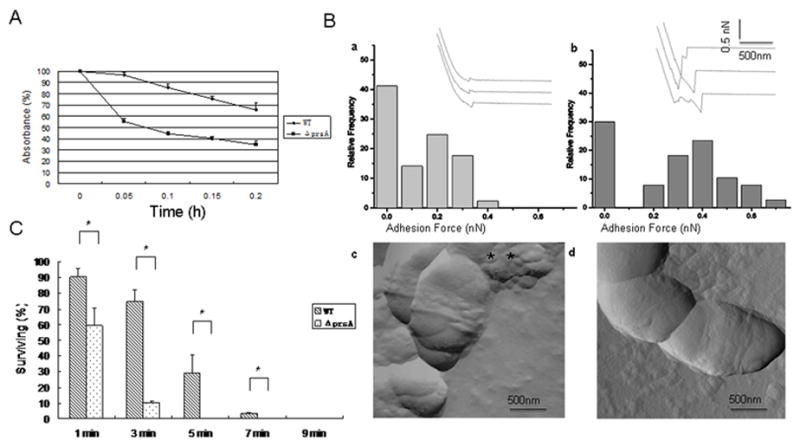
Cell surface characteristics. (A) Hydrophobicity: Adherence of S. mutans UA140 wild type and its prsA-deficient derivative to hexadecane in the BATH assay. Results are expressed as percentage absorbance of the aqueous phase after hexadecane treatment relative to the initial absorbance. Each point represents the mean of three independent experiments. (B) AFM analysis: Adhesion histograms and representative force curves (inset) recorded with hydrophobically modified tips on S. mutans UA140 (a) and the prsA-deficient strain (b) using a maximum applied force of 1nN. The surface topographies of wild type (c) and mutant strain (d) were also observed (amplitude images) using high-resolution AFM imaging. Two biological replicates were performed and representative image are shown. (C) Resistance of S. mutans UA140 and the prsA-deficient strain to sonication at a constant frequency of 22 kHz. Results are expressed as percentage of viable cells after sonication relative to untreated cells. Each point represents the mean ± SD of two independent measurements. The asterisk indicates that prsA-deficient strains were significantly less resistant to sonication than wild type at the same treatment time point (Student’s t test p value <0.05).
Sonication was used as a measure of the degree of physical cell membrane integrity. Results showed that, after 5 min sonication treatment, the prsA mutant suffered a drastic reduction (about 300-fold) in viability compared to the wild type (~3-fold) (Fig 2C), indicating that deletion of the prsA gene renders cells more sensitive to sonication-induced lysis.
The prsA-deficient strain displays reduced early biofilm formation and forms overnight biofilms with aberrant architecture
Our data revealed that lack of prsA resulted in altered cell surface characteristics. Since surface properties are important for cell adherence and biofilm formation, we further investigated the effect of PrsA on these phenotypes. Bacterial counts showed an almost 100-fold reduction in the number of attached prsA-deficient cells compared to the wild-type (Fig 3A). Interestingly, when sucrose was replaced by glucose, both strains formed similar thin biofilms and the bottom of the well was not evenly covered with cells.
Figure 3.
