Abstract
Genetic and microarray analyses have provided useful information in the area of plant and pathogen interactions. Pseudomonas syringae pv. tomato DC3000 (Pst) causes bacterial speck disease in tomato. Previous studies have shown that changes in response to pathogen infection at transcript level are variable at different time points. This study provides not only proteomic changes between a resistant and a susceptible genotype, but also information on changes between an early and a late time point. Using iTRAQ quantitative proteomics approach, we have identified 2,369 proteins in tomato leaves, and 477 of them were determined to be responsive to Pst inoculation. Unique and differential proteins after each comparison were further analyzed to provide information about protein changes and the potential functions they play in the pathogen response. This information is applicable not only to tomato proteomics, but also adds to the repertoire of now available for crop proteomic analysis and how it changes in response to pathogen infection.
Keywords: iTRAQ, proteomics, tomato, Pseudomonas, defense response
Introduction
The study of pathogen response and defense in crop species is of essential importance as its applications are directly related to agricultural production. Especially, Pseudomonas syringae has been noted for its wide range of host specificity [1]. Among them, Pseudomonas syringae pv. tomato DC3000 (Pst) causes speck disease in tomato (Solanum lycopersicum), a crop having both nutritional and economical value[2].
Both Pst and tomato have co-evolved to counteract the efforts of plant innate immunity response or the pathogen, respectively[3]. Pathogen infection and recognition occur in a series of responses. First, plants utilize pattern recognition receptors to recognize pathogen associated molecular patterns (PAMPs) such as flagellin and lipopolysaccharaide. In PAMP triggered immunity (PTI), a signal cascade involves reactive oxidative species (ROS) production, activation of mitogen activated protein-kinases, cell wall fortification, and pathogen response (PR) gene transcription[4, 5]. However, Pst can attenuate the PTI responses by injecting effector proteins (e.g., AvrPto and AvrPtoB) into the plant cells to inhibit the basal defense responses[6]. In recognition of these effector proteins, plants have evolved to express resistance genes encoding R proteins, which are able to recognize Pst effector proteins and signal a hypersensitive response (HR) and pathogen resistance gene expression[7, 8]. This defense response is called effector triggered immunity (ETI), characteristic of an incompatible response[9]. Plants that do not recognize effector proteins are susceptible and have a compatible reaction with the infecting pathogen. A compatible interaction with a hemibiotroph such as Pseudomonas is characterized by increased bacterial growth, increased levels of hormones such as ethylene and jasmonic acid (JA), and necrotic cell death[10]. Incompatible reactions are characterized by HR involving a decrease in bacterial growth, programmed cell death, an increase in salicylic acid (SA) used to induce pathogenesis-related proteins, and systemic acquired resistance response. ROS is produced in a biphasic nature[11, 12]in both compatible and incompatible reactions after inoculation with pathogens [13]. Although it has been noted that components of both responses are shared[14], some responses, such as JA and SA induced responses, are antagonistic and result in different responses to bacterial infection[15]. In a compatible reaction, necrotic cell death serves as a means of nutrition to the hemibiotroph, while programmed cell death during an incompatible reaction is a controlled means to restrict bacterial growth[13].
In this study, we used the tomato cultivar, Rio-Grande (RG) which has two nearly homogenic genotypes: RG-PtoR (PtoR) expresses the R gene Pto that results in an incompatible interaction with the bacteria[16], while RG-prf3 (prf3), characterized by a 1kb deletion in Prf3, exhibits a susceptible phenotype and allows compatible development of speck disease[17]. When Pto, a serine/threonine kinase, and Prf3, a nucleotide binding site leucine rich repeat (NBS-LRR) protein are present, they form a Pto-Prf complex and detect effector proteins AvrPto and AvrPtoB[18, 19]. This initiates a resistance response characterized by activation of signaling cascades such as MAPK and ROS and defense gene transcription[4, 20, 21]. Previously, microarray[8, 22] and individual protein analysis have been focused on the interaction between Pst and tomato, but the information is fragmented and incomplete. Although global proteomics profiling in plant pathogen interaction has been done in Arabidopsis thaliana[23] and this information is useful in analyzing proteomic changes in defense responses to Pst, no such high volume proteomics information exists for tomato pathogen interactions in RG-PtoR (resistant) and RG-prf3 (susceptible). Here, we examined the proteomic profile for the interaction between tomato and Pst using iTRAQ LC-MS/MS technology. In particular, we compared proteomic changes between the two genotypes PtoR and prf3 for both early and late time points. Such analyses help to elucidate the effects and genetic differentiation involved with pathogen response in different genotypes and time points in respect to gene expression differences at the proteomic level. Therefore, we sought to determine: 1) how proteome changes in response to pathogen at different time points 2) how proteomic changes in response to pathogen differ between a compatible and an incompatible response, and 3) whether chosen proteomic changes correlate with transcript changes.
