Abstract
Arrestins are dynamic proteins that move between cell compartments triggered by stimulation of G-protein-coupled receptors. Even more dynamically in vertebrate photoreceptors, arrestin1 (Arr1) moves between the inner and outer segments according to the light conditions. Previous studies have shown that the light-driven translocation of Arr1 in rod photoreceptors is initiated by rhodopsin through a phospholipase C/protein kinase C (PKC) signaling cascade. The purpose of this study is to identify the PKC substrate that regulates the translocation of Arr1. Mass spectrometry was used to identify the primary phosphorylated proteins in extracts prepared from PKC-stimulated mouse eye cups, confirming the finding with in vitro phosphorylation assays. Our results show that Bardet–Biedl syndrome 5 (BBS5) is the principal protein phosphorylated either by phorbol ester stimulation or by light stimulation of PKC. Via immunoprecipitation of BBS5 in rod outer segments, Arr1 was pulled down; phosphorylation of BBS5 reduced this co-precipitation of Arr1. Immunofluorescence and immunoelectron microscopy showed that BBS5 principally localizes along the axonemes of rods and cones, but also in photoreceptor inner segments, and synaptic regions. Our principal findings in this study are threefold. First, we demonstrate that BBS5 is post-translationally regulated by phosphorylation via PKC, an event that is triggered by light in photoreceptor cells. Second, we find a direct interaction between BBS5 and Arr1, an interaction that is modulated by phosphorylation of BBS5. Finally, we show that BBS5 is distributed along the photoreceptor axoneme, co-localizing with Arr1 in the dark. These findings suggest a role for BBS5 in regulating light-dependent translocation of Arr1 and a model describing its role in Arr1 translocation is proposed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1403-4) contains supplementary material, which is available to authorized users.
Keywords: Arrestin, Cilia, BBS5, BBSome, Translocation
Introduction
The function of most cells reflects the carefully choreographed expression and interaction of the cellular components. Many cells partition these elements so that the interactions can be regulated and coordinated according to the cellular demands. Primary cilia are specialized organelles used by cells for sensing a host of environmental cues, including the detection of photons, sensing of chemicals, and monitoring mechanical stress [1]. These sensory structures play integral roles in a variety of cell functions, including cell development, phototransduction, and olfaction. Because of this sensory function, transduction molecules are selectively partitioned into these ciliated structures where their interactions can be regulated and coordinated. Rod photoreceptors are an extreme example of such partitioning, with a cilium that is highly elaborated into a membrane-rich outer segment that is specialized for capturing photons. Accordingly, the photoreceptor outer segment contains all of the elements of the enzyme cascade required for transducing the energy of photons into a change in membrane potential, whereas the photoreceptor inner segment provides the synthetic and metabolic resources necessary to support the function of the outer segment.
Although this cellular partitioning into the outer segments holds true for most of the phototransduction cascade proteins in rods and cones, there are a few exceptions wherein the proteins are translocated between the inner and outer segments through the transition zone of the cilia according to the photic environment of the retina. The most dramatic of these protein changes in rods are arrestin1 (Arr1) and the alpha subunit of the visual G-protein transducin, which nearly quantitatively reverse their localization between the inner and outer segments as the cells move between light and dark [2–4]. For Arr1, dark adaptation leads to concentration of the protein in the inner segment portion of the photoreceptor, whereas light adaptation promotes translocation to the outer segments. The functional consequences of this translocation are not fully resolved. One hypothesis put forward is that the translocation of Arr1 extends the dynamic range of rods, providing a more rapid quenching of activated rhodopsin in higher light flux [5], thus reducing response amplitude and accelerating recovery. Another hypothesis is that translocation of Arr1 to the rod outer segments in light prevents metabolic rundown of rod photoreceptors from excessive activation of the phototransduction signaling cascade under lighting conditions that are beyond the response range of rods [6].
The mechanism regulating Arr1 translocation in photoreceptors appears to be complicated. Several well-designed studies have clearly established that the ciliary structure connecting the inner segment to the outer segment does not represent a significant barrier to movement of cytosolic proteins, and that the rates of Arr1 translocation that have been measured can be accounted for by diffusion [7–9]. Thus, it is simple to explain the partitioning of Arr1 to the outer segment in the light by high-affinity binding of Arr1 to photoactivated rhodopsin. Support for this hypothesis is provided by transgenic mice with modified levels of rhodopsin in which the fraction of Arr1 translocating to the outer segment is directly proportional to the level of expressed rhodopsin [10]. However, Arr1 translocation into the outer segments is not simply controlled by diffusion, but requires the microtubule cytoskeleton [11, 12] and its initiation by light involves a signaling cascade. The existence of this signaling cascade was first suggested by observations showing that Arr1 did not translocate until a level of illumination was reached that activated approximately 3 % of the rhodopsin, at which level of illumination 30-fold more molecules of Arr1 moved to the outer segments than the number of activated rhodopsin molecules [5]. Subsequent investigations into the nature of this signaling event implicated phospholipase C (PLC) and protein kinase C (PKC) activation in initiating Arr1 translocation, showing that pharmacological activation of PLC and PKC could initiate Arr1 translocation, even in the absence of light [13].
In this study, we attempted to extend these observations to identify the PKC substrate that might be involved in regulating Arr1 translocation. We hypothesized that the signaling cascade that initiates Arr1 translocation, i.e., activation of protein kinase C through phospholipase C, phosphorylates a protein that could regulate Arr1 translocation. We show that one target of PKC is Bardet–Biedl syndrome 5 (BBS5) protein, which is a component of the supramolecular BBSome complex essential for formation of primary cilia. Here, we show that BBS5 directly interacts with Arr1 in vitro, and that phosphorylation of BBS5 leads to a reduction in its binding of Arr1. Further, light and electron microscopy revealed the co-distribution of Arr1 and BBS5 along the microtubules of the axoneme in dark-adapted rod and cone photoreceptors. To our knowledge, our observations provide the first evidence that any of the BBSome proteins are regulated by phosphorylation and identify a novel interaction partner for both Arr1 and for one of the BBSome elements.
Materials and methods
Phosphorylation in eye cups
C57BL/6 mice were dark adapted overnight, and eye cups prepared from euthanized animals under dim red light by excising the entire eye globe from the mouse, and then dissecting away the cornea and lens in Tris-buffered saline (15 mM Tris pH 7.0, 100 mM NaCl, 1 mM EGTA, 10 mM MgCl2, 5 % DMSO). Each eyecup was incubated with 50 μCi 32P-γATP/10 μM ATP for 15 min at 37 °C, to allow penetration of the label and then subsequently exposed to 100 μM phorbol-12,13-diacetate (Sigma-Aldrich, St. Louis, MO, USA) for 15 min or to diffuse illumination (1,000 lux) for 15 min. Retinas were removed from the eye cups and homogenized in fresh Tris buffer, and centrifuged (5 min, 16,000 × g) to obtain an aqueous soluble fraction. Extracts were separated on either 4–15 or 12 % SDS-PAGE gels (Bio-Rad, Hercules, CA, USA) and autoradiographed. Samples for mass spectrometry were prepared without radioactive phosphate, and were fractionated by anion exchange chromatography (diethylaminoethyl-sephacel resin, Sigma-Aldrich, St. Louis, MO, USA), eluting with a 100–500 mM NaCl gradient. Fractions were separated by SDS-PAGE and phosphorylated species were detected by immunoblotting using anti-phosphoserine and anti-phosphothreonine antibodies (AB1603 and MA1-26593, respectively, EMD, Millipore, Billerica, MA, USA).
For experiments using Xenopus eye cups, eyes from transgenic tadpoles expressing arrestin–GFP fusion protein [14] were prepared as described above except that one eye from each animal was placed in tadpole Ringers solution (10 mM NaCl, 0.15 mM KCl, 0.2 mM CaCl2, 0.1 mM MgCl2), with 32P-γATP and the contralateral eye was placed in tadpole Ringer’s solution without radioactive phosphate. Each pair of eyes was subjected to 15 min of white light illumination at defined intensity (0–1,000 lux), measured with a calibrated photometer. The non-radioactive eye was then immediately placed into methanolic paraformaldehyde for fixation [14], and the eye in radioactive solution was homogenized in SDS-containing sample loading buffer [15] and separated on SDS-PAGE for autoradiography. Arr1 distribution in rods in cryosections from the fixed eye was imaged and quantified as previously described [13].