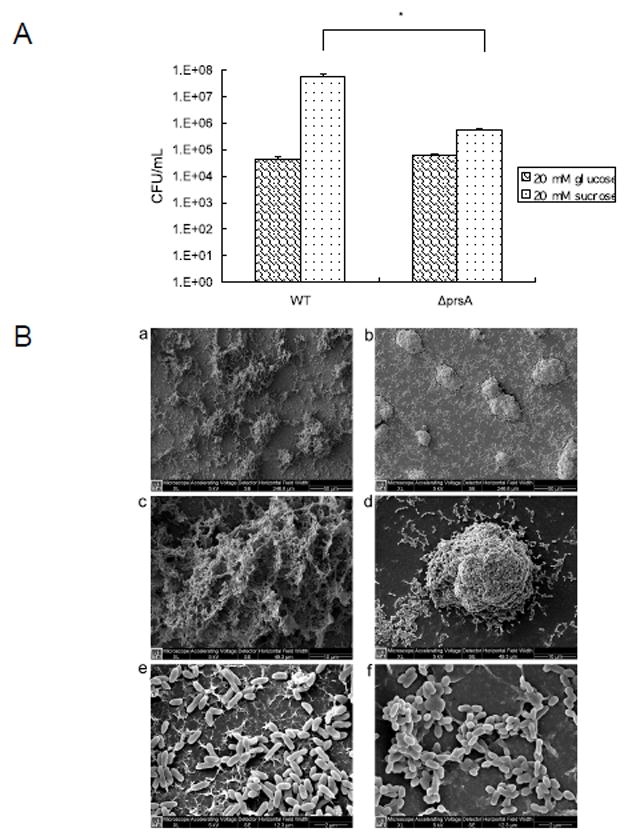
Biofilm formation characteristics. (A) Early Biofilm formation of S. mutans UA140 wild type and the prsA-deficient strains in minimal defined medium supplemented with glucose or sucrose. Each data point is the average of triplicate samples, and the error bars correspond to the standard deviations. The asterisk indicates that there was significantly less prsA mutant cells attached to the well of a 24-well flat-bottomed polystyrene microtiter plate than wild type in the presence of sucrose (Student’s t test p value <0.05). (B) Scanning electron micrographs of S. mutans 16 h-biofilms formed on glass surfaces. S. mutans UA140 wild-type biofilms (a, c, e); prsA-deficient strain biofilms (b, d, f). Magnifications, ×1000 (a, b), ×5000 (c, d) and ×20000 (e, f).
SEM analysis of biofilms grown on the surfaces of glass in defined medium with sucrose revealed that wild-type biofilms presented a uniform, sieve-like appearance with thick and compact layers of cells. In contrast, the mutant strain formed more compact microcolonies compared to its parent strain.
The prsA-deficient strain shows reduced insoluble glucan and mutacin IV production as well as the heterologous protein GFP-SpaP
Since PrsA has been shown to be involved in the post-export of a variety of exoproteins in Bacillus (Jacobs et al., 1993; Vitikainen et al., 2001), we suspected that the deletion of prsA might affect the profile of cell wall/ membrane proteins in S. mutans as well. As shown in Fig 4A, the prsA-deficient strain displayed an altered cell wall/ membrane protein profile compared to the wild type, with some protein bands showing increased intensity, while others exhibiting an obvious reduction. Since the effect of PrsA on cell wall/ membrane proteins is relatively general, we chose several known exoproteins for further characterization.
Figure 4.
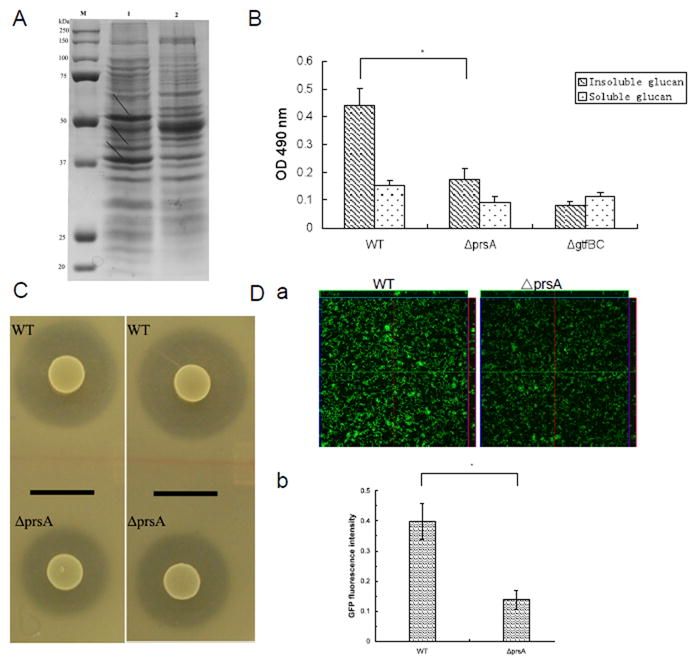
Analysis of PrsA-dependent phenotypes. (A) SDS-PAGE (10%) analysis of cell cell wall/ membrane proteins from S. mutans UA140 and the prsA-deficient strain. Lanes: M, prestained protein markers (Bio-rad); 1, S. mutans UA140; 2, UA140 prsA-deficient strain. Arrows indicate differentially expressed cell wall/ membrane proteins. (B) Glucan production of S. mutans UA140 wild type and the prsA-deficient strains in TH medium supplemented with 100 mM sucrose. S. mutans UA140 gtfBC-deficient strain was used as a negative control. Each data point is the average of triplicate samples, and the error bars indicate standard deviations. The asterisk indicates that prsA-deficient strain produced significantly less insoluble glucan than wild type (Student’s t test p value <0.05). (C) Drop inoculum deferred antagonism assay for S. mutans UA140 wild type and the prsA-deficient strains. The clear zone indicates mutacin IV production. The indicator strains are S. sanguinis (left panel) and S. gordonii (right panel) respectively. (D) GFP fluorescent signals of 3-h biofilms of S. mutans wild-type and the prsA-deficient, both carrying surface-displayed GFP-SpaP fusion protein. (a) CLSM analysis of surface-expressed GFP fluorescent signals within biofilms. (b) Quantification of GFP fluorescent signals within biofilms. The fluorescence intensities of S. mutans wild-type and the prsA-deficient biofilms were normalized to the number (CFU counts) of bacteria present in the well. The GFP expression percentage was calculated as the amount of fluorescence signal of prsA-deficient strain of vs its parent strain.
SEM imaging analysis revealed that the prsA mutant biofilm had less extracellular matrix compared to that of the wild type (Fig 3B). In S. mutans, cell wall associated glucosyltransferases (GTF) are responsible for synthesizing glucan, one of the main components of the extracellular matrix. Since deletion of prsA could potentially affect translocation of GTF proteins to the cell surface and thus alter glucan production, we determined the glucan production ability in both strains. Our results showed that the prsA-deficient strain produced less insoluble glucan compared to the wild-type (Fig 4B), while no difference in soluble glucan production was observed.
Furthermore, the effect of the prsA deletion on mutacin IV, a well-known secreted peptide bacteriocin was investigated. The deferred antagonism assay showed that the inhibition zone produced by the prsA-deficient strain was 49.67±4.20% and 50.39±4.52% of the one produced by the wild type using S. gordonii DL1 and S. sanguinis ATCC 10556 as indicator strains (Fig 4C).
To investigate whether PrsA facilitates the folding of exported heterologous proteins, we constructed derivatives of S. mutans wild type and prsA mutant, both displaying a GFP protein on the cell surface via a fusion to SpaP. Our previous study showed that fusion of GFP to SpaP resulted in surface localization and efficient folding of GFP with proper function (unpublished). Therefore, the post-export folding of heterologous protein GFP can be analyzed based on the fluorescence intensity of fused GFP. By quantifying image stacks of three randomly chosen biofilm spots, we found that the fluorescence intensity and thus the GFP surface display was reduced in the prsA mutant biofilm (> 3-fold) compared to the wild-type (Fig 4D).
DISCUSSION
S. mutans secretes numerous proteins (enzymes)/peptides that play crucial roles in competing with other bacteria and establishing itself within oral cavity via biofilm formation. The expression and activity of these exoproteins have been shown to be regulated at multiple levels, including post-translational as well as translocation regulation. In this study, we report the identification of PrsA, a predicted foldase that is involved in the post-export of a variety of proteins including membrane-associated proteins in S. mutans.
