Abstract
Traditional drug targets have historically included signaling proteins that respond to small-molecules and enzymes that use small-molecules as substrates. Increasing attention is now being directed towards other types of protein targets, in particular those that exert their function by interacting with nucleic acids or other proteins rather than small-molecule ligands. Here, we systematically compare existing examples of inhibitors of protein–protein interactions to inhibitors of traditional drug targets. While both sets of inhibitors bind with similar potency, we find that the inhibitors of protein–protein interactions typically bury a smaller fraction of their surface area upon binding to their protein targets. The fact that an average atom is less buried suggests that more atoms are needed to achieve a given potency, explaining the observation that ligand efficiency is typically poor for inhibitors of protein– protein interactions. We then carried out a series of docking experiments, and found a further consequence of these relatively exposed binding modes is that structure-based virtual screening may be more difficult: such binding modes do not provide sufficient clues to pick out active compounds from decoy compounds. Collectively, these results suggest that the challenges associated with such non-traditional drug targets may not lie with identifying compounds that potently bind to the target protein surface, but rather with identifying compounds that bind in a sufficiently buried manner to achieve good ligand efficiency, and thus good oral bioavailability. While the number of available crystal structures of distinct protein interaction sites bound to small-molecule inhibitors is relatively small at present (only 21 such complexes were included in this study), these are sufficient to draw conclusions based on the current state of the field; as additional data accumulate it will be exciting to refine the viewpoint presented here. Even with this limited perspective however, we anticipate that these insights, together with new methods for exploring protein conformational fluctuations, may prove useful for identifying the “low-hanging fruit” amongst non-traditional targets for therapeutic intervention.
Introduction
The majority of modern drugs modulate the function of a relatively small number of protein targets that include enzymes, G-protein coupled receptors (GPCRs), ion channels, transporters, and nuclear hormone receptors 1, 2. With the exception of proteases, each of these broad classes of protein have evolved to bind a cognate small-molecule, and most therapeutics disrupt activity by competitively binding to the same region of the protein surface as the natural interaction partner. These surface binding pockets are typically deep and present a well-defined shape that often complements the natural substrate 3. In such cases, mimicry of the natural substrate (or transition state) may serve as an attractive starting point for designing new inhibitors 4. Given that new inhibitors often have similar chemical properties as endogenous ligands, identification of one or more natural ligands with drug-like physicochemical characteristics can also be used to infer the “druggability” of a new protein target 5.
In contrast to these “traditional” protein targets for therapeutic intervention, there are also a tantalizing number of well-validated “non-traditional” potential targets. These proteins have evolved to bind not small-molecules but rather other macromolecules, and include targets involved in protein–DNA interactions (e.g., 6, 7), protein–RNA interactions (e.g., 8, 9), and protein–protein interactions (e.g., 10, 11). The interaction surfaces of these proteins are often large and flat, lacking a deep pocket suitable for small-molecule binding 3. Given the size of the natural substrate in these cases, examples of mimicry by small molecules leading to potent inhibitors are few 12-17, and even in these successful cases the resulting inhibitors tend to be larger than typical orally available drugs 18, 19.
Structure-based virtual screening methods offer a means to directly identify novel inhibitory compounds that complement the target protein surface 20; these methods are not limited by the requirement for template compound(s) implicit to ligand-centric (mimicry) approaches 21. In the simplest terms, virtual screening requires some method for sequentially positioning each candidate compound from a library at its most likely position on the protein surface (i.e., “docking”), followed by a subsequent discrimination step (i.e., “scoring”) to rank each of the resulting complexes based on their likelihood of showing the desired activity.
The historical focus on inhibiting proteins evolved to bind a cognate small-molecule (both enzymes and proteins involved in signaling) has led to an understanding of many structural features exhibited by such complexes 22. These insights have been facilitated in part by databases such as MOAD 23, 24, which have enabled comparisons to reveal subtle differences in enzyme versus non-enzyme classes of “traditional” drug targets 25.
The question now arises whether the same structural features apply to inhibitors that bind at “non-traditional” sites (i.e., those not evolved for small-molecule binding). To identify any systematic differences between these two broad classes of targets is critical, since structure-based virtual screening methods absolutely require these insights to appropriately rank docked complexes and select the most promising compounds for experimental characterization. Since very few examples of direct inhibitors of nucleic acid binding sites have been described, here we instead focus on small-molecule inhibitors of protein–protein interactions. While the widespread impression in the field is that small-molecule inhibitors of protein–protein interactions tend to be larger and contribute less binding affinity per atom than inhibitors of traditional drug targets, this viewpoint is predicated largely on a study that described a relatively small number of examples 26. Here we seek to carry out a more thorough quantitative evaluation of properties that distinguish each class of inhibitor, then ask how these differences affect the performance of virtual screening tools when applied to protein–protein interaction sites.
Results
Extent of ligand burial in inhibitory complexes
For this study we compiled a set of 21 unique protein–protein interaction sites for which a crystal structure has been solved in complex with a small-molecule inhibitor (Table 1), which we will refer to as the “PPI set.” We compared properties of these representative complexes to those of the Astex Diverse Set from Gold 27, which contains crystal structures of proteins of pharmaceutical or agrochemical interest each bound to a small-molecule inhibitor with drug-like chemical properties. The chemical properties of the ligands in the PPI set are comparable to those of the Astex set (the construction and composition of both sets are described in the Methods section).
Table 1. Inhibitors bound to protein interaction sites, the PPI set.