Mass spectrometry
For protein identification by mass spectrometry, in-gel trypsin digestion was performed. Briefly, relevant protein bands were cut out of the gel and diced into 1.5–3.5-mm cubes. Each sample was then de-stained, reduced by dithiothreitol, alkylated by iodoacetamide, and incubated overnight at 37 °C with trypsin in 25 mM ammonium bicarbonate buffer (pH 8.0). Peptides were extracted by serial addition and collection of 5 % formic acid (FA) in H2O, 50 % acetonitrile/45 % H2O/5 % FA, and 95 % acetonitrile/5 % FA. Supernatant was vacuum centrifuged to dryness. Prior to LC–MS analysis, samples were re-suspended in 97.5 % H2O/2 % acetonitrile/0.5 % FA. Chromatography was performed using a Nano-LC Ultra 2D+ (Eksigent, Dublin, CA, USA) equipped with a Proteopep 2 IntegraFrit trapping column (100 μm i.d. × 2.5 cm; C18, 5 μm, 300 Ǻ) and a Proteopep 2 IntegraFrit analytical column (75 μm i.d. × 10 cm; C18, 5 μm, 300 Ǻ, New Objective, Woburn, MA, USA). Samples were loaded onto the trap column at 2 μl/min (solvent A) for 12 min, after which a valve was switched to include the analytical column. Peptides were then eluted with a gradient (300 nl/min) of 2 % B to 45 % B over 50–80 min (Solvent A: 97.5 % H2O, 2 % acetonitrile, 0.5 % formic acid; Solvent B: 1.5 % H2O, 98 % acetonitrile, 0.5 % formic acid). Nano-LC effluent was analyzed on-line by positive-ion micro-electrospray with a linear ion trap (LTQ XL) or LTQ OrbiTrap XL (Thermo Fisher Corp, Pittsburgh, PA, USA) with “top-5 data-dependent” acquisition. Resulting data was searched against the UniProt Mus musculus FASTA database (Concatenated Random) with MASCOT (Matrix Science). Identified peptides and proteins were validated and visualized with Scaffold 3.6 (Proteome Software, Portland, OR) at a 2 % false-positive rate.
Immunoblotting
Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membrane (EMD, Millipore, Billerica, MA, USA). Proteins were detected on the blots using the following primary antibodies: BBS5 [polyclonal antibody (Proteintech, Chicago, IL, USA) or monoclonal antibody (prepared as described below)], arrestin1 (polyclonal antibody gift from Paul Hargrave), creatine kinase B (Santa Cruz Biotechnology, Dallas, TX, USA), and glutathione-S-transferase (Rockland Immunochemicals, Gilbertsville, PA, USA). Immunoreactive bands were detected with anti-rabbit or anti-mouse secondary antibodies conjugated to alkaline phosphatase, using nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate substrate (Life Technologies Corp, Carlsbad, CA, USA) or enzyme-linked chemiluminescence (WesternBreeze, Life Technologies Corp, Carlsbad, CA, USA).
Protein expression and purification
Recombinant BBS5 protein was expressed and purified from BBS5 cDNA amplified by reverse-transcription PCR from poly(A)+ RNA isolated from murine retina. Flanking EcoRI sites were incorporated on the 5′ and 3′ ends, including a His(6) tag after the initiating ATG. The cDNA was cloned into pET-28a and pGEX-4T-1 for expression as a His-tagged protein or as a fusion protein with glutathione-S-transferase (GST), respectively. His-tagged BBS5 and BBS5–GST fusion were purified from inclusion bodies over His-GraviTrap columns (GE Healthcare, Piscataway, NJ, USA) in 6 M guanidine hydrochloride (GHC). For immunoprecipitation experiments, the BBS5–GST protein was refolded by first dialyzing the protein in drip dialysis buffer (0.1 M Tris pH 7.0, 0.4 M l-arginine, 2 mM EDTA, 500 mM NaCl) with 1 M GHC, and then subsequently diluting the GHC by slowly dripping in drip dialysis buffer without GHC until the final concentration of GHC reached 0.1 M. The refolded protein was then dialyzed into 20 mM HEPES (pH 7.5) with 5 mM MgCl2, 1 mM CaCl2, and 100 mM NaCl. For samples requiring GST protein, GST was expressed from the pGEX-4T-1 vector, purified over GST GraviTrap (GE Healthcare, Piscataway, NJ, USA), and dialyzed in the above HEPES buffer.
Antibody preparation
For monoclonal antibody production, 8 to 12 week-old female BALB/c mice (Jackson Laboratory, Bar Harbor, ME, USA) were injected intraperitoneally with 50 μg purified His-tagged BBS5 in Sigma Adjuvant System oil (Sigma-Aldrich, St. Louis, MO, USA). Two booster immunizations of 50 μg of the same antigen in Sigma-Aldrich Adjuvant System were given, and 3 days before spleens were taken for fusion, an injection of 50 μg His-tagged BBS5–GST antigen in saline was administered intraperitoneally. Hybridoma cell lines were prepared by fusion of SP2/0 mouse myeloma cells with the splenocytes from the immunized mice using 50 % polyethylene glycol 1500 (Roche Applied Science, Indianapolis, IN, USA). The culture supernatants from the resulting hybridomas were screened for anti-BBS5 activity via ELISA. Positive cultures were expanded, subcloned, isotyped (Southern Biotech, Birmingham, AL, USA), and further characterized by immunoblots, immunohistocytochemistry, and immunoprecipitation (see Supplemental Fig. 1 for validation of the anti-BBS5 monoclonal antibody).
Immunoprecipitation
Aqueous-soluble extracts were prepared from bovine rod outer segments. Briefly, purified rod outer segments (prepared in dim red light according to [16]), were suspended in LAP200N buffer (50 mM HEPES pH 7.4, 200 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 10 % glycerol, 0.05 % NP-40) with 1 mM ATP and 1× phosphatase inhibitor (Thermo Scientific, Asheville, NC, USA) and were either maintained in the dark (30 min), exposed to light for 30 min, or treated with 100 μM phorbol-12,13-diacetate for 30 min. The outer segments were then sonicated and centrifuged (30 min, 40,000 × g) to produce a cleared extract. This extract was incubated with protein G-coated magnetic beads (Life Technologies Corp, Carlsbad, CA, USA) loaded with purified anti-BBS5 monoclonal antibody, washed with LAP200N buffer, and eluted with 0.1 M glycine (pH 2.5). Control beads were loaded with anti-actin monoclonal antibody (Sigma-Aldrich, St. Louis, MO, USA). Eluates were analyzed by immunoblotting, detecting Arr1 in the samples with an anti-Arr1 polyclonal antibody and BBS5 with an anti-BBS5 polyclonal antibody (ProteinTech, Chicago, IL, USA).
For immunoprecipitation of BBS5 in vitro, magnetic beads were loaded with anti-GST antibody (Rockland Immunochemicals, Gilbertsville, PA, USA). For fluorimetric quantitation, fluorescently labeled Arr1 was prepared using purified His-tagged Arr1 with an S199C substitution to generate a reactive sulfhydryl group (prepared as previously described [17]), and labeled with AlexaFluor 546-maleimide (Life Technologies Corp, Carlsbad, CA, USA). Samples containing 0.6 μM BBS5–GST and 1.2 μM arrestin–Alexa546 were incubated with the antibody-coated beads in LAP200N buffer for 4–16 h at 4 °C. After washing with LAP200N buffer, proteins were eluted with 0.1 M glycine (pH 2.5), neutralized with 1.5 M Tris (pH 8.8) and fluorescence quantified (PTI QM-1 steady-state fluorescence spectrophotometer, 530 nm excitation, 550–580 nm emission, Birmingham, NJ, USA). Control samples included reactions with no BBS5–GST protein or with 0.6 μM GST. Averages of fluorescence emission peaks from immunoprecipitated Arr1 were statistically compared to control samples containing GST (Student’s t test).