The auto-aggregation phenotype, as well as increased cell surface hydrophobicity and reduced resistance to mechanical breakage indicated a substantial change in the cell surface structure and properties upon deletion of the prsA gene. This was further corroborated by our AFM data showing the “saw tooth-like” force rupture patterns between hydrophobic AFM probes and prsA-deficient strains (Fig. 2B), which often reflects the unfolding of cell surface-bound proteins (Rief et al., 1998; Hu et al., 2011). As demonstrated in other Gram-positive bacteria such as Bacillus and Lactococcus, PrsA acts as a chaperone to assist the folding and stability of exported proteins (Lindhom et al., 2006; Wahlström et al., 2003). Thus, the presence of unfolded proteins on the cell surface of the PrsA-deficient S. mutans derivative would be consistent with a role of PrsA as a cell surface chaperone in S. mutans similar to findings in other species. The notion that the PrsA of S. mutans has roles similar to the ones extensively researched especially in B. subtilis is further sustained by the observed differences in the cell wall/ membrane protein profile of the prsA mutant compared with its parent strain (Fig. 4A). In a recent proteome analysis, PrsA-depleted B. subtilis cells were found to differentially secrete almost 200 proteins (Hyyryläinen et al., 2010).
Further phenotypic analysis of the PrsA deficient S. mutans derivative indicated that one group of proteins affected by PrsA function are the glycosyl transferases of S. mutans. The prsA-deficient strain exhibited a significant decrease in insoluble glucan production which resulted in reduced sucrose-dependent adhesion and biofilm formation (Fig. 3). It is well known that the majority of dental biofilm matrix is rich in polysaccharides (Paes Leme et al., 2006), of which glucan is one of the main components. These glucan-rich matrix could provide binding sites that promote accumulation of microorganisms on the tooth surface and further establishment of pathogenic biofilms (Koo et al., 2010). S. mutans encodes three GTFs. GtfB synthesizes mostly insoluble glucan, GtfC forms a mixture of insoluble and soluble glucan, while GtfD produces predominantly soluble glucan (Paes Leme et al., 2006). The insoluble glucan contributes to the glue-like characteristics of dental biofilms (Xiao et al., 2012). S. mutans strains deficient in insoluble glucan production are essentially non-cariogenic in a rodent model (Yamashita et al., 1993). While deletion of prsA resulted in a near 60% reduction in insoluble glucan production, it did not affect soluble glucan production, suggesting that PrsA might be involved in the post-export and/ or function of GTF B and C, but not GTF D.
Mutacin is a known secreted bacteriocin and has been implicated as virulence factor of S. mutans (Kuramitsu, 1993). S. mutans strain UA140 could produce both mutacin I and IV (Qi et al., 2001). Our results indicated reduced mutacin IV in the prsA mutant. This correlates well with previous related studies in other Gram-positive bacteria, where PrsA has been shown to be involved in the post-export of a variety of virulence factors. However there was no difference in mutacin I between the wild type and the prsA mutant (data not shown). In Bacillus species, α-amylase, β-glucannase and lipoproteinβ-lactamase were exported in a PrsA dependent manner (Jacobs et al., 1993; Vitikaninen et al., 2005), while over-expression of PrsA has been shown to increase the secretion of major bacillary exoenzymes (Kontinen & Sarvas, 1993). In Group A streptococcus, genomic disruption of prsA decreased the production of enzymatically active streptococcal pyrogenic exotoxin (SpeB) but not the level of the pro-SpeB zymogen (Ma et al., 2006).
The function of heterologous protein is mainly limited by the post-translational events including the inefficient translocation and folding of proteins and protease degradation (Tjalsma et al., 2004) and PrsA has been suggested to facilitate the folding of exported proteins into their final, mature conformation (Jacobs et al., 1993). In this study, we found that the fluorescence signal of a surface displayed GFP-SpaP fusion protein was reduced in the S. mutans strain lacking prsA. This result is consistent with observations that there was a positive correlation between the level of PrsA and the amount of a surface-localized fusion protein consisting of S. mutans SpaP and the pertussis toxin S1 fragment following expression in the heterologous host S. gordonii (Davis et al., 2011). Furthermore, the level of PrsA was found linearly proportional to protein secretion rate in B. subtilis (Vitikainen et al., 2001); while overproduction of PrsA could increase stability of the amylase and the protective antigen from B. subtilis (Kontinen & Sarvas, 1993; Vitikaninen et al., 2005; Williams et al., 2003), and prevent degradation of proteins on the cell surface of L. lactis (Drouault et al., 2002).