Potency is taken from reported Kd or Ki values where available; if unavailable, IC50 values were used instead. In two cases (“-NR-”), no measure of potency has been reported. θlig indicates the fraction of ligand SASA exposed in the complex, as defined in Equation 1.
| Protein target | PDB id | Inhibitor molecular weight (Da) | Potency (μM) | Ligand efficiency (kcal/mol per non-hydrogen atom) | θlig |
|---|---|---|---|---|---|
| ZipA | 1y2f | 424 | 12 | 0.22 | 0.57 |
| HPV E2 | 1r6n | 608 | 0.18 | 0.22 | 0.56 |
| XIAP-BIR3 | 1tft | 535 | 0.005 | 0.29 | 0.52 |
| HIV-gp41 | 2kp8 | 580 | 14 | 0.15 | 0.51 |
| S100B | 3gk1 | 279 | 80 | 0.29 | 0.49 |
| IL-2 | 1pw6 | 534 | 6 | 0.2 | 0.47 |
| PCNA | 3vkx | 651 | 3 | 0.33 | 0.47 |
| Grb2-SH2 | 3in7 | 556 | -NR- | -NR- | 0.46 |
| Menin | 4gq4 | 415 | 0.022 | 0.39 | 0.45 |
| VHL | 3zrc | 410 | 5.4 | 0.24 | 0.45 |
| TNFα | 2az5 | 548 | 22 | 0.16 | 0.45 |
| calmodulin | 1ctr | 408 | 1 | 0.29 | 0.43 |
| clathrin | 4g55 | 475 | 12 | 0.24 | 0.42 |
| Mdm2 | 4erf | 478 | 0.0004 | 0.4 | 0.41 |
| SHANK PDZ | 3o5n | 304 | 17.2 | 0.29 | 0.40 |
| integrin | 2vc2 | 523 | -NR- | -NR- | 0.40 |
| BRD4 | 2yel | 423 | 0.0155 | 0.33 | 0.39 |
| calpain | 1alw | 308 | 0.3 | 0.68 | 0.38 |
| Bcl-xL | 1ysi | 552 | 0.036 | 0.27 | 0.38 |
| HIV integrase | 4e1n | 438 | 0.019 | 0.33 | 0.37 |
| WDR5 | 3ur4 | 383 | 0.45 | 0.31 | 0.34 |
It has been noted anecdotally that inhibitors of protein-protein interactions bind at flatter regions of the protein surface than do inhibitors of “traditional” drug targets 3,26 To systematically characterize whether the ligand is less buried upon binding, we defined a parameter θlig that quantifies the fraction of ligand solvent accessible surface area (SASA)28 that remains exposed upon binding:
| (Eqn. 1) |
where SASAprotein is the SASA of the protein with the ligand removed, SASAligand is the SASA of the ligand with the protein removed, and SASAcomplex is the total SASA of the protein-ligand complex. We note that SASAprotein and SASAligand are each computed directly from the structures that comprise the complex, and not from other unbound crystal structures.
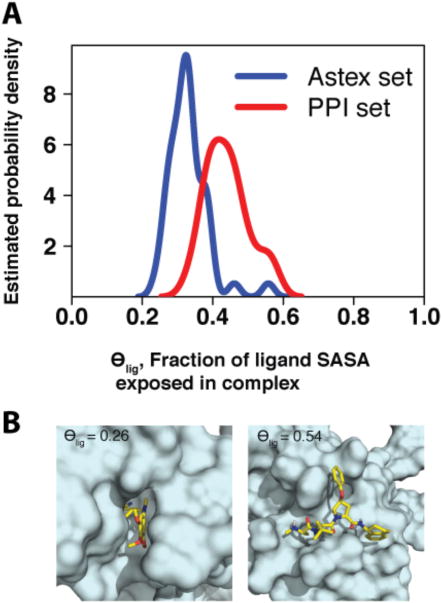
We computed θlig for each complex in the Astex set and the PPI set; the results of this comparison are presented in Figure 1. It is immediately evident that the bound inhibitors at protein interaction sites retain more exposed surface area than their counterparts which bind at sites evolved for small-molecule binding (high θlig for traditional targets); this observation further holds for other analogous sets of druglike complexes (DUD-E 29 and SB2010 30, Figure S1). Among the members of the Astex set, we find that the class of protein targets with the highest θlig values is serine proteases (βII tryptase, factor Xa, factor VIIa, thrombin, urokinase; see Table S1). These enzymes contribute four of the ten highest θlig values in the Astex set, corresponding to statistically significant enrichment of this target class (p < 0.01). Although they are enzymes, the natural substrates of serine proteases are proteins rather than small-molecules; for this reason, it is perhaps unsurprising that θlig values for such complexes resemble those associated with protein interaction sites more than those associated with traditional drug targets.
Figure 1.
(A) Distribution of θlig values in the PPI and Astex data sets. Ligands bound at protein interaction sites (red, median value 0.45) tend to be more exposed than drug-like compounds (blue, median value 0.33), with a difference of means that is statistically significant (p < 10-6). As described in the text, proteases are an exception in the latter set. (B) Representative examples of complexes with low and high θlig (PDB IDs 1s3v and 1tft, respectively).
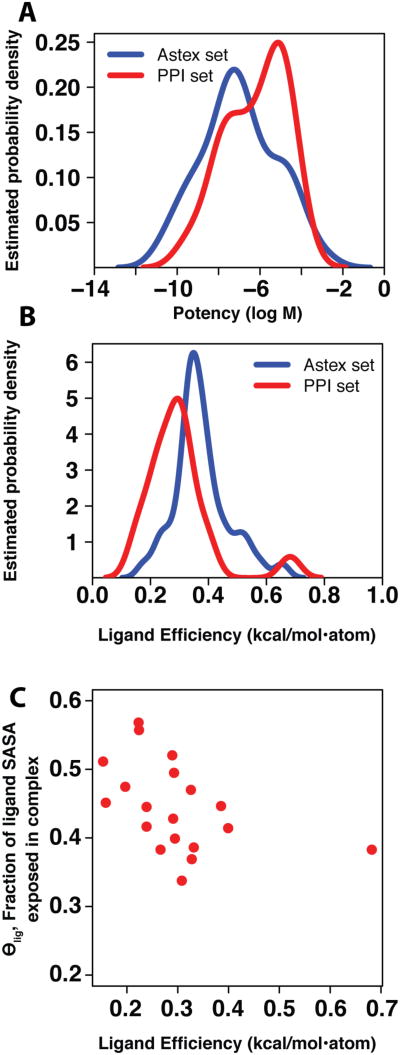
Since the set of physical forces that underlie binding must be the same for both classes of complexes, we next asked how this difference in exposed surface area influences binding affinity. We compared the potency of each complex, and found the distributions from the two sets to be essentially the same (Figure 2a). Due to the larger compound size required to achieve this potency (Figure S2), however, we find that inhibitors acting at protein interaction sites bind with less ligand efficiency (binding energy per non-hydrogen atom 31) than orally active compounds acting on traditional targets (Figure 2b). A single outlier with high ligand efficiency in the PPI set is evident, corresponding to an inhibitor of calpain (1alw in Table 1, with ligand efficiency 0.68 kcal/mol per non-hydrogen atom). Further inspection reveals this to be partially an artifact of the way in that ligand efficiency is defined, since this inhibitor contains a bromine atom: while the potency of this compound is high given its size, the ligand efficiency is further exaggerated because it is normalized using the number of non-hydrogen atoms rather than molecular weight. Excluding this outlier, we observe a clear relationship between θlig and ligand efficiency in the PPI set (Figure 2c), but not in the Astex set (Figure S3). Due to the variation in the molecular weight of the compounds within each set, this relationship is not apparent when simply examining potency as a function of θlig (Figure S4).