Phosphorylation of BBS5 in vitro
Phosphorylation of refolded BBS5–GST protein was attempted in vitro using either protein kinase C (PKC, Promega Corp, Madison, WI, USA), cAMP-dependent protein kinase (PKA, Promega Corp, Madison, WI, USA), cGMP-dependent protein kinase (PKG, Promega Corp, Madison, WI, USA), casein kinase I (CKI, Promega Corp, Madison, WI, USA), casein kinase II (CKII, Promega Corp, Madison, WI, USA), or calcium/calmodulin-dependent protein kinase II (CAMKII, Life Technologies Corp, Carlsbad, CA, USA). Reactions were performed with 3 μM BBS5–GST, 10 μM ATP/32P-γATP, and 20–1,000 U protein kinase in 20 mM HEPES pH 7.5 with 5 mM MgCl2, 1 mM CaCl2, and 100 mM NaCl. Reactions were incubated at 30 °C for 20 min. To remove autophosphorylated kinases, the reactions were immunoprecipitated with anti-GST antibody as described above prior to SDS-PAGE separation and autoradiography.
Immunohistochemistry
Fluorescence microscopy was performed on eyes of adult wild-type C57BL/6 mice as described previously [18]. Briefly, cryosections were placed on poly-l-lysine-pre-coated coverslips and incubated for 20 min at RT with 0.01 % Tween 20 in phosphate-buffered saline (PBS). After a PBS washing step, sections were incubated with blocking solution (0.5 % cold-water fish gelatin plus 0.1 % ovalbumin in PBS) and incubated for 2 h followed by overnight incubation with primary antibodies diluted in blocking solution at 4 °C. After washing with PBS, sections were incubated with secondary antibodies conjugated to AlexaFluor 488 or AlexaFluor 568 (Life Technologies Corp, Carlsbad, CA, USA) in PBS and with DAPI (Sigma-Aldrich, St. Louis, MO, USA) to stain the DNA of the cell nuclei. Sections were mounted in Mowiol 4.88 (Carl Roth GmbH, Karlsruhe, Germany), and imaged with a Leica DM-6000B microscope. Images were obtained with a charge-coupled device camera (DFC 360FX, Leica, Wetzlar, Germany) and processed with Adobe Photoshop CS. Fluorescence confocal microscopy was performed on cryosections of Xenopus retina as previously described [14]. Primary antibodies used were anti-BBS5 mouse monoclonal (isotype IgG2a, see above and Supplemental Fig. 1), anti-BBS5 rabbit polyclonal (ProteinTech, Chicago, IL, USA), anti-Arr1 rabbit polyclonal (gift from Paul Hargrave), and anti-Xenopus arrestin4 (xCAR#2-41, IgG1 [19]). Secondary antibodies utilized were anti-mouse IgG1–AlexaFluor 488, anti-mouse IgG2a–AlexaFluor 647, and anti-rabbit–AlexaFluor 594. To demonstrate specificity of the monoclonal antibody for BBS5, the antibody was preabsorbed with excess purified His-tagged BBS5 prior to addition to retinal sections. Images were acquired using an UltraVIEW VoX, 3D spinning disk laser confocal microscope, equipped with a Yokogawa CSU-X1 spinning disk scanner (PerkinElmer, Waltham, MA, USA), and images processed with Volocity 3D Imaging software (PerkinElmer, Waltham, MA, USA).
Immunoelectron microscopy
For immunoelectron microscopy, we applied previously described protocols for pre-embedding labeling [20, 21] and post-embedding immunogold labeling [22]. Ultrathin sections were cut on a Leica Ultracut S microtome. Ultrastructural analysis was performed using a Tecnai 12 BioTwin transmission electron microscope (FEI, Eindhoven, Netherlands). Images were obtained with a charge-coupled device SIS Megaview3 SCCD camera (Olympus, Shinjuka, Tokyo, Japan) and processed with Adobe Photoshop CS.
Results
Identification of BBS5 as a phosphoprotein
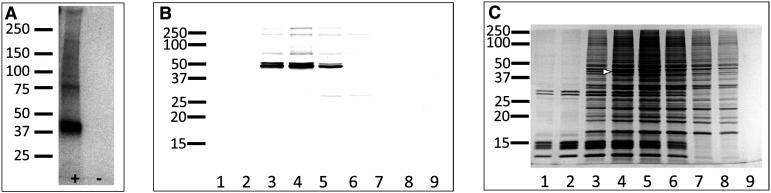
Because of previous findings showing that PLC/PKC signaling was involved in initiating Arr1 translocation in rod photoreceptors [13], we attempted to extend these observations by identifying potential PKC substrates in the photoreceptors. To accomplish this goal, dark-adapted murine retinal eye cups were pre-incubated with 32P-γATP and then stimulated with 100 μM phorbol ester to activate PKC. Under these conditions, the primary phosphorylated species in the aqueous-soluble fraction is a band at approximately 40 kDa, with several minor bands at higher molecular weights (Fig. 1a). To identify this protein, a soluble extract was prepared from multiple murine retinas stimulated with 100 μM phorbol ester, fractionated by anion exchange chromatography, and probed for phosphorylated species with anti-phosphoserine/phosphothreonine proteins by immunoblotting (Fig. 1b). The predominant immunoreactive bands were excised for mass spectrometric identification (Fig. 1c). Table 1 shows the results obtained from the analysis of fractions 4 and 5, showing all proteins for which a minimum of four peptides was identified in duplicate samples. Of the four proteins identified in both samples, only Bardet–Biedl syndrome 5 (BBS5) and creatine kinase B meet the criteria of being represented in both samples and being within 10 kDa of the expected 40-kDa size identified for the phosphoprotein. To discriminate between these two proteins, immunoblots were performed on parallel samples, comparing the size of the phosphoprotein with that of BBS5 and creatine kinase B (Fig. 2). These results clearly show that the BBS5 immunoreactive band precisely corresponds to the phosphoprotein, whereas creatine kinase B is significantly larger.
Fig. 1.
Retinal proteins phosphorylated by PKC stimulation with phorbol ester. a Autoradiogram of murine retinal extract prepared after stimulating dark-adapted eye cups with 100 μM phorbol-12,13-acetate in 5 % DMSO (+) or with 5 % DMSO alone (−) in the presence of 32P-γATP; retinal homogenates were separated on 4–15 % gradient PAGE. b Immunoblot of retinal extract fractionated on DEAE resin, probed with anti-phosphoserine/phosphothreonine antibodies; fractions 1 through 9 were separated on 12 % SDS-PAGE. c Fractions 1 through 9 were run in parallel to those in “b” and were stained with Coomassie brilliant blue; white arrowhead indicates the band excised for mass spectrometric analysis; molecular mass indicators are shown to the left in kilodaltons
Table 1.
Results of mass spectrometric analysis of phosphoproteins isolated from phorbol-ester stimulated murine retina
| Identified protein | Accession number | MW (kDa) | # Peptides (sample 1) | # Peptides (sample 2) |
|---|---|---|---|---|
| Bardet–Biedl syndrome 5 | A2AUC6|A2AUC6_MOUSE | 37 | 7 | 7 |
| Creatine kinase B-type | Q04447|KCRB_MOUSE | 43 | 6 | S |
| Vimentin | P20152|VIME_MOUSE | 54 | 4 | 8 |
| Brain acid soluble protein 1 | Q91XV3|BASP1_M0USE | 22 | 7 | 5 |
Replicate samples were submitted for LC–MS/MS. Results are shown in which both samples returned a minimum of four peptides matching any particular protein
Fig. 2.