GTFs contain amino-terminal signal peptide which is thought to direct these proteins to the Sec secretory pathway (Navarre & Schneewind, 1999); while mutacin might be translocated by a specific ATP-binding transporter (van Belkum et al., 1997). The involvement of PrsA in GTF and mutacin IV production as demonstrated in this study suggested that, like foldase homologues identified in many other gram positive bacteria, PrsA may work closely with cellular export machineries in assisting the folding of a variety of exoproteins in S. mutans.
Acknowledgments
We thank H. Kuramitsu from the University of Buffalo (NY, USA) for kindly providing the gtfBC-deficient derivative of S. mutans UA140. This work was supported by a grant from the National Institute of Health (NIH-1-R01-DE020102) and a grant from the Natural Sciences Foundation of China (30672322). Dr. Wenyuan Shi wishes to disclose his potential Conflict of Interests here as he serves as a part time chief science officer of C3 Jian Inc, a California-based biotechnology company which is developing technologies to eliminate S. mutans within human oral cavities.
References
- Ahimou F, Denis FA, Touhami A, Dufrene YF. Probing microbial cell surface charges by atomic force microscopy. Langmuir. 2002;18:9937–9941. [Google Scholar]
- Alsteens D, Dague E, Rouxhet PG, Baulard AR, Dufrêne YF. Direct measurement of hydrophobic forces on cell surfaces using AFM. Langmuir. 2007;23:11977–11979. doi: 10.1021/la702765c. [DOI] [PubMed] [Google Scholar]
- Davis E, Kennedy D, Halperin SA, Lee SF. Role of the cell wall microenvironment in expression of a heterologous SpaP-S1 fusion protein by Streptococcus gordonii. Appl Environ Microbiol. 2011;77:1660–1666. doi: 10.1128/AEM.02178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon JK, Fuerst JA, Hayward AC, Davis GHG. A comparison of five methods for assaying bacterial hydrophobicity. J Microbiol Method. 1986;6:13–19. [Google Scholar]
- Drouault S, Anba J, Bonneau S, Bolotin A, Ehrlich SD, Renault P. The peptidyl-prolyl isomerase motif is lacking in PmpA, the PrsA-like protein involved in the secretion machinery of Lactococcus lactis. Appl Environ Microbiol. 2002;68:3932–3942. doi: 10.1128/AEM.68.8.3932-3942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substance. Anal Chem. 1956;28:350–356. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- Dunning DW, McCall LW, Powell WF, et al. SloR modulation of the Streptococcus mutans acid tolerance response involves the GcrR response regulator as an essential intermediary. Microbiol. 2008;154:1132–1143. doi: 10.1099/mic.0.2007/012492-0. [DOI] [PubMed] [Google Scholar]
- Fekkes P, Driessen AJ. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JDF, Ting YT, Jack RW, Tagg JR, Heng NCK. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA 159: Elucidation of the antimicrobial repertoire by genetic dissection. Appl Environ Microbiol. 2005;71:7613–7617. doi: 10.1128/AEM.71.11.7613-7617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ulstrup J, Zhang J, Molin S, Dupres V. Adhesive properties of Staphylococcus epidermidis probed by atomic force microscopy. Physical chemistry chemical physics : PCCP. 2011;13:9995–10003. doi: 10.1039/c0cp02800b. [DOI] [PubMed] [Google Scholar]
- Hyyryläinen HL, Bolhuis A, Darmon E, et al. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol Microbiol. 2001;41:1159–1172. doi: 10.1046/j.1365-2958.2001.02576.x. [DOI] [PubMed] [Google Scholar]
- Hyyryläinen HL, Marciniak BC, Dahncke K, et al. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol. 2010;77:108–127. doi: 10.1111/j.1365-2958.2010.07188.x. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Andersen JB, Kontinen V, Sarvas M. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzyme synthesized either with or without pro-sequences. Mol Microbiol. 1993;8:957–966. doi: 10.1111/j.1365-2958.1993.tb01640.x. [DOI] [PubMed] [Google Scholar]