Figure 2.
(A) Distribution of potency values in the Astex and PPI sets. Our set of ligands bound at protein interaction sites (red, median value 1.0 μM) have similar potency to drug-like compounds (blue, median value 0.07 μM). No statistically significant difference in means is observed (p = 0.105). (B) Distribution of ligand efficiencies in the Astex and PPI sets. The ligand efficiency of inhibitors bound at protein interaction sites (red, median value 0.29 kcal/mol•atom) tends to be lower than the ligand efficiency of drug-like compounds (blue, median value 0.36 kcal/mol•atom), and the difference in the means is statistically significant (p < 0.007, or p < 0.0002 upon removal of the single bromine-containing outlier described in the text). (C) The relationship between θlig and ligand efficiency in the PPI set. As expected, there is a negative correlation between these properties, with a statistically significant non-zero Spearman rank correlation coefficient (p < 0.006).
These results are in agreement with a previous report drawn from a much smaller set of protein targets 26, and are also consistent with our observation that the bound inhibitors at protein interaction sites retain more exposed surface area: an average atom buries less hydrophobic surface area upon binding, so the contribution of an average atom to the binding energy—the very definition of ligand efficiency—is expected to be lower 31.
Properties of complexes produced by virtual screening
We have shown above that our collection of inhibitors binding at protein interaction sites have similar potency as a traditional drug-like set, but that affinity is achieved through a structurally distinct mode of interaction. It is therefore critical to examine how well modern energy functions perform in these contrasting regimes.
To evaluate the ability of a representative energy function to distinguish known inhibitors from a large number of “decoy” compounds, we carried out a mock virtual screening experiment using the FRED software package 32-34. Starting from the ZINC database 35, we constructed a “decoy library” of 10,000 compounds with chemical properties (molecular weight and XlogP) matched to the inhibitors in the Astex and PPI sets. To eliminate potential challenges associated with sampling, we used OMEGA 36-38 to build up to 300 conformers of each compound to be used for docking. We also included the active conformer of the known inhibitor (taken from the crystal structure of the protein–ligand complex) in our virtual screen. The exact active conformer of an inhibitor is generally not present in a screening library, and adding it to the set should, at least in theory, simplify the screening problem. As an indicator of how favorably a protein's known inhibitor is scored, we use its rank relative to the members of the decoy library. As an important caveat, we note that the decoy compounds are not necessarily inactive. Nonetheless, we expect that even if the decoy library does contain a small fraction of compounds that are active, the known inhibitor should rank with these among the top scoring compounds.
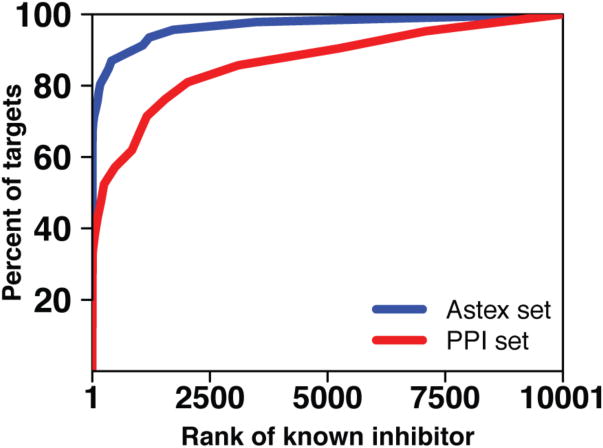
We carried out this screening experiment for each complex in the Astex set and the PPI set. Within each set we collected, at increasing values of a rank threshold, the fraction of targets for which the known inhibitor was ranked better than the threshold (Figure 3). As can be appreciated from this figure, a perfect method, which would rank the known inhibitor at the top of the list for every target, would lead to a vertical rise at the very left of the curve. In contrast, a completely random method would be expected to rank the known inhibitor in the top 1% for 1% of the targets, in the top 10% for 10% of the targets, etc., leading to a curve that follows the diagonal of the plot area.
Figure 3.
To test the ability of virtual screening tools to distinguish known inhibitors for the protein targets in our test sets, we embedded each inhibitor in a set of 10,000 “decoy” compounds and screened this library against each protein target. For each target, we sorted the docked scores for each member of the screening library and determined the rank of the known inhibitor. In this figure, we plot a cumulative histogram of the percent of protein targets for which the known inhibitor is ranked better than the threshold value indicated on the x-axis. Rather than comparing results for individual protein targets, this aggregate representation allows comparison of performance between the two test sets (Astex and PPI). Here we find that this virtual screening tool has difficulty identifying known inhibitors that bind at protein interaction sites (red) relative to its performance in the drug-like set (blue), and that the difference in the mean rank of the known inhibitor between the two sets is statistically significant (p < 10-6).