The phosphoprotein is BBS5. a Autoradiograph of aqueous extracts prepared from dark-adapted murine eye cups incubated with 32P-γATP and stimulated with 100 μM phorbol-12,13-acetate; samples were separated on 4–15 % gradient SDS-PAGE. b Immunoblot of same soluble extract probed with anti-BBS5 antibody. c Western blot of same soluble extract probed with anti-creatine kinase B antibody
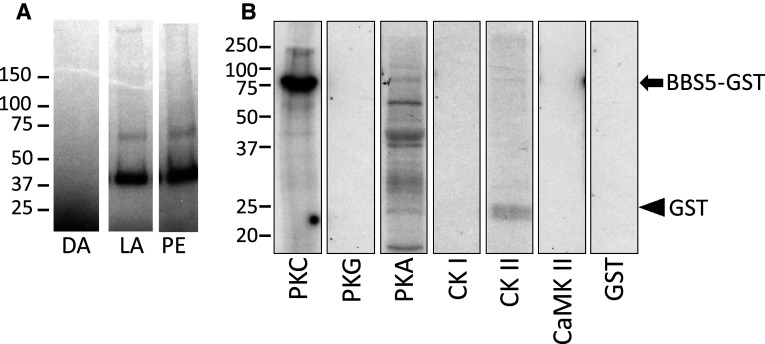
To verify that our findings using phorbol ester stimulation of retina have physiological relevance and to demonstrate broader species relevance, we also examined the phosphorylation of proteins in response to light stimulation. For this experiment, dark-adapted Xenopus tadpole eyes were pre-incubated with 32P-γATP and then exposed to either light (1,000 lux) or 0.1 mM phorbol ester, and the aqueous-soluble fraction analyzed (Fig. 3a). In both cases, light stimulation and phorbol ester treatment of the eyes led to phosphorylation of the same band at approximately 40 kDa.
Fig. 3.
Phosphorylation of BBS5 in situ and in vitro. a Autoradiogram of aqueous-soluble extracts prepared from Xenopus tadpole eyes incubated with 32P-γATP, and maintained in the dark (DA), or stimulated with either 15-min light exposure (LA) or with 0.1 mM phorbol-12,13-acetate for 15 min (PE); samples were separated on 4–15 % gradient SDS-PAGE. b Autoradiogram of BBS5/GST fusion protein incubated with the indicated protein kinases in the presence of 32P-γATP. BBS5/GST (black arrow) is phosphorylated only in the presence of PKC. The final lane shows absence of phosphorylation of purified GST (black arrowhead) in the presence of PKC. Molecular mass standards are indicated to the left (kDa). PKC protein kinase C, PKG cGMP-dependent protein kinase, PKA cAMP-dependent protein kinase, CKI casein kinase I, CKII casein kinase II, CaMKII calcium/calmodulin-dependent protein kinase II
Although phorbol esters canonically activate PKC, there are examples where phorbol esters can activate other protein kinases [23–25]. To verify that BBS5 is phosphorylated by PKC, we expressed and purified BBS5 as a fusion protein with GST and then attempted to phosphorylate the BBS5/GST protein in vitro with various protein kinases (Fig. 3b). In this experiment, only PKC led to robust phosphorylation of the BBS5 fusion protein. Importantly, note that the GST protein by itself is not phosphorylated by PKC, indicating that the phosphorylation of the BBS5–GST fusion protein is on the BBS5 portion of the fusion and not on the GST portion.
Distribution of BBS5 in retina
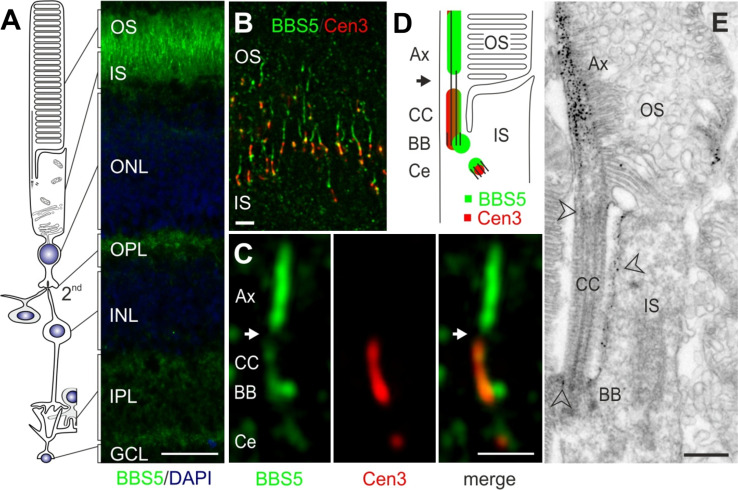
To better understand the potential function of BBS5, we examined its distribution in retina particularly focusing on its photoreceptor localization. In longitudinal sections across the entire mouse retina, BBS5 immunoreactivity was prominent over the boundary between the inner and outer segments, but was also evident in the outer and inner segments, as well as the inner and outer plexiform layers, and ganglion cell layer (Fig. 4a). At higher magnification, and utilizing co-staining with centrin-3 antibody as a centriolar/ciliary marker, it is clear that BBS5 reactivity localized not only to the basal body and the adjacent centriole but also to the outer segment axoneme (Fig. 4b–d). Additional faint labeling of BBS5 was present in the connecting cilium. Curiously, there was a gap in BBS5 immunoreactivity between the connecting cilium (transition zone) and the basal part of the axoneme. Using immunoelectron microscopy to visualize BBS5 localization, a similar distribution is revealed (Fig. 4e). At this level of resolution, it is clear that the most abundant location of BBS5 reactivity is along the axonemal portion of the cilium extending into outer segments. Labeling is nearly absent from the connecting cilia of the photoreceptor cells, which is consistent with previous findings on other ciliary proteins using the pre-embedding labeling protocol [26]. Interestingly, there is significant immunoreactivity in the periciliary region of the inner segments, particularly along the surface adjacent to the connecting cilium.
Fig. 4.
Localization of BBS5 protein in murine retina. a Indirect immunofluorescence of longitudinal cryosections through unfixed mouse retina stained for BBS5 polyclonal antibody (green) and DAPI as a marker for the nuclei. BBS5 protein is abundant in the photoreceptor layer including the photosensitive outer segment (OS), the connecting cilium (CC), in the inner segment (IS), and the synaptic compartments of the outer (OPL) and inner plexiform layer (IPL), and ganglion cell layer (GCL). Bar 25 μm; outer nuclear layer (ONL); inner nuclear layer (INL). b Merged image of indirect immunofluorescence double labeling of longitudinal cryosections through mouse photoreceptor cell OS and IS, with antibodies to BBS5 (green) and the ciliary marker centrin-3 (red) demonstrates BBS5 localization in the ciliary apparatus of photoreceptor cells. Bar 1 μm. c High-magnification image of immunofluorescence double labeling of BBS5 and centrin-3 in the region of the photoreceptor cell CC. BBS5 reactivity partially co-localized with centrin-3 at the basal body (BB) and the adjacent centriole (Ce) and localized to the outer segment axoneme (Ax); white arrow indicates an apparent gap in BBS5 localization. Bar 1 μm. d Schematic illustration of the connecting cilium area of a rod photoreceptor cell. Centrin-3 markings are shown in red, BBS5 markings are shown in green. e Immunoelectron microscopic localization of BBS5 in longitudinal sections through a part of a mouse rod photoreceptor cells. BBS5 is abundantly localized to the Ax. Arrowheads indicate additional labeling in the CC and in the BB region of the cilium as well as the periciliary membrane in the apical part of the IS. Bars 0.5 μm
BBS5 localization under different lighting conditions
Considering that BBS5 phosphorylation is stimulated by light, and considering that one of the effects of PKC activation is to initiate the translocation of Arr1 from the inner segments to the outer segments, we investigated the distribution of BBS5 and Arr1 in both light- and dark-adapted conditions. In dark-adapted Xenopus retina, BBS5 strongly localized to the inner segment, axonemes of the outer segments, and outer plexiform layer of both rod and cone photoreceptors (Fig. 5a). We note that Xenopus photoreceptors appear to have more BBS5 signal in the inner segments than in mouse (compare to Fig. 4). Following light adaptation, the distribution of BBS5 appeared qualitatively the same, although with perhaps an intensification of BBS5 reactivity over the axonemes and in the outer plexiform layer (Fig. 5c). We also note an apparent change in the distribution of BBS5 along the axoneme, with BBS5 appearing more uniformly distributed along the axoneme in light-adapted rods (Fig. 5c), whereas the distribution is somewhat punctate in rod photoreceptors that are dark adapted (Fig. 5a). Note that during light adaptation, there is retinomotor movement of the cone cell bodies proximally to join the cell body region of the rods, a phenomenon noted previously in fish and amphibian retinas by other researchers (reviewed in Burnside and Nagle [27]). Similar to BBS5 distribution, in dark-adapted retina Arr1 is predominantly localized to the inner segments, outer plexiform layer, and axonemes of the outer segments (Fig. 5a′′). Particularly noteworthy is the nearly precise co-localization of the immunoreactivity of both Arr1 and BBS5 along the axonemes (Fig. 5a′). In contrast to BBS5, however, during light adaptation, Arr1 relocalizes in rods to the outer segments (Fig. 5c′′). Indirect immunofluorescence analysis of BBS5 distribution in the mouse retina under these same light conditions confirmed the data obtained in Xenopus (data not shown). Immunoelectron microscopy analysis confirms the fluorescent immunohistochemical findings in rodents and Xenopus photoreceptor cells (Fig. 6). In dark-adapted photoreceptor cells, Arr1 remains concentrated in the axoneme of the outer segment (Fig. 6a, a′, c, d) where BBS5 is also found (Fig. 6b).