- Kelly C, Evans P, Bergmeier L, et al. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- Kontinen VP, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Coordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu HK. Virulence factors of mutans streptococci: role of molecular gentics. Crit Rev Oral Biol Med. 1993;4:159–176. doi: 10.1177/10454411930040020201. [DOI] [PubMed] [Google Scholar]
- Lindholm A, Ellmen U, Tolonen-Martikainen M, Palva A. Heterologous protein secretion in Lactococcus lactis is enhanced by the Bacillus subtilis chaperone-like protein PrsA. Appl Microbiol Biotechnol. 2006;73:904–914. doi: 10.1007/s00253-006-0551-y. [DOI] [PubMed] [Google Scholar]
- Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bryant AE, Salmi DB, et al. Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J Bacteriol. 2006;188:7626–7634. doi: 10.1128/JB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VR, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol. 2006;188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias VR, Beveridge TJ. Lipoteichoic acid is a major component of the Bacillus subtilis periplasm. J Bcteriol. 2008;190:7414–7418. doi: 10.1128/JB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Kreth J, Shi W, Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Molecular Microbiol. 2005;57:960–969. doi: 10.1111/j.1365-2958.2005.04733.x. [DOI] [PubMed] [Google Scholar]
- Muller M, Koch HG, Beck K, Schafer U. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog Nucl Acid Res Mol Biol. 2001;66:107–157. doi: 10.1016/s0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA. The role of sucrose in cariogenic dental biofilm formation-new insight. J Dent Res. 2006;85:878–887. doi: 10.1177/154405910608501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Gautel M, Schemmel A, Gaub HE. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J. 1998;75:3008–30014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvas M, Harwood CR, Bron S, van Dijl JM. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:311–327. doi: 10.1016/j.bbamcr.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Sharma S, Cross SE, French S, et al. Influence of substrates on Hepatocytes: a nanomechanical study. J Scanning Probe Microscopy. 2009;4:7–16. [Google Scholar]
- Tjalsma H, Antelmann H, Jongbloed JD, et al. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev. 2004;68:207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum MJ, Worobo RW, Stiles ME. Double-glycine-type leader peptides direct secretion of bacteriocins by ABC transporters: colicin V secretion in Lactococcus lactis. Mol Microbiol. 1997;23:1293–1301. doi: 10.1046/j.1365-2958.1997.3111677.x. [DOI] [PubMed] [Google Scholar]
- Vitikainen M, Pummi T, Airaksinen U, et al. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of alpha-amylase in Bacillus subtilis. J Bacteriol. 2001;183:1881–1890. doi: 10.1128/JB.183.6.1881-1890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitikaninen M, Hyyryläinen HL, Kivimäki A, Kontinen VP, Sarvas M. Secretion of heterologous proteins in Bacillus subtilis can be improved by engineering cell components affecting post-translocational protein folding and degradation. J Appl Microbiol. 2005;99:363–375. doi: 10.1111/j.1365-2672.2005.02572.x. [DOI] [PubMed] [Google Scholar]
- Wahlström E, Vitikainen M, Kontinen VP, Sarvas M. The extracytoplasmic folding factor PrsA was thought to be required for protein secretion only in the presence of the cell wall, in Bacillus subtilis. Microbiol. 2003;149:569–577. doi: 10.1099/mic.0.25511-0. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol. 2002;68:1196–1203. doi: 10.1128/AEM.68.3.1196-1203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, Rees ML, Jacobs MF, et al. Production of Bacillus anthracis protective antigen is dependent on the extracellular chaperone, PrsA. J Biol Chem. 2003;278:18056–18062. doi: 10.1074/jbc.M301244200. [DOI] [PubMed] [Google Scholar]
- Xiao J, Klein MI, Falsetta ML, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Tsukioka Y, Nakano Y, Tomihisa K, Oho T, Koga T. Biological functions of UDP-glucose synthesis in Streptococcus mutans. Microbiol. 1998;144:1235–1245. doi: 10.1099/00221287-144-5-1235. [DOI] [PubMed] [Google Scholar]