The first notable observation is that FRED performs exceptionally well for traditional targets (the Astex set) in this intentionally “easy” experiment. Using illustrative thresholds, the known inhibitor is ranked within the top 2% of the library (within the top 200 of 10,000 compounds) for 80% of the protein targets in the Astex set, suggesting that indeed the known inhibitor is near optimal given the scoring function. In contrast, the known inhibitor is ranked in the top 2% of the library for only 50% of the targets in the PPI set. As shown in Figure 3, this difference in performance holds irrespective of the threshold applied—in other words, performance on the Astex set is superior regardless of whether one counts “successes” as protein targets for which the known inhibitor is ranked in the top 5%, top 10%, top 25%, etc. Given the design of this experiment, the dramatically worse performance for the PPI set points strongly to a deficiency in discrimination of the correctly bound inhibitor from bound decoy compounds if the known inhibitor retained a large fraction of exposed surface area upon binding (high θlig).
To eliminate the possibility that this difference in performance originated from a bias in the composition of our decoy library (e.g., our library could be comprised of decoys that are viable inhibitors of protein interaction sites but easy to rule out as enzyme inhibitors), we carried out an analogous experiment in which we used the DUD-E server 29 to build a separate set of 50 decoy compounds for each target, matched to the chemical properties of the known inhibitor. We observe the same performance difference between the Astex set and the PPI set in this experiment (Figure S5) as in the previous screening experiment. The observed difference in performance between these two sets of target proteins holds when another software package, DOCK 6.6 39, is used to carry out the virtual screen (Figure S6), indicating that particular details specific to FRED are not responsible for the observed differences.
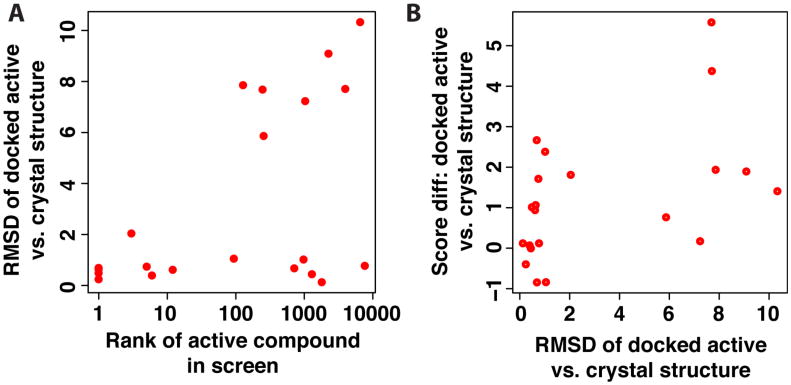
Since this screening experiment was explicitly designed to be “easy” with respect to sampling (for example, by including the active conformer taken from the crystal structure), we expected that the poorer performance on the PPI set stemmed from difficulty assigning the correctly docked pose a suitably favorable score. To test this, we collected the RMSD of the top-scoring docked pose of the active compound relative to its position in the crystal structure. We find the active compounds that (correctly) scored amongst the very top of the library collection were inevitably correctly docked (low RMSD) (Figure 4a); this is unsurprising, since one might expect that correctly predicting the pose is usually a pre-requisite for correctly identifying an active compound. Of the active compounds which did not score in the top 1% of the library, approximately half were correctly docked (RMSD less than 2 Å) with the other half dramatically mis-docked (RMSD greater than 5 Å). We then scored each of the crystal structures of the same complexes, and computed the difference in score relative to the docked pose. While in a few cases the crystal structures scored slightly better that nearly-correct docked poses, each of the mis-docked poses scored better than the crystal structure (Figure 4b, high RMSD points all have positive score differences). Collectively these results suggest that the screening challenges presented in the PPI set lie not with sampling, but rather with assigning the correctly docked ligand a suitably favorable score: relative to decoy compounds, and also relative to mis-docked poses of the ligand.
Figure 4.
(A) For each of the active compounds in the PPI set, the root mean square deviation (RMSD) of the docked active compound is computed relative to the crystal structure, and reported as a function of the rank of the active compound in the virtual screening experiment. Approximately half the compounds that were not ranked highly were mis-docked (high RMSD), while the other half were correctly docked but still did not rank well relative to the decoy compounds (low RMSD but high rank). (B) The difference in score between the docked active compound and the crystal structure is shown as a function of the RMSD. The mis-docked structures scored better than the crystal structure in all cases (positive score differences), suggesting that the energy function did not provide the correct relative ranking of these two poses.
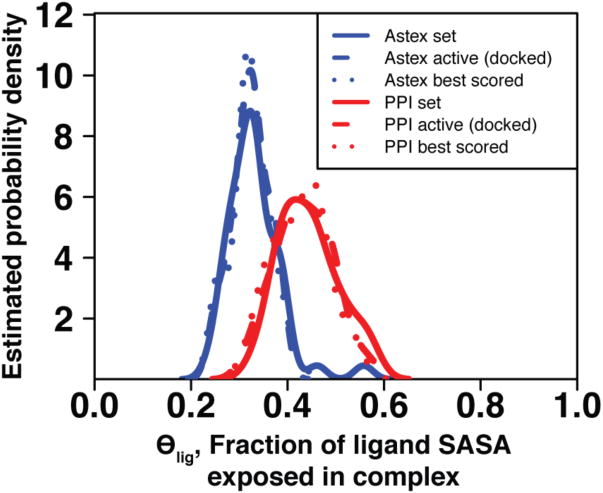
We then selected the top-ranked compound from each screen, whether it was the known inhibitor or a decoy, and evaluated θlig in the docked complex. Intriguingly, the distributions of these values match the corresponding distributions from complexes solved with the known inhibitors (Figure 5). In other words, though the known inhibitors from the PPI set were often not highly ranked in this screening experiment, the highly ranked decoys nonetheless retained a large fraction of exposed surface area upon binding (high θlig). Thus, the discrimination problem in the PPI set is not a bias in favor of low θlig complexes over high θlig complexes, but rather a failure to discriminate the known (high θlig) inhibitor from decoy compounds that also have high θlig values. In contrast, the more extensive burial of compounds in the Astex set may lend additional clues for correctly identifying the cognate inhibitor, explaining the relative ease in correctly identifying inhibitors for these targets. These observations further suggest that the protein conformation itself is the primary determinant of θlig values in inhibitory complexes: unless a pocket suitable for extensive ligand burial is present on the protein surface, it is simply not possible to have an inhibitor with low θlig. This constraint, together with the poor ligand efficiency associated with high θlig values, suggests limited druggability of these particular protein conformations.