2 Materials and Methods
2.1 Plant material
Seedlings of PtoR and prf3 were germinated in MetroMix 500 soil (BWI Companies, Apopka, USA) in the growth chamber (160 µmol photons m−2s−1 with a photoperiod of 22°C for 8 hours (day) and 20°C for 16 hours (night), 70% relative humidity). After a week seedlings of similar size were then transplanted to four-inch diameter pots and grown for an additional three weeks in the homogenous conditions. Three biological replicates were grown in a randomized complete block design. Plants were inoculated at fourth week. The first fully expanded leaf was used for protein extraction.
2.2 Pseudomonas preparation and inoculation
A single colony of Pst was grown on Kings B Medium (KBM) for overnight at 28°C. The bacteria inoculum was prepared by scraping the cultured bacteria from the KBM plate into 20ml H2O. An OD600 of 0.03 (~106 cfu/ml) was prepared in 1L inoculation buffer (10mM MgCl2 with 400µl Silwett-70). The control inoculum for the mock inoculation contained the same components as the bacteria inoculum but with no addition of bacteria. Four week old plants were inoculated by dipping into the above inoculation solution for 30 seconds. The plants were covered and placed into the growth chamber until the chosen time point of observation.
2.3 Determination of time points for proteomic study
In order to determine which time points should be used for examining proteomic changes, we used two different methods, DAB staining[24] and bacterial growth assay[25]. The DAB staining detects ROS by forming a dark brown polymerization product in reaction with H2O2 in the presence of peroxidase[26]. For this staining, the fully expanded leaf of two genotypes from a series of times points was placed in 0.1% DAB stain (pH 3.8) epidermal side up, and vacuum infiltrated for 15 min. After removal of chlorophyll, the images were taken with a 12× Optical Zoom 25mm EQUIV 36–432 (Panasonic Inc., Japan).
The bacteria growth assay was performed as described in a previous study[25]. A disc (1cm2) was obtained from each leaf sample and ground in 10mM MgCl2. The solution was diluted and spotted onto a KBM Rifampicin (4µg/ml) plate. Colonies were counted for three replicates, and then ANOVA was performed to see whether there is significant statistical difference among different time points of two genotypes and Tukey-Kramer HSD was performed using JMP Pro 9.0.2 (SAS Institute, Cary, NC, USA) to determine whether there are differences in levels of bacterial growth across time points.
2.4 Protein extraction, digestion, and iTRAQ labeling LC-MS/MS
Three biological replicates of control and Pst inoculated leaves were prepared for the proteomics experiments. Proteins were extracted and quantified following a previous method[27]. For each sample, 100µg protein was dissolved in a dissolution buffer (AB Sciex Inc., Foster City, CA, USA). The samples were reduced, alkylated, trypsin-digested, and labeled following the manufacturer’s instructions for the iTRAQ Reagents 8-plex kit (AB Sciex Inc.). Mock inoculated lines (PtoR 4hai, PtoR 24hai, prf3 4hai, and prf3 24hai) were labeled with iTRAQ tags 113, 115, 117, and 119; and Pst inoculated lines (PtoR 4 hours after inoculation (hai), PtoR 24hai, prf3 4hai, and prf3 24hai) were labeled with tags 114, 116, 118, and 121, respectively (Supporting Information Fig. S1). After combining the eight labeled samples, an off-line 2D LC-MS/MS method with strong cation exchange (SCX) chromatography as a first step was used to fractionate the proteome as previously described[27]. The fractionated tryptic peptides were separated with a Dionex C18 PepMap HPLC column (San Francisco, CA, USA), and analyzed using a hybrid quadrupole-TOF QSTAR Elite MS/MS system (AB Sciex Inc., Framingham, MA, USA)[28] and TripleTOF™ 5600 (AB Sciex Inc., USA)[29].