Fig. 5.

Immunolocalization of BBS5 in Xenopus distal retina. a Cryosections of dark-adapted Xenopus retina were stained with anti-BBS5 monoclonal (green) and anti-Arr1 rabbit polyclonal antibody (a′′; red). The merge image (a′) indicates co-localization of BBS5 and Arr1 along the axoneme and inner segments (IS) of the rod photoreceptors (white arrowheads mark two axonemes that are stained for both Arr1 and BBS5). b Additional cells that also stained for BBS5 (white arrow in both a, b), but not for Arr1, were identified as cones by staining for arrestin4 using a monoclonal anti-cone arrestin4 with an isotype-specific secondary (green). c In cryosections of light-adapted Xenopus retina, BBS5 (green) remains along the axonemes and inner segments, whereas Arr1 (red) principally localizes to the rod outer segments (OS). d A control section stained using BBS5 monoclonal (green) pre-absorbed to heterologously expressed BBS5 protein demonstrates specificity of the monoclonal antibody. Scale bar 20 μm
Fig. 6.
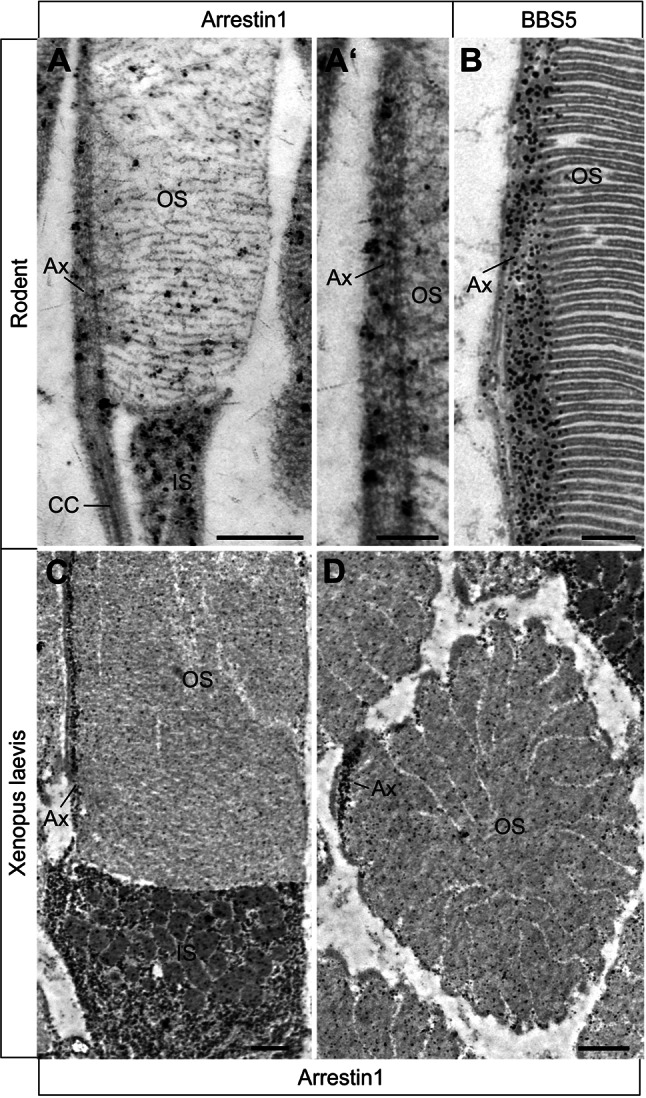
Immunoelectron microscopic localization of Arr1 and BBS5 in dark-adapted rod photoreceptor cells. (a, a′) Immunoelectron microscopic localization of Arr1 in longitudinal sections of rodent rods at the axonemal region shows Arr1 is predominantly found in the inner segment (IS) and along the axoneme (Ax) of the outer segment (OS). b BBS5 labeling on a longitudinal section of through a portion of OS, showing heavy labeling by anti-BBS5 along the Ax. c, d Immunoelectron microscopic localization of Arr1 in dark-adapted Xenopus rod photoreceptor cells. c In longitudinal section, Arr1 immunoreactivity is principally localized to the IS and along the axoneme (Ax) of the OS. d In a cross section through the OS, Arr1 reactivity is significantly concentrated at the axoneme. Bars a 0.5 μm, a′ 0.2 μm, b 0.2 μm, c, d 1 μm
BBS5 interaction with Arr1
Considering the co-localization of Arr1 with BBS5 that we observed along the axoneme, we investigated whether there might be a direct interaction between Arr1 and BBS5. To address this question, we performed immunoprecipitation of BBS5 from extracts prepared from dark-adapted bovine rod outer segments (Fig. 7). In this pull down, Arr1 co-precipitated with beads loaded with anti-BBS5 antibody (lanes 1 and 2), but not in the control samples loaded with anti-actin antibody (lanes 7 and 8). Arr1 also co-precipitated with BBS5 in extracts prepared from outer segments that were either exposed to light or treated with 100 μM phorbol ester (lanes 3–6), although the amount of Arr1 was reduced by nearly 50 % (ρ < 0.05) compared to extracts prepared from outer segments maintained in the dark.
Fig. 7.
Arrestin1 co-immunoprecipitates with BBS5 in rod outer segments. a Arr1 was detected in immunoblots of samples of bovine rod outer segment lysates immunoprecipitated with anti-BBS5 antibody. In the upper panel, ROS extracts were immunoprecipitated with BBS5 monoclonal antibody (lanes 1–6) or with an anti-actin monoclonal antibody (lanes 7–8). ROS extracts were either maintained in constant dark (lanes 1–2), exposed to light (lanes 3–4), or exposed to 0.1 mM phorbol ester (lanes 5–6); ROS extracts for the control samples were maintained in the dark (lanes 7–8). The lower panels are immunoblots of the ROS lysate input used in each sample, using anti-Arr1 polyclonal antibody to detect Arr1 input levels (middle blot) and BBS5 input levels (lower blot). b Quantitation of the relative amounts of Arr1 co-precipitated with BBS5 in each of the three treatments in the upper panel of a; samples exposed to light or phorbol ester were statistically compared to the dark samples, with statistical significance (ρ < 0.05) indicated by an asterisk (n = 4)
The experiment illustrated in Fig. 7 demonstrates that BBS5 and Arr1 associate in outer segments, but does not necessarily indicate whether this interaction is direct or is part of a complex, incorporating one or more scaffolding intermediates. To address this question, we performed immunoprecipitation in an in vitro reconstituted assay, using BBS5–GST fusion protein purified from expression in bacteria and fluorescently labeled Arr1 purified from expression in yeast (Fig. 8). Under these conditions, BBS5–GST was very effective at pulling down Arr1, particularly when compared to the GST control that contained just GST without BBS5 fusion. We also immunoprecipitated Arr1 with BBS5–GST protein that was pre-phosphorylated with PKC. Phosphorylated BBS5–GST immunoprecipitated significantly less Arr1 than unphosphorylated BBS5 fusion protein (ρ < 0.05), reducing pulldown of Arr1 by 25 %.