Figure 5.
The distribution of ligand solvent exposure for the best-scoring compound in each test set is similar to the analogous distribution for the experimentally-derived complexes of known inhibitors in the corresponding test set. Within a set distributions are very similar for crystal structures of known inhibitors, the known inhibitor docked back to the corresponding protein target, and the top-scoring compound from among 10,000 “decoy” compounds.
We next sought to determine whether this conclusion applies to alternate conformations of these particular protein interaction sites. We have recently described an approach for exploring protein fluctuations enriched in conformations containing surface pockets suitable for small molecule binding 40. We therefore carried out the same experiment described above, this time screening against either the unbound protein structure, the structure of the protein in complex with its protein partner, or a structure generated via biased simulations. We find similar distributions of θlig for the top-scoring complex when screening against these conformations as well (Figure 6), suggesting that these too lack pockets suitable for extensive ligand burial that are required for complexes with low θlig. On the basis of this result, computational screening against these alternate protein conformations is unlikely to yield inhibitors with dramatically higher ligand efficiency.
Figure 6.
The distribution for the extent of ligand solvent exposure (θlig) for the best-scoring compound in a screen against PPI targets is similar regardless of whether the protein conformation used in docking corresponds to an inhibitor-bound structure (red), the unbound structure (green), a protein–bound structure (brown), or a structure generated from simulations biased towards pocket-containing conformations 40 (pink).
Discussion
Previous analyses have shown that known to be inhibitors of protein–protein interactions can be systematically distinguished from drug-like compounds on the basis of size and shape 41, 42, and also that the deep pockets present on traditional drug targets (sites evolved for small-molecule binding) are typically absent at protein interaction sites 3, 43. Here, we demonstrate the differences in the components of the inhibitory complexes—the compound and the protein target—are manifest in the structural features of the complexes themselves.
Our results further demonstrate that modern virtual screening methods typically are less suited for identifying inhibitors of protein interactions than for identifying inhibitors of traditional drug targets. The difficulty in discriminating active compounds from decoy compounds may be due to an incomplete representation of the underlying physical forces that govern binding in this alternate structural regime; or, more simply, these methods may have been parameterized for optimal performance when applied to traditional drug targets at the expense of performance on non-traditional targets.
Nonetheless, the results from the virtual screening experiments described here demonstrate that the protein conformations tested can only harbor a relatively exposed ligand, because they present relatively flat surface pockets. This feature of bound complexes does not preclude identification of potently binding compounds; rather, it simply implies that such compounds will require many atoms to achieve this potency, and therefore may violate Lipinski's “rule-of-five” criteria for oral availability 18, 19. For enzymes, the rule-of-five compliance of the natural endogenous substrate has been shown to be a good predictor of druggability 5, since inhibitors may occupy similar chemical space. For protein targets not naturally evolved to bind small-molecules, the natural binding partner cannot be used to draw such inferences. Collectively, our results do not suggest that protein interaction sites are necessarily less “bindable” than traditional drug targets, but rather that the size of compounds required for the desired potency may make protein interaction sites intrinsically less “druggable.”
Our conclusions are also highly complementary to those reached in a previous study comparing inhibitory complexes of enzyme versus non-enzyme drug targets 25 through a survey of the MOAD database 23. Both types of target are “traditional” by our definition, in that both have evolved to bind small-molecules. Interestingly, the authors of this study find higher ligand efficiency in non-enzyme complexes, and propose that the origin of this difference stems from the fact that non-enzyme ligands (signaling molecules such as hormones) are typically more “encapsulated” than their enzyme counterparts 25. This suggestion is in direct agreement with our observations: ligand efficiency of inhibitors bound at protein interaction sites (median value 0.29 kcal/mol•atom) is lower than that of inhibitors of these “traditional” enzyme and non-enzyme targets (median 0.36 and 0.41 kcal/mol•atom, respectively 25), in keeping with the limited degree to which the protein surface can accommodate these ligands.
While inhibitors bound to the protein interaction sites studied here can only achieve limited burial (and thus poor ligand efficiency), it is important to note that the protein conformation was fixed throughout these docking experiments. This restriction of the protein surface, in turn, may have influenced our observed recapitulation of θlig values. Achieving improved ligand efficiency for ligands binding to protein interaction sites is expected to require that the protein undergo conformational changes to induce formation of alternate pocket shapes, but it is unclear a priori what range of pocket shapes are available at a given protein surface. A number of methods have recently been described to preferentially explore alternate conformations of protein surfaces suitable for small-molecule binding 40, 44, 45. While our studies presented here did not find that such conformations would be likely to bind a small-molecule with high ligand efficiency, such an approach may nonetheless prove useful in rapidly screening protein interaction sites for those targets which are not only “bindable”, but also “druggable” in this sense. Alternatively, fragment screening approaches 46—rapidly gaining popularity in campaigns targeting protein interaction sites 47—may also provide a route for identifying moieties that bind with high ligand efficiency. The existence of such fragments may imply that the protein surface can provide surface pockets suitable for extensively burying at least some ligands.
The number of crystal structures of distinct protein interaction sites bound to small-molecule inhibitors accumulated in the literature to date remains small (Table 1), and thus this study provides a snapshot of the field at present from these available examples. Accordingly, we cannot rule out design bias in these set of examples, or insufficient exploration of chemical space in this relatively new field. It will be interesting to learn, over the upcoming years, whether the challenges described in this study can be circumvented through new approaches. Meanwhile, the number of examples of small-molecules directly inhibiting binding at other types of recognition sites is even fewer. Nonetheless, we anticipate that inhibitors of these other “challenging” targets, such as protein–nucleic acid interactions, may share the same poor ligand efficiency—and thus limited druggability—as we describe here for inhibitors of protein–protein interactions. In order to access these many diverse and biologically critical targets, then, we may need to carefully select proteins amenable for complexes with low θlig, target allosteric sites that can accommodate extensively buried ligands, or develop new approaches to improve the delivery and oral accessibility for compounds outside the “rule-of-five” chemical space.