2.5 iTRAQ LC-MS/MS data analysis
The MS/MS data were processed by a thorough search considering biological modification and amino acid substitution against Solanum lycopersicum database (315,012 entries based on six frames; http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=tomato, Release 13.0) under the Paragon™ algorithm[30] using ProteinPilot v.4.2 software (AB Sciex Inc.). After searching MS/MS spectra against this database, the results were combined into each group. Plant species, fixed modification of methylmethane thiosulfate-labeled cysteine, fixed iTRAQ modification of amine groups in the N-terminus and lysine, and variable iTRAQ modifications of tyrosine were considered. The ProteinPilot cutoff score was set to 1.3 (a confidence level of 95%), and the false discovery rate (FDR) was estimated against concatenated databases (Supporting Information Table S1).
Functional analysis of proteins for each genotype and time point combination was conducted using GO annotation (http://www.geneontology.org/) with Fisher’s exact test based on FDR (p-value ≤0.05)[31]. Blast2GO level 3 filtering was used to examine unique protein changes during comparison analysis. For protein quantification, we only considered MS/MS spectra that were unique to a particular protein and where the sum of the signal-to-noise ratio for all of the peak pairs was >9 (software default settings, AB Sciex Inc.). The accuracy of each protein ratio was calculated by the ProGroup analysis in the software whether the protein is significantly differentially expressed[30]. To be identified as being significantly differentially expressed, a protein should be quantified with at least three MS/MS spectra and used a fold change >1.2 or <0.8 with a p-value <0.05 of at least three peptides. In addition, a meta-analysis was performed using Fisher’s combined probability test[32] to determine whether there are differences in between control and experimental group. Accession numbers were compared between each genotype and treatment, and unique proteins were identified. A protein was considered shared if the accession number was identified in the two genotypes or time points. A protein is considered to be expressed differently if it has an increase in expression in one component of the comparison and a decrease in the other. A change in expression was determined in comparison with the corresponding controls.
2.7 Quantitative real-time PCR