Fig. 8.
Arrestin1 directly interacts with BBS5. Fluorescently labeled Arr1 (S199C-Alexa546) was incubated with either BBS5–GST (BBS5), phosphorylated BBS5–GST (p-BBS5), GST, or with no GST-containing protein (control), and pulled down with anti-GST-coated magnetic beads and fluorescence quantified. Asterisk indicates statistically significant difference between samples (ρ < 0.05), n = 8. Inset shows an immunoblot of the immunoprecipitated samples from one replicate probed for Arr1 to validate the fluorimetric quantitation
BBS5 phosphorylation and Arr1 translocation
To better understand if BBS5 phosphorylation might have any bearing on Arr1 translocation, we undertook a study of the effects of lighting intensity on BBS5 phosphorylation in situ using transgenic Xenopus tadpoles expressing GFP-tagged Arr1 [14]. Eye cups were prepared from tadpoles and either incubated in 32P-γATP or simply maintained in tadpole Ringer’s solution, and were exposed to increasing intensities of lighting for 15 min, ranging from 0 to 5,000 lux. The radiolabeled eyes were processed for SDS-PAGE and autoradiography and the contralateral eye fixed for confocal microscopy. Figure 9a shows that there was no detectable phosphorylation of the 40-kDa BBS5 band at 0–10 lux; whereas there was increasing and prominent phosphorylation of this band at higher lighting intensities. In parallel eyes that were processed for confocal imaging, translocation of the Arr1–GFP fusion protein from the inner segment to the outer segment was initiated at the same lighting intensity as stimulated the phosphorylated of BBS5 (Fig. 9b, c).
Fig. 9.
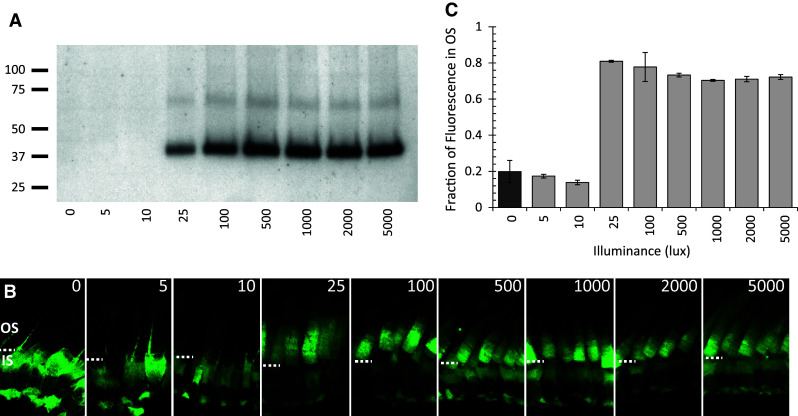
Phosphorylation of BBS5 correlates with the same light intensity that initiates translocation of Arr1 to the outer segments. a One eye from each Arr1–GFP tadpole was incubated with 32P-γATP and was exposed to increasing intensities of light (0–5,000 lux) for 15 min, homogenized, separated by SDS-PAGE, and autoradiographed; molecular mass standards (in kDa) are indicated to the left. b The contralateral eye was exposed to the same lighting and then fixed for imaging of GFP fluorescence in the photoreceptors by confocal microscopy. The dashed white line indicates the boundary between the outer segments (OS) and inner segments (IS). c Quantification of Arr1 translocation to the outer segments; the fraction of GFP fluorescence in the OS was measured and averaged from a minimum of 15 photoreceptors in two separate sections from each of two eyes
Discussion
Principal findings
Our most significant findings in this study are threefold. First, we show that BBS5 is a member of the phosphoproteome of the retina, being phosphorylated in a light-dependent manner by PKC. This mass spectrometric identification was validated by immunoblotting, and also by using heterologously expressed BBS5 to show that this protein is a suitable substrate for phosphorylation by PKC in vitro. The phosphorylation of BBS5 by both light and PKC agonist (phorbol ester) establishes this post-translational modification as being physiologically relevant. Furthermore, its identification in both mice and frogs indicates that this regulation of BBS5 is likely to be a feature common to a broad range of vertebrate organisms. Bardet–Biedl syndrome 5 protein is one member of a complex of proteins termed the BBSome, a complex of at least eight highly conserved proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBIP10) that principally localize to primary cilia [28]. Studies in model organisms indicate that the BBSome plays a key role in ciliary transport of proteins, regulating access of proteins to the cilium [29–31]. In photoreceptors, it is clear that the BBSome plays a similar role in both ciliary biogenesis and ciliary protein trafficking [28, 32]. A particular role for BBS5 in this process has not yet been ascribed, although recent studies have shown that BBS5 is one of the components added later to the core elements of the BBSome complex during its assembly [33]. Phosphorylation of BBS5 has not been reported previously and its effect on the association of BBS5 with other BBSome components is now an obvious track for investigations.
Our findings localizing BBS5 to the basal body/pericentriolar region in photoreceptors are consistent with these published findings. However, in our studies, we find that in addition to the previously noted basal body localization, BBS5 is also concentrated along the axoneme of both rod and cone photoreceptors. This is a new finding for the distribution of a BBSome component in photoreceptors. Interestingly, in a mouse model lacking BBS4, there is a misalignment of disc membranes along the axoneme [34], perhaps consistent with a role for the BBSome proteins along the axoneme in directing disc alignment.
Our third principal finding is the demonstration that Arr1 directly interacts with BBS5, adding to the growing list of interacting partners for Arr1. In addition to its well-established interaction with rhodopsin [35], Arr1 has also been previously shown to interact with multiple protein partners, including N-ethylmaleimide-sensitive factor [36], enolase1 [37], tubulin [38, 39], apoptosis signal-regulating kinase 1, kinases in the mitogen-activated protein kinase pathway [40, 41], calmodulin [42], and ubiquitin ligases [41, 43–45]. This novel binding partner for Arr1 is significant not only for understanding the full spectrum of Arr1 interactions but also because of the key role played by the ciliary structure in rod and cone photoreceptors. Our data suggest that the axoneme not only serves as a structural element that is used by photoreceptors for maintaining proper organization of the outer segment disc stacks or a transport track for longitudinal molecular delivery of outer segment molecules [12, 46], but it also appears that the axonemal cytoplasm serves as a reservoir for Arr1 in the dark, maintaining a concentration of Arr1 in the outer segments that can rapidly diffuse to the disc membranes in response to light exposure when BBS5 is phosphorylated.
Relationship to translocation
This project was initiated with the intent of discovering how Arr1 translocation might be triggered by PKC activation [13]. Our discovery of BBS5 as a light-stimulated PKC target suggests that this phosphorylation step might be a key step in the regulation of Arr1 translocation. Although our findings are not conclusive in this regard, the evidence supporting this interpretation is several-fold. First, we show that BBS5 is located along the axoneme, precisely co-localizing with Arr1. One longstanding mystery of Arr1 localization in rod photoreceptors has been its strong localization not only in the inner segment of dark-adapted rods but also along the axoneme, particularly visible in the larger amphibian retinas [11, 14, 47]. The direct interaction between Arr1 and BBS5 shown by our findings suggests that BBS5 may serve as a binding partner for Arr1 along the axonemal structure, which is a non-membrane delimited structure that extends nearly to the apical end of the outer segment. Second, the observation that the affinity of Arr1 to BBS5 is reduced by PKC-dependent phosphorylation of BBS5 is consistent with the reduced localization of Arr1 to the axoneme observed in light-adapted rods in which BBS5 would be phosphorylated. This finding suggests that phosphorylation of BBS5 may promote the dissociation of Arr1 from the axoneme. Finally, our findings show a direct correlation between the illumination intensity that initiates BBS5 phosphorylation and that which initiates Arr1 translocation to the outer segments. Interestingly, in our in vitro reconstituted pull-down assays using heterologously expressed BBS5 and Arr1, the effect of BBS5 phosphorylation on Arr1 pull down was not as great as in the assay using endogenous BBS5 and Arr1 in outer segments. Although there could be many explanations, one possibility to consider is that Arr1 may be binding to BBS5 along the axoneme in association with other proteins in a complex, such that phosphorylation of BBS5 may have broader effects on this complex than just directly on its interaction with Arr1. The possibility of such a complex is under further investigation. In total, these findings provide a compelling link for BBS5 in process of Arr1 translocation in rods.