Methods
Protein data sets
To build the PPI set, we merged entries from the 2P2I 48 and TIMBAL 49 databases and then supplemented this collection with additional recent examples from our own curation of the literature. We eliminated from consideration any complexes with inhibitors of molecular weight outside the range 200 Da – 675 Da, to discard small fragments and large peptide-like compounds. We further excluded structures containing proteins with co-factors, proteins closely related in sequence to other members of our set, and all examples of covalently bound inhibitors. In cases where more than one suitable inhibitor-bound structure has been solved, we retained the structure in complex with the most potent ligand; we have carried out the experiments described here with other complexes as well and find similar results (data not shown). Our set contains compounds with molecular weight range 279 – 651 Da (mean: 468, standard deviation: 102 Da) and XlogP range of -4.9 – 6.5 (mean: 2.6, standard deviation: 2.9). Overall our set contains structures of 21 non-redundant proteins, each in complex with a unique inhibitor bound at a protein interaction site (Table 1): to our knowledge this represents the largest such collection reported to date.
The Astex diverse set contains 85 protein–ligand complexes 27, intended as examples of drug-like complexes (indeed, 23 of the ligands are approved drugs). For appropriate comparisons to our PPI set, we refined this set by removing examples containing a protein cofactor or second ligand at the binding site (typically NADPH, FAD, ATP, heme, or metal ions). We again removed any complexes with inhibitors outside the molecular weight range 200 Da – 650 Da. Overall the Astex set we used in these studies consisted of 46 binary protein–ligand complexes (Table S1), with molecular weight range 208 – 576 Da (mean: 362, standard deviation: 89 Da) and XlogP in the range -3.8 – 5.4 (mean: 1.9, standard deviation: 2.0).
Molecular properties were calculated using the MolProp toolkit version 2.1.5 (OpenEye Scientific Software, Santa Fe, NM) 50.
Computational screening
Starting from the ZINC database of commercially available compounds 35, we compiled a set of 10,000 randomly selected compounds with molecular weight between 200 and 750 Da. These compounds served as “decoy” compounds for the screening experiment (Figure 3).
In a separate experiment, we instead used the DUD-E server 29 to generate a set of 50 “decoy” compounds specifically matched to the known inhibitor of the intended target; in other words, each protein target was screened against a custom compound library built to match the chemical properties of the known inhibitor of this protein target (Figure S5).
For each decoy compound as well as for the known inhibitor, we used the OMEGA version 2.4.3 software 36-38 to generate up to 300 conformers, using default parameters. Charges were added using the QuacPac version 1.5.0 toolkit (OpenEye Scientific Software, Santa Fe, NM) 51. As described earlier, we included the ligand conformation observed in the crystal structure among the conformers for the known inhibitor.
We carried out computational docking using the FRED version 3.0.0 software 32-34, a part of the OEDocking suite (OpenEye Scientific Software, Santa Fe, NM). The binding site for each protein was defined based on the bound ligand in the crystal structure, using the OEDocking ′receptor_setup′ utility. All FRED docking runs were carried out using the default parameters and the resulting poses were ranked using the Chemgauss4 score.
Additional docking calculations were carried out using DOCK version 6.6 39. The molecular surface for each protein target was calculated using the dms program, and spheres were generated in the active site using the sphgen program (both are available as DOCK utilities). All spheres within 10 Å of the bound ligand from the crystal structure were retained. A grid was generated and the ligand was docked using the default parameters; then the grid score was used to rank each docked complex.
The same set of decoy compounds were used for the experiment using FRED and the analogous experiment using DOCK.
Alternate protein conformations
Alternate pocket-containing protein conformations were generated via biased simulations as described elsewhere 40. Briefly, we applied a biasing potential proportional to the size of the surface pocket, and carried out Monte Carlo simulations adapted from refinement protocols used in comparative modeling applications. This methodology is implemented in the Rosetta software suite 52, and is freely available for academic use (www.rosettacommons.org).
Simulations were carried out for the following list of proteins (with PDB code for the unbound starting structure used in the simulation and the “target” residue at which the biasing potential was applied): Bcl-xL (1r2d, 141), BRD4 (2oss, 146), FKBP12 (2ppn, 26), HPV E2 (1r6k, 33), IL-2 (1m47, 42), HIV integrase (3l3u, 178), Mdm2 (1z1m, 61), menin (4gpq, 278), XIAP-BIR3 (1f9x, 308), ZipA (1f46, 85). In each case 1000 output structures were generated, and virtual screening was then performed using the lowest-energy pocket-containing conformation.
Statistical analysis
Probability densities were estimated using standard kernel techniques in the R statistical computing environment 53.
The statistical significance of differences in means between distributions was evaluated using a standard two-tailed nonparametric permutation test. Briefly, this test works by combining the two data sets in question, D1 and D2 (with n1 and n2 observations, respectively) into a single data set D (with n = n1 + n2 elements). A pair of data sets, D1′ and D2′, are then created by random sampling from D. These data sets contain the same number of elements as the original data sets (i.e., n1 and n2), but are constructed from a random partitioning of the original observations. For a given randomized pair, we can calculate the difference in means as tR = | μ(D1′) - μ(D2′) |, where μ(X) is the sample mean of data set X. This procedure is repeated to generate N randomized replicates, producing a distribution of tR values. This represents an estimate of the distribution of differences in means, given the null hypothesis that D1 and D2 are drawn from distributions of the same mean.
This distribution is then compared to the observed difference in means, tobs = | μ(D1) - μ(D2) |, in order to estimate a p-value of that observed difference. In the set of N random replicates, we calculate the number of replicates Ngeq where tR ≥ tobs. The p-value is then defined as p = Ngeq / (N + 1). For all p-values quoted in this work, the test outlined above was performed using the “twotpermutation” method implemented in the DAAG package in R 53, with N = 106 replicates.