To investigate whether gene expression is correlated between transcript and protein level, we employed the relative quantitation in real-time RT-PCR (qRT-PCR) for 20 genes selected based on proteomic data (Supporting Information Table S2). VeriQuest™ SyBr with Fluorescein kit (Affymetrix Inc., Santa Clara, CA, USA) was used with CFX90 (Bio-Rad, Hercules, CA, USA) as previously described[27]. For each reaction three technical and three biological replicates were included. Relative expression of the target genes was calculated using the comparative CT method (Applied Biosystems Inc., Framingham, USA). The differences in Ct values (ΔCt) between the target gene and endogenous control were calculated to normalize the differences in the cDNA concentrations for each reaction[27]. The ANOVA test was performed to assess whether there is difference in gene expression level between inoculated and control sample.
3. Results and Discussion
3.1 Oxidative status and bacterial growth in leaves after inoculation
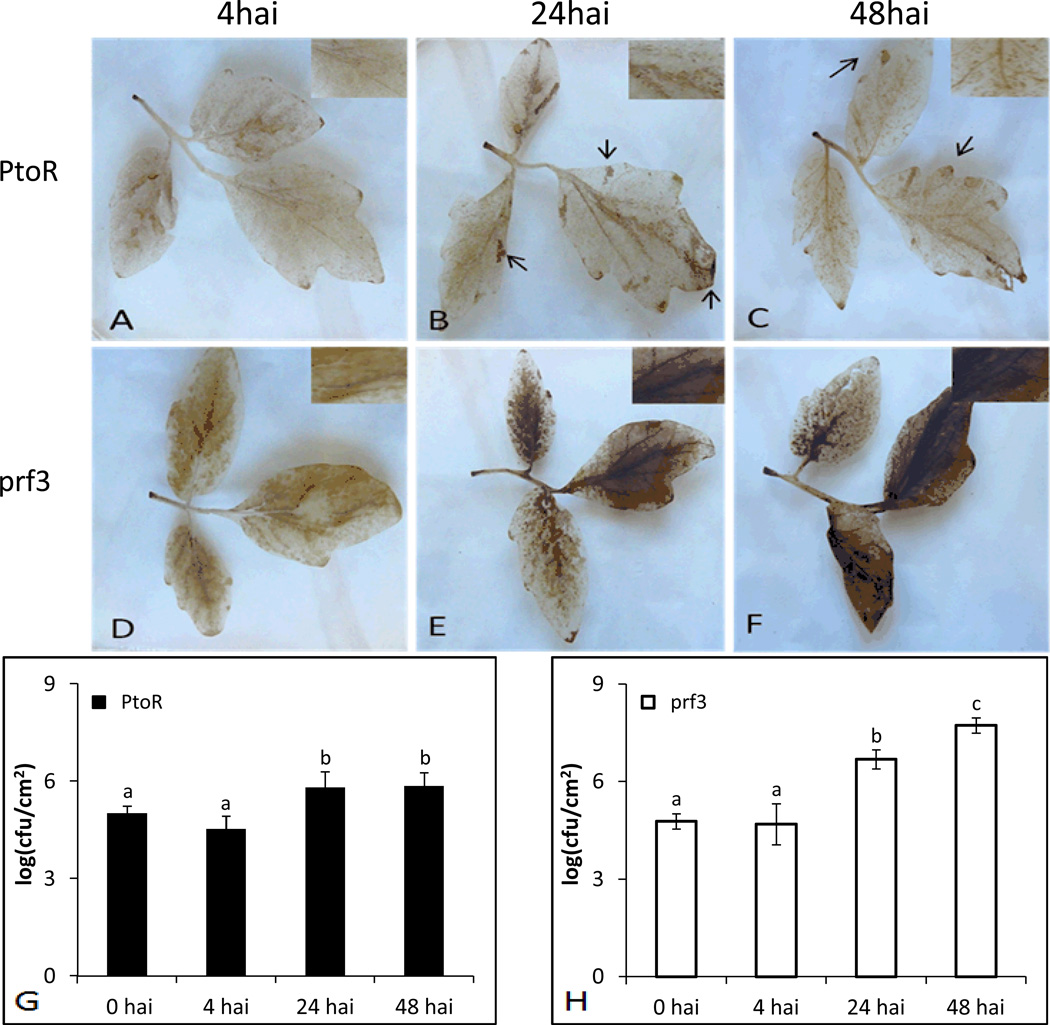
To mimic natural bacterial infection of tomato, we chose a dipping method in which leaves are not damaged. DAB staining was used to evaluate H2O2 levels in infected tomato leaves from the two genotypes (Supporting Information Fig. S2). Both PtoR and prf3 genotypes showed a light DAB staining at 4hai (Fig. 1A and D) that dissipated over the course of the infiltration period until 24hai (Fig. 1 B and E). At 24hai, PtoR had isolated spots on the leaf that stained suggesting oxidative bursts (Fig. 1B), while prf3 inoculated stained leaves showed large regions of DAB staining (Fig. 1E), indicating higher levels of H2O2 in the cells and apoplast. In particular, these results showed two increases in H2O2; an initial increase at 4hai and a larger increase at 24hai, which showed different levels of staining between the two genotypes. Controlled H2O2 production in PtoR suggests structured programmed cell death mechanisms. The dark staining produced in prf3 24hai suggests a lack of H2O2 control, i.e., HR in order to confine the bacteria. Previous data indicated that a decrease in H2O2 is observable in susceptible genotypes[5]. However, under our inoculation conditions, an increase in staining was observed in prf3 24hai, when compared to PtoR 24hai. To test whether a darker stain was due to an increase in the number of bacteria, we performed a bacterial growth assay to examine the growth trends between the two genotypes.
Figure 1.
DAB staining of leaves and bacterial growth assay between genotypes and time points after Pst inoculation. Zoomed in areas show specific H2O2 staining in cells or a flooding of H2O2 in the apoplast. Arrows indicate areas of HR. Letters a, b and c indicate a significant change (p-value <0.05) as determined by ANOVA. (A) PtoR 4hai; (B) PtoR 24hai; (C) PtoR 48hai; (D) prf3 4hai; (E) prf3 24hai; (F) prf3 48hai; (G) bacterial growth compared amongst time points in PtoR; (H) bacterial growth compared amongst time points in prf3.