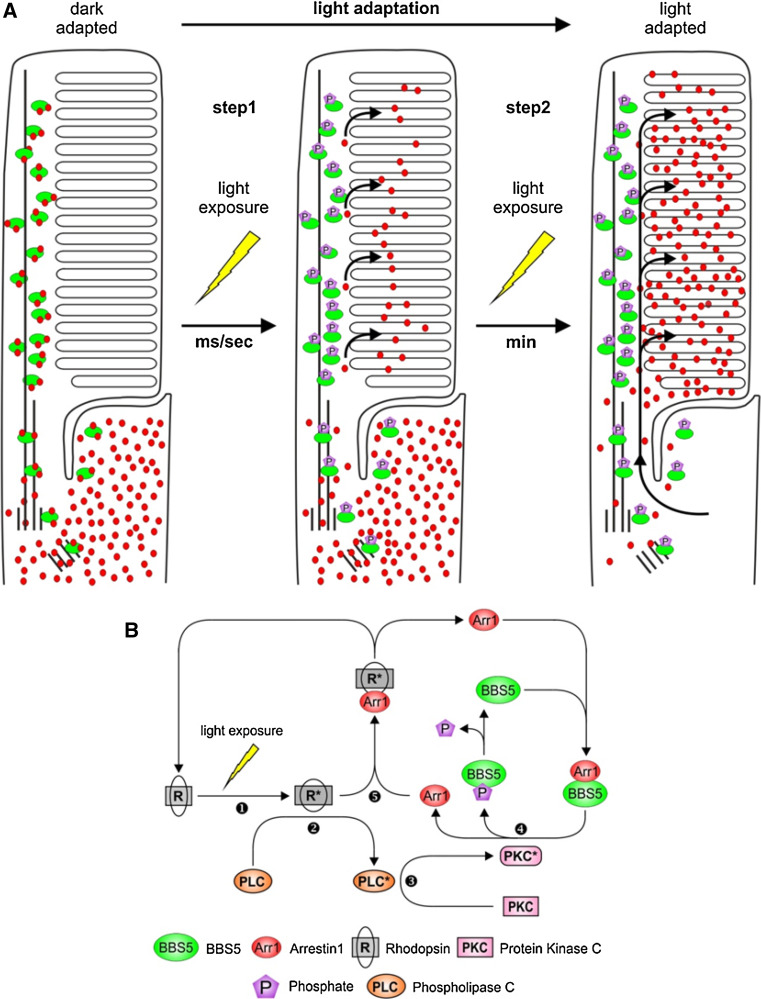
Thus we propose the following model for light-dependent translocation of Arr1 (Fig. 10). In the dark-adapted condition, Arr1 localizes to the inner segments and axonemes as a consequence of multiple factors, including steric occlusion of Arr1 tetramers and direct binding of Arr1 to multiple interaction partners, such as enolase1, NSF, tubulin, and BBS5; the localization to the axoneme in the outer segments is a consequence of binding to BBS5. In our model, the light-driven translocation of Arr1 to the outer segment is a two-step process. In the first step, light activation of the phospholipase C/PKC signaling cascade leads to phosphorylation of BBS5, which reduces the affinity of Arr1 for its axonemal association, thus permitting Arr1 to diffuse throughout the outer segment disks and then bind to light-activated rhodopsin. In response to a dim flash in the dark-adapted state, this fast recruitment of Arr1 from the axonemal pool of Arr1 would assure the rapid quenching of activated rhodopsin, independent from recruiting molecules of Arr1 from the pool of Arr1 in the inner segment, which would certainly take much longer. In the second step of Arr1 translocation, Arr1 moves rapidly from its pool in the inner segment by diffusion, unrestricted by phosphorylated BBS5, to bind to light-activated phospho-rhodopsin for which it has a high affinity. This model explains the earlier observation that PKC agonists lead to a temporary release of Arr1 into the outer segments of dark-adapted retinas, but which does not remain in the outer segments [13], presumably because there is no light-activated rhodopsin to which to bind. One consequence of this model is that binding of Arr1 to BBS5 during dark adaptation could potentially retard the flow of Arr1 to the inner segment; however, because the amount of Arr1 greatly exceeds that of BBS5, the pool of BBS5 would quickly become saturated. Further, it is also possible that the dephosphorylation of BBS5 is slower than the deactivation/dephosphorylation of rhodopsin such that the bulk of Arr1 translocates to the inner segment prior to the dephosphorylation of BBS5. Future investigations will focus on testing this model. Although there are no knockout models of BBS5 currently available, knock down of gene expression or specifically altering the interaction between Arr1 and BBS5 are rational approaches for ascertaining the role of this interaction in regulating Arr1 translocation.
Fig. 10.
Model for BBS5-dependent arrestin1 localization in rod photoreceptor cells in response to illumination. a In dark-adapted rods, Arr1 (red dots) predominantly localizes to an Arr1 pool in the inner segment (IS). In outer segment (OS), Arr1 is retained by binding to non-phosphorylated BBS5 (green ovals) along the axoneme. In the first step, upon light exposure, BBS5 is phosphorylated and releases Arr1 into the OS disks for shutting down activated rhodopsin signaling; in the subsequent second step, the bulk of Arr1 in the photoreceptor moves by diffusion from the inner segment to the outer segment, driven by binding to activated rhodopsin. b Schematic representation of the signal cascade leading to Arr1 release throughout the OS. Light activation of rhodopsin (R) (1), signaling through an undefined G-protein, leads to activation of phospholipase C (PLC) (2) and protein kinase C (PKC) (3), which phosphorylates BBS5 along the axoneme (4). Arr1 has reduced affinity to phospho-BBS5 and can now diffuse throughout the OS and can interact with activated rhodopsin (R*) (5)
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. Validation of BBS5 monoclonal antibody. A) Immunoblot of His(6)-tagged BBS5 (lane 1) and BBS5-GST fusion protein (lane 2) probed with anti-BBS5 monoclonal antibody; parallel gel stained with Coomassie brilliant blue showing the purified His-BBS5 (lane 3) and BBS5-GST (lane 4). B) Aqueous soluble extracts were prepared from Xenopus and mouse whole retina homogenates, separated on 12 % SDS-PAGE, and either transferred to PVDF membrane for immunoprobing with anti-BBS5 monoclonal antibody (lanes 1 and 2) or stained with Coomassie brilliant blue (lanes 3 and 4); molecular mass standards indicated in kilodaltons. (DOCX 568 kb)
Acknowledgments
This research was supported by grants from the National Eye Institute (EY014864, EY006225, EY021721), Research to Prevent Blindness, Inc., University of Florida Faculty Enhancement Opportunity grant, C.M. Overstreet Retinal Eye Disease ARMD Research Fund, EU FP7/2009/241955 “SYSCILIA” and the FAUN Foundation, Nuremberg, Germany.