Supplementary Material
Table S1. Inhibitors bound to traditional targets, a subset of the Astex set. Potency is taken from reported Kd or Ki values where available; if unavailable, IC50 values were used instead. In several cases (“-NR-”), no measure of potency has been reported. θlig indicates the fraction of ligand SASA exposed in the complex, as defined in Equation 1.
Figure S1 (complements Figure 1) The distribution of the extent of inhibitor solvent exposure (θlig) is similar across a number of drug-like sets: Astex 27(blue), DOCK 54(magenta), DUD-E 29(brown), and SB2010 30(green).
Figure S2 (complements Figure 2) The distribution of molecular weights for the inhibitors in each set underlies the observed difference in ligand efficiencies. Inhibitors binding at protein interaction sites (red, median value 475 Da) are typically larger than their drug-like counterparts (blue, median value 355 Da), and the difference in the means is statistically significant (p < 10-4).
Figure S3 (complements Figure 2) The relationship between θlig and ligand efficiency for both the PPI set and the Astex set. While there is a statistically significant negative correlation between these properties for the PPI set (as noted in Figure 2c), no statistically significant correlation exists for the Astex set (p = 0.27).
Figure S4 (complements Figure 2) The relationship between θlig and potency for both the PPI set and the Astex set. No statistically significant correlation exists between these properties for either set (p = 0.33 for the PPI set, p = 0.50 for the Astex set).
Figure S5 (complements Figure 3) A known inhibitor was embedded in a custom set of 50 “decoy” compounds selected by the DUD-E server to match the physical properties of the known inhibitor. FRED exhibits superior ability to identify the known drug-like inhibitors from the decoy compounds (blue), relative to inhibitors that bind at protein interaction sites (red), and the difference in the means is statistically significant (p < 0.002).
Figure S6 (complements Figure 3) The observation that virtual screening at protein interaction sites performs less well than for drug-like compounds holds for other docking software as well. DOCK 6.6 was used to identify a known inhibitor embedded in a custom set of 50 “decoy” compounds selected by the DUD-E server to match the physical properties of the known inhibitor. DOCK 6.6 exhibits superior ability to identify the known drug-like inhibitors from the decoy compounds (blue), relative to inhibitors that bind at protein interaction sites (red), though this difference is not statistically significant (p = 0.11).
Acknowledgments
We are grateful to Andrea Bazzoli for critically reading the manuscript and offering helpful suggestions. We are grateful to OpenEye Scientific Software (Santa Fe, NM) for providing an academic license for the use of FRED, OMEGA, MolProp, and QuacPac. We are grateful to the developers of the DOCK software for providing an academic license for the use of this program. This work was supported by a grant from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM099959), the National Science Foundation through TeraGrid allocation TG-MCB130049, and the Alfred P. Sloan Fellowship (J.K.).
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews Drug discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nature reviews Drug discovery. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 3.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug discovery today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Naylor E, Arredouani A, Vasudevan SR, Lewis AM, Parkesh R, Mizote A, Rosen D, Thomas JM, Izumi M, Ganesan A, Galione A, Churchill GC. Identification of a chemical probe for NAADP by virtual screening. Nature chemical biology. 2009;5:220–226. doi: 10.1038/nchembio.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauman EB, Rai BK, Huang ES. Structure-based druggability assessment--identifying suitable targets for small molecule therapeutics. Current opinion in chemical biology. 2011;15:463–468. doi: 10.1016/j.cbpa.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 7.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan P, Baltimore D. Sam68 is required for both NF-kappaB activation and apoptosis signaling by the TNF receptor. Mol Cell. 2011;43:167–179. doi: 10.1016/j.molcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muto J, Imai T, Ogawa D, Nishimoto Y, Okada Y, Mabuchi Y, Kawase T, Iwanami A, Mischel PS, Saya H, Yoshida K, Matsuzaki Y, Okano H. RNA-binding protein Musashi1 modulates glioma cell growth through the post-transcriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PLoS One. 2012;7:e33431. doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its alpha receptor. Science. 2005;308:1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 11.Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J Biol Chem. 2003;278:23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- 12.Cochran AG. Antagonists of protein-protein interactions. Chem Biol. 2000;7:R85–94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 13.Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2008;27(Suppl 1):S149–157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart KD, Huth JR, Ng TI, McDaniel K, Hutchinson RN, Stoll VS, Mendoza RR, Matayoshi ED, Carrick R, Mo H, Severin J, Walter K, Richardson PL, Barrett LW, Meadows R, Anderson S, Kohlbrenner W, Maring C, Kempf DJ, Molla A, Olejniczak ET. Non-peptide entry inhibitors of HIV-1 that target the gp41 coiled coil pocket. Bioorg Med Chem Lett. 2010;20:612–617. doi: 10.1016/j.bmcl.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 15.Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JA. Beta-hairpin peptidomimetics: design, structures and biological activities. Accounts of chemical research. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]
- 17.Harker EA, Daniels DS, Guarracino DA, Schepartz A. Beta-peptides with improved affinity for hDM2 and hDMX. Bioorg Med Chem. 2009;17:2038–2046. doi: 10.1016/j.bmc.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 19.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 20.Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH. Structure-based virtual screening for drug discovery: a problem-centric review. The AAPS journal. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tresadern G, Bemporad D, Howe T. A comparison of ligand based virtual screening methods and application to corticotropin releasing factor 1 receptor. J Mol Graph Model. 2009;27:860–870. doi: 10.1016/j.jmgm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Seifert MH. Targeted scoring functions for virtual screening. Drug Discov Today. 2009;14:562–569. doi: 10.1016/j.drudis.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 24.Smith RD, Hu L, Falkner JA, Benson ML, Nerothin JP, Carlson HA. Exploring protein-ligand recognition with Binding MOAD. J Mol Graph Model. 2006;24:414–425. doi: 10.1016/j.jmgm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Carlson HA, Smith RD, Khazanov NA, Kirchhoff PD, Dunbar JB, Jr, Benson ML. Differences between high- and low-affinity complexes of enzymes and nonenzymes. Journal of medicinal chemistry. 