Both genotypes showed similar colony forming units at 4hai, but significantly different growth at 24hai (Fig. 1G and H). No significant changes in bacterial growth from 24hai to 48hai were observed in PtoR (Fig. 1G), suggesting a defense response to control the bacteria population in PtoR at later time points after infection. However, prf3 had a significant increase in bacterial growth beginning at 24hai and continued to 48hai (Fig. 1H). This increase indicates that prf3 is not able to efficiently control bacterial growth because of the defect in PtoR-Prf protein complex formation. In the prf3 mutant, pathogens seem to be able to take advantage of the loss of resistance proteins and multiply during the infection process. Our results are consistent with previous results comparing bacterial growth in resistant and susceptible lines after inoculation with Pst[7, 33]. Based on DAB staining and bacterial growth results (Fig. 1, Supporting Information Fig. S2), 4hai and 24hai were chosen as two representative early and late treatment time points for further proteomic analysis.
3.2 Overall proteome changes in pathogen response
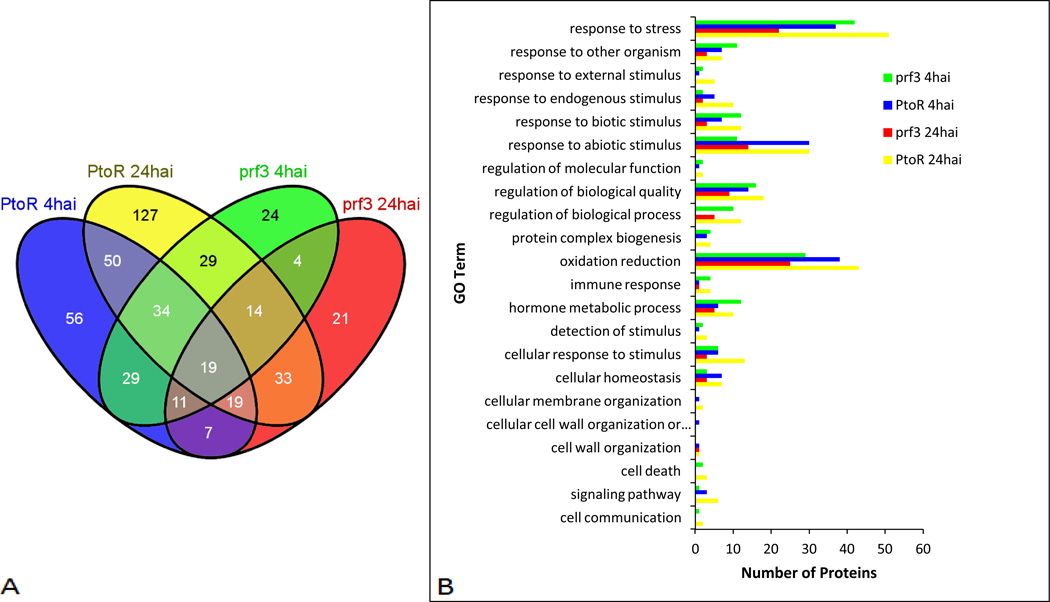
Proteomics analysis was performed in an effort to examine proteome changes in tomato genotypes PtoR and prf3 after the early and late responses (Fig. 2A) to Pst infection using iTRAQ LC-MS/MS for protein identification and quantification (Supporting Information Fig. S3). A total of 2369 proteins were identified (Supporting Information Table S1), and 477 of the proteins were quantified as having significant changes in expression (Table 1, Fig. 2A). We observed 444 total proteins with an increase in expression (>1.2) and 91 proteins with a decrease in expression (< 0.8) relative to their respective controls (Supporting Information Table S1). Among them, there are 225, 325, 164 and 128 Pst infection-responsive proteins identified in PtoR 4hai, PtoR 24hai, prf3 4hai, and prf3 24hai, compared to their respective controls in pathogen response (Fig. 2A, Supporting Information Table S3). Biological processes were identified for proteins with a change in expression using Blast2GO software[31] (Fig. 2B, Supporting Information Table S3). The processes include response to other organism, cell death, oxidation reduction, cell membrane organization, and hormone metabolic process (Fig. 2B). Two genotypes and two time points were used to examine response differences between the incompatible (PtoR) and compatible (prf3) interaction and between the early (4hai) and late (24hai) responses (Fig. 2A). A total of 380, 244, 277 and 355 proteins were found to be differentially expressed between PtoR 4hai and 24hai, between prf3 4hai and 24hai, between PtoR and prf3 at 4hai, and between PtoR and prf3 at 24hai, respectively (Table 1, Supporting Information Table S3). These proteins, which include unique proteins and shared proteins in different comparisons, provide a comprehensive view of proteome level responses to Pst infection.
Figure 2.
Proteomic changes between time points and genotypes. (A) Venn diagram showing unique and shared proteins between time points and genotypes. (B) Biological process analysis for unique proteins with a significant change in expression for both time points and genotypes. Processes are representative of Blast2Go level 3 sorting. Proteins may be identified in more than one process.
Table 1.
Proteomic changes in tomato leaves after Pseudomonas infection between different time points (4hai and 24hai) and genotypes (PtoR and prf3).
| Expression pattern | Time point comparisons | Genotype comparisons | |||
|---|---|---|---|---|---|
| Component 1a | Component 2a | PtoR_4hai PtoR_24hai |
prf3_4hai prf3_24hai |
PtoR_4hai prf3_4hai |
PtoR_24hai prf3_24hai |
| ↑ | - | 77b (20.3)c | 112 (45.9) | 114 (41.1) | 217 (61.1) |
| ↑ | ↑ | 94 (24.7) | 27 (11.1) | 88 (31.8) | 65 (18.3) |
| - | ↑ | 161 (42.4) | 58 (23.8) | 48 (17.3) | 17 (4.8) |
| - | ↓ | 15 (3.9) | 22 (9.0) | 5 (1.8) | 13 (3.7) |
| ↓ | - | 5 (1.3) | 4 (1.6) | 17 (6.1) | 23 (6.5) |
| ↓ | ↓ | 1 (0.3) | 1 (0.4) | 0 (0.0) | 5 (1.4) |
| ↑ | ↓ | 13 (3.4) | 19 (7.8) | 1 (0.4) | 14 (3.9) |
| ↓ | ↑ | 14 (3.7) | 1 (0.4) | 4 (1.4) | 1 (0.3) |
| 380 (100) | 244 (100) | 277 (100) | 355 (100) | ||
Component in the analysis being examined and trend in expression being examined (↑ increased; ↓ decreased, - not detected). In each comparison, component 1 is listed first, and component 2 is listed second.
Number of proteins
Percent of the total proteins identified.
3.3 Proteomic changes in early and late responses to pathogen infection
3.3.1 Changes in PtoR
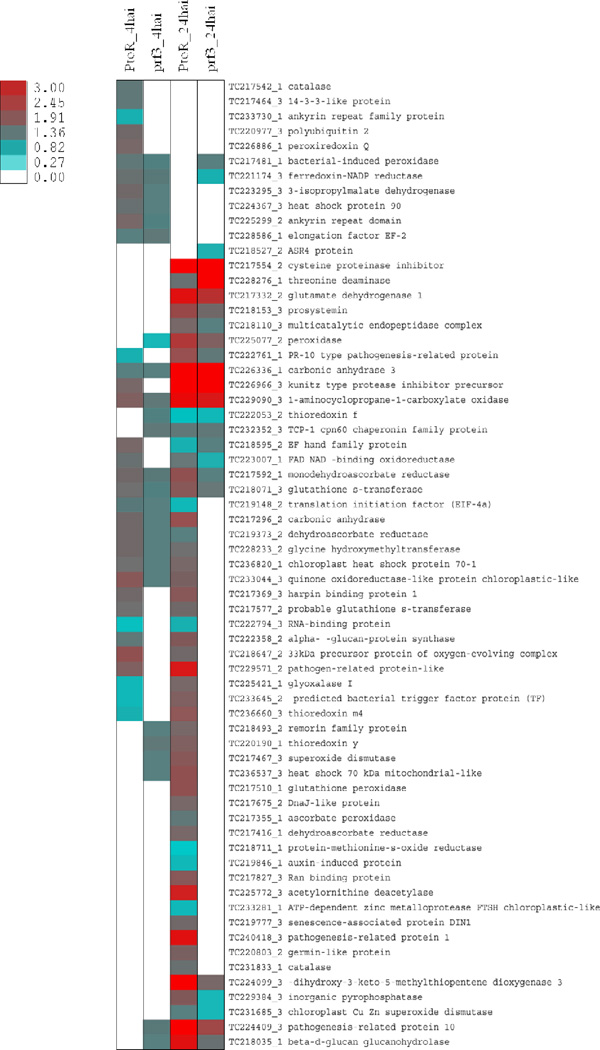
When protein profiles at 4hai and 24hai were compared (Fig. 2A), the PtoR plants showed different infection-responsive proteins patterns (Table 1). At 4hai, 82 unique responsive proteins (77 with an increase and 5 with a decrease), approximately 22% of the total proteins identified (380)) were observed; while 176 unique proteins (161 increased and 15 decreased; approximately 46% of the total identified proteins (380)), were observed at 24hai (Table 1, Supporting Information Table S3). Interestingly, 27 proteins were shared between the two time points, but had a difference in trends of expression (Table 1, Fig. 2, Supporting Information Table S3). An increase in the number of infection responsive proteins at 24hai suggests that although a transcriptional change occurs at 4hai in response to pathogen attack[8], a larger proteomic change occurs at 24hai than at 4hai in PtoR. These protein changes may play essential roles in developing the resistance phenotype of PtoR. Interesting proteins are further described in their functional context in subsequent sections of this manuscript (Figs. 2B and 3).
Figure 3.
Heatmap of defense associated protein expression. Unique and shared protein with significant expression changes between genotypes and time points were used. White spaces are representative of a protein that was identified but did not have a significant change in expression relative to the control.
3.3.2 Changes in prf3
A comparison between prf3 4hai and 24hai showed a change in the number of proteins that were increasing and decreasing relative to their respective controls. Unlike in PtoR, the susceptible prf3 has a reverse trend in protein changes. Here we observed an increase in the number of proteins with a change in expression at 4hai instead of the later 24hai (Table 1, Fig. 2). In prf3 4hai, we identified 116 unique proteins (112 with an increase and 4 with a decrease), approximately 48% of the total proteins (244) identified in this comparison (Table 1, Supporting Information Table S3). In prf3 24hai, we identified 80 unique proteins (58 with an increase and 22 with a decrease; 35% of the total proteins (244)) (Table 1, Supporting Information Table S3). Twenty shared proteins with different trends in expression were identified in both time points. Out of the shared proteins with different changes in expression, 19 of the proteins increased in prf3 4hai and decreased in prf3 24hai (Table 1, Supporting Information Table S3). A change in the number of unique proteins identified at each time point in the prf3 suggests a difference in the response to pathogen infection, which contributes to the development of the susceptible phenotype of prf3 in Pst infection.
3.4 Proteomic changes between PtoR and prf3 genotypes
3.4.1. Changes at 4hai
Previous research has shown transcriptional changes between the resistant and susceptible genotypes occur at 4hai[8]. Here we examined if there were any changes in the proteomes between the two genotypes. We identified 131 proteins (114 with an increase and 17 with a decrease; about 47% of total proteins (277)) in PtoR. Fifty three proteins (48 with an increase and 5 with a decrease; about 19% of total proteins (277)), were observed in prf3 4hai. Only five proteins with different trends in expression were shared between the two genotypes (Table 1, Fig. 2, Supporting Information Table S3). The difference in the number of proteins observed between genotypes at the early stage of infection provides information about proteins potentially involved in early compatible and incompatible responses to pathogen infection.
3.4.2 Changes at 24hai
A comparison between genotypes at 24hai was made to examine protein changes between genotypes in a late response to pathogen infection (Fig. 2A). Here we observe a higher number of unique proteins identified in PtoR 24hai when compared to prf3 24hai (Table 1, Fig. 2, Supporting Information Table S3). In PtoR 24hai, we identified 230 unique proteins (217 with an increase and 13 with a decrease), approximately 68% of the proteins identified in the comparison. Only 40 unique proteins (17 with an increase and 23 with a decrease) were observed in prf3 24hai. Fifteen proteins were shared between the two genotypes but had different trends in expression (Table 1, Fig. 2, Supporting Information Table S3). The distinct differences in protein levels highlight in addition to early stage responses the late stage protein changes in PtoR contribute positively to the establishment of the disease resistance phenotype.
3.5 Biological processes of unique proteins involved in the response to pathogen
Biological processes of unique proteins identified from each comparison (Fig. 2, Supporting Information Table S3) were examined to further understand the changes occurring at each time point and between genotypes. We identified unique proteins involved in response to stress, response to other organism, response to abiotic and biotic stimulus, oxidation reduction, regulation of biological quality, membrane organization, and cell death.
When PtoR 4hai was examined, it was observed to have the second highest numbers of unique proteins with a change in expression (after PtoR 24hai) in processes including response to endogenous stimulus, oxidation reduction, and signaling (Fig. 2B). However, the number of proteins involved in cellular homeostasis, response to abiotic stimulus, and response to other organism were the same as PtoR 24hai (Fig. 2B). Interestingly, no proteins annotated as being involved in cell death were identified in PtoR 4hai. The cell wall organization process included an alpha-glucan protein synthase (TC222358_2), a transferase previously observed to be involved in cell wall synthesis and callose deposition[32–34]. This process is known to be an early defense mechanism to reduce pathogen infection[5, 35–37]. Most research in callose deposition is performed in Arabidopsis; however, here we show an increase in this protein during an incompatible interaction in tomato in response to Pst (Fig. 3). A glutathione-S-transferase involved in reducing oxidative stress[38] has previously been described as increasing in PTI and ETI[39]. In this study when genotypes were compared at 4hai (Fig. 2A), a probable glutathione-S-transferase (TC217577_2) was observed in PtoR (Fig. 3, Supporting Information Table S3). This protein had an increase in expression relative to the control (Fig. 3). Because this protein is unique to PtoR 4hai in this comparison, it may serve as a biomarker for incompatible interactions at this time point.
In PtoR 24hai, more proteins with a change in expression were identified as being involved in oxidation reduction, cellular response to stimulus, response to stress, and signaling (Fig. 3). PR1, a pathogen-related protein, is known to be involved in late response to pathogen infection[40]. We identified this protein (TC240418_3) in PtoR 24hai when compared to both PtoR 4hai and prf3 24hai (Fig. 3); this suggests a late pathogen defense response is occurring at this time point. Methionine-S-oxide reductase (TC218711_1) is involved in the repair of proteins that have undergone oxidative damage [41]. We identified this protein to be decreased in PtoR 24hai (Fig. 3, Supporting Information Table S3). Previous research has shown an increase in gene expression of this protein in PtoR 4hai[8]. However, it is not clear whether the decrease of this protein at 24hai was due to transcriptional down regulation or protein turnover. Nevertheless, an increase in the number of proteins with changes in expression at this time point suggests that the biological processes these proteins were identified as being involved in are important for plant defense.
We also identified biological processes in which prf3 4hai had more proteins categorized as having a change in expression. Those processes included response to other organisms and the hormone metabolic process. Processes such as response to biotic stimulus, protein complex biogenesis, and immune response had the same number of proteins as identified in PtoR 24hai (Fig. 2B). Other processes such as oxidation reduction and response to stress also included proteins in prf3 4hai that had changes in expression. Interestingly, a remorin family protein (TC218493_2), previously annotated as being involved in control of plant infection was identified in prf3 4hai when compared to both PtoR 4hai and prf3 24hai; an increase in this protein was observed (Fig. 3, Supporting Information Table S3). Over expression of remorin has been shown to limit the infection process of Potato Virus X in potato, but a decrease in remorin in a legume decreased symbiotic bacterial infection[42]. Here the increase of remorin may be part of a basal defense response at 4hai and does not play a role in increasing bacterial infection (Fig. 1). This proposal is supported by the increase in expression of glycine hydroxymethyl transferase (TC228233_2) (Fig. 3, Supporting Information Table S3), associated with immune response at 4hai. This enzyme was previously identified as being involved in controlling bacteria-dependent cell death[43]. The biological processes identified in prf3 4hai as well as the increase in proteins when compared to prf3 24hai suggests more proteins related to pathogen response and basal defense have changes in expression in prf3 4hai (Fig. 2B and 3); however, the numbers of proteins involved were still lower in most processes than those identified in PtoR