References
- 1.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 2.Brann MR, Cohen LV. Diurnal expression of transducin mRNA and translocation of transducin in rods of rat retina. Science. 1987;235:585–587. doi: 10.1126/science.3101175. [DOI] [PubMed] [Google Scholar]
- 3.Broekhuyse RM, Tolhuizen EFJ, Janssen APM, Winkens HJ. Light induced shift and binding of S-antigen in retinal rods. Curr Eye Res. 1985;4:613–618. doi: 10.3109/02713688508999993. [DOI] [PubMed] [Google Scholar]
- 4.Philp NJ, Chang W, Long K. Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett. 1987;225:127–132. doi: 10.1016/0014-5793(87)81144-4. [DOI] [PubMed] [Google Scholar]
- 5.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elias R, Sezate S, Cao W, McGinnis J. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol Vis. 2004;10:672–681. [PubMed] [Google Scholar]
- 7.Calvert PD, Schiesser WE, Pugh EN. Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. J Gen Physiol. 2010;135:173–196. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci USA. 2012;109:203–208. doi: 10.1073/pnas.1115109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peet JA, Bragin A, Calvert PD, Nikonov SS, Mani S, Zhao X, Besharse JC, Pierce EA, Knox BE, Pugh EN. Quantification of the cytoplasmic spaces of living cells with EGFP reveals arrestin-EGFP to be in disequilibrium in dark adapted rod photoreceptors. J Cell Sci. 2004;117:3049–3059. doi: 10.1242/jcs.01167. [DOI] [PubMed] [Google Scholar]
- 10.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson JJ, Orisme W, Fellows J, McDowell JH, Shelamer CL, Dugger DR, Smith WC. A role for cytoskeletal elements in the light-driven translocation of proteins in rod photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:3988–3998. doi: 10.1167/iovs.05-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reidel B, Goldmann T, Giessl A, Wolfrum U. The translocation of signaling molecules in dark adapting mammalian rod photoreceptor cells is dependent on the cytoskeleton. Cell Motil Cytoskelet. 2008;65:785–800. doi: 10.1002/cm.20300. [DOI] [PubMed] [Google Scholar]
- 13.Orisme W, Li J, Goldmann T, Bolch S, Wolfrum U, Smith WC. Light-dependent translocation of arrestin in rod photoreceptors is signaled through a phospholipase C cascade and requires ATP. Cell Signal. 2010;22:447–456. doi: 10.1016/j.cellsig.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson JJ, Tam BM, Moritz OL, Shelamer CL, Dugger DR, McDowell JH, Hargrave PA, Papermaster DS, Smith WC. Arrestin migrates in photoreceptors in response to light: a study of arrestin localization using an arrestin-GFP fusion protein in transgenic frogs. Exp Eye Res. 2003;76:553–563. doi: 10.1016/S0014-4835(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.McDowell JH. Preparing rod outer segment membranes, regenerating rhodopsin, and determining rhodopsin concentration. Meth Neurosci. 1993;15:123–130. doi: 10.1016/B978-0-12-185279-5.50013-3. [DOI] [Google Scholar]
- 17.McDowell JH, Smith WC, Miller RL, Popp MP, Arendt A, Abdulaeva G, Hargrave PA. Sulfhydryl reactivity demonstrates different conformational states for arrestin, arrestin activated by a synthetic phosphopeptide, and constitutively active arrestin. Biochemistry. 1999;38:6119–6125. doi: 10.1021/bi990175p. [DOI] [PubMed] [Google Scholar]
- 18.Wolfrum U. Centrin in the photoreceptor cells of mammalian retinae. Cell Motil Cytoskelet. 1995;32:55–64. doi: 10.1002/cm.970320107. [DOI] [PubMed] [Google Scholar]
- 19.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Functional specialization of beta2–arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maerker T, van Wijk E, Overlack N, Kersten FFJ, McGee J, Goldmann T, Sehn E, Roepman R, Walsh EJ, Kremer H, Wolfrum U. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- 21.Sedmak T, Sehn E, Wolfrum U, Roger DS (2009) Immunoelectron microscopy of vesicle transport to the primary cilium of photoreceptor cells. In: Methods in cell biology. Academic Press, New York, pp 259-272 [DOI] [PubMed]
- 22.Wolfrum U, Liu X, Schmitt A, Udovichenko IP, Williams DS. Myosin VIIa as a common component of cilia and microvilli. Cell Motil Cytoskelet. 1998;40:261–271. doi: 10.1002/(SICI)1097-0169(1998)40:3<261::AID-CM5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Hambleton J, Weinstein SL, Lem L, DeFranco AL. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages. Proc Natl Acad Sci USA. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson KR, Becker KP, Facchinetti MM, Hannun YA, Obeid LM. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane: extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA) J Biol Chem. 2002;277:35257–35262. doi: 10.1074/jbc.M203033200. [DOI] [PubMed] [Google Scholar]
- 25.Crusius K, Auvinen E, Alonso A. Enhancement of EGF- and PMA-mediated MAP kinase activation in cells expressing the human papillomavirus type 16 E5 protein. Oncogene. 1997;15:1437–1444. doi: 10.1038/sj.onc.1201312. [DOI] [PubMed] [Google Scholar]
- 26.Sedmak T, Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol. 2010;189:171–186. doi: 10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burnside B, Nagle B. Retinomotor movements of the photoreceptors and retinal pigment epithelium: mechanisms and regulation. In: Osborne N, Chader G, editors. Progress in Retinal Research. New York: Pergamon Press; 1983. pp. 67–109. [Google Scholar]
- 28.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 29.Jin H, Nachury MV. The BBSome. Curr Biol. 2009;19:R472–R473. doi: 10.1016/j.cub.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Mockel A, Perdomo Y, Stutzmann F, Letsch J, Marion V, Dollfus H. Retinal dystrophy in Bardet–Biedl syndrome and related syndromic ciliopathies. Prog Retin Eye Res. 2011;30:258–274. doi: 10.1016/j.preteyeres.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, Hu J. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol. 2012;14:950–957. doi: 10.1038/ncb2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC. Intrinsic protein–protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet–Biedl syndrome protein complex, the BBSome. J Biol Chem. 2012;287:20625–20635. doi: 10.1074/jbc.M112.341487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kϋhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 36.Huang S-P, Brown BM, Craft CM. Visual arrestin 1 acts as a modulator for N-ethylmaleimide-sensitive factor in the photoreceptor synapse. J Neurosci. 2010;30:9381–9391. doi: 10.1523/JNEUROSCI.1207-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith WC, Bolch S, Dugger DR, Li J, Esquenazi I, Arendt A, Benzenhafer D, McDowell JH. Interaction of arrestin with enolase1 in photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:1832–1840. doi: 10.1167/iovs.10-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 39.Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song X, Gurevich EV, Gurevich VV. Cone arrestin binding to JNK3 and Mdm2: conformational preference and localization of interaction sites. J Neurochem. 2007;103:1053–1062. doi: 10.1111/j.1471-4159.2007.04842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2–arrestin interaction but inhibits arrestin ubiquitination. Biochem. 2011;50:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song X, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roepman R, Wolfrum U. Protein networks and complexes in photoreceptor cilia. Subcell Biochem. 2007;43:209–235. doi: 10.1007/978-1-4020-5943-8_10. [DOI] [PubMed] [Google Scholar]
- 47.McGinnis JF, Matsumoto B, Whelan JP, Cao W. Cytoskeleton participation in subcellular trafficking of signal transduction proteins in rod photoreceptor cells. J Neurosci Res. 2002;67:290–297. doi: 10.1002/jnr.10120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Validation of BBS5 monoclonal antibody. A) Immunoblot of His(6)-tagged BBS5 (lane 1) and BBS5-GST fusion protein (lane 2) probed with anti-BBS5 monoclonal antibody; parallel gel stained with Coomassie brilliant blue showing the purified His-BBS5 (lane 3) and BBS5-GST (lane 4). B) Aqueous soluble extracts were prepared from Xenopus and mouse whole retina homogenates, separated on 12 % SDS-PAGE, and either transferred to PVDF membrane for immunoprobing with anti-BBS5 monoclonal antibody (lanes 1 and 2) or stained with Coomassie brilliant blue (lanes 3 and 4); molecular mass standards indicated in kilodaltons. (DOCX 568 kb)