2008;51:6432–6441. doi: 10.1021/jm8006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 27.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. Journal of medicinal chemistry. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 28.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. Journal of molecular biology. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 29.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. Journal of medicinal chemistry. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee S, Balius TE, Rizzo RC. Docking validation resources: protein family and ligand flexibility experiments. J Chem Inf Model. 2010;50:1986–2000. doi: 10.1021/ci1001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. P Natl Acad Sci USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGann M. FRED pose prediction and virtual screening accuracy. Journal of chemical information and modeling. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 33.McGann M. FRED and HYBRID docking performance on standardized datasets. Journal of computer-aided molecular design. 2012;26:897–906. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 34.FRED version 3.0.0. OpenEye Scientific Software; S. F., NM: http://www.eyesopen.com, In. [Google Scholar]
- 35.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: A Free Tool to Discover Chemistry for Biology. Journal of chemical information and modeling. 2012 doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. Journal of chemical information and modeling. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawkins PC, Nicholls A. Conformer generation with OMEGA: learning from the data set and the analysis of failures. Journal of chemical information and modeling. 2012;52:2919–2936. doi: 10.1021/ci300314k. [DOI] [PubMed] [Google Scholar]
- 38.OMEGA version 2.4.3. OpenEye Scientific Software; S. F., NM: http://www.eyesopen.com, In. [Google Scholar]
- 39.Brozell SR, Mukherjee S, Balius TE, Roe DR, Case DA, Rizzo RC. Evaluation of DOCK 6 as a pose generation and database enrichment tool. Journal of computer-aided molecular design. 2012;26:749–773. doi: 10.1007/s10822-012-9565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DK, Karanicolas J. Druggable protein interaction sites are more predisposed to surface pocket formation than the rest of the protein surface. PLoS computational biology. 2013;9:e1002951. doi: 10.1371/journal.pcbi.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neugebauer A, Hartmann RW, Klein CD. Prediction of protein-protein interaction inhibitors by chemoinformatics and machine learning methods. Journal of medicinal chemistry. 2007;50:4665–4668. doi: 10.1021/jm070533j. [DOI] [PubMed] [Google Scholar]
- 42.Villoutreix BO, Labbe CM, Lagorce D, Laconde G, Sperandio O. A leap into the chemical space of protein-protein interaction inhibitors. Curr Pharm Des. 2012;18:4648–4667. doi: 10.2174/138161212802651571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keil M, Exner TE, Brickmann J. Pattern recognition strategies for molecular surfaces: III. Binding site prediction with a neural network. Journal of computational chemistry. 2004;25:779–789. doi: 10.1002/jcc.10361. [DOI] [PubMed] [Google Scholar]
- 44.Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lerner MG, Meagher KL, Carlson HA. Automated clustering of probe molecules from solvent mapping of protein surfaces: new algorithms applied to hot-spot mapping and structure-based drug design. Journal of computer-aided molecular design. 2008;22:727–736. doi: 10.1007/s10822-008-9231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 47.Valkov E, Sharpe T, Marsh M, Greive S, Hyvonen M. Targeting protein-protein interactions and fragment-based drug discovery. Top Curr Chem. 2012;317:145–179. doi: 10.1007/128_2011_265. [DOI] [PubMed] [Google Scholar]
- 48.Bourgeas R, Basse MJ, Morelli X, Roche P. Atomic analysis of protein-protein interfaces with known inhibitors: the 2P2I database. PLoS One. 2010;5:e9598. doi: 10.1371/journal.pone.0009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des. 2009;74:457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 50.MolProp version 2.1.5. OpenEye Scientific Software; S. F., NM: http://www.eyesopen.com, In. [Google Scholar]
- 51.QuacPac version 1.5.0. OpenEye Scientific Software; S. F., NM: http://www.eyesopen.com, In. [Google Scholar]
- 52.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 54.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Inhibitors bound to traditional targets, a subset of the Astex set. Potency is taken from reported Kd or Ki values where available; if unavailable, IC50 values were used instead. In several cases (“-NR-”), no measure of potency has been reported. θlig indicates the fraction of ligand SASA exposed in the complex, as defined in Equation 1.
Figure S1 (complements Figure 1) The distribution of the extent of inhibitor solvent exposure (θlig) is similar across a number of drug-like sets: Astex 27(blue), DOCK 54(magenta), DUD-E 29(brown), and SB2010 30(green).
Figure S2 (complements Figure 2) The distribution of molecular weights for the inhibitors in each set underlies the observed difference in ligand efficiencies. Inhibitors binding at protein interaction sites (red, median value 475 Da) are typically larger than their drug-like counterparts (blue, median value 355 Da), and the difference in the means is statistically significant (p < 10-4).
Figure S3 (complements Figure 2) The relationship between θlig and ligand efficiency for both the PPI set and the Astex set. While there is a statistically significant negative correlation between these properties for the PPI set (as noted in Figure 2c), no statistically significant correlation exists for the Astex set (p = 0.27).
Figure S4 (complements Figure 2) The relationship between θlig and potency for both the PPI set and the Astex set. No statistically significant correlation exists between these properties for either set (p = 0.33 for the PPI set, p = 0.50 for the Astex set).
Figure S5 (complements Figure 3) A known inhibitor was embedded in a custom set of 50 “decoy” compounds selected by the DUD-E server to match the physical properties of the known inhibitor. FRED exhibits superior ability to identify the known drug-like inhibitors from the decoy compounds (blue), relative to inhibitors that bind at protein interaction sites (red), and the difference in the means is statistically significant (p < 0.002).
Figure S6 (complements Figure 3) The observation that virtual screening at protein interaction sites performs less well than for drug-like compounds holds for other docking software as well. DOCK 6.6 was used to identify a known inhibitor embedded in a custom set of 50 “decoy” compounds selected by the DUD-E server to match the physical properties of the known inhibitor. DOCK 6.6 exhibits superior ability to identify the known drug-like inhibitors from the decoy compounds (blue), relative to inhibitors that bind at protein interaction sites (red), though this difference is not statistically significant (p = 0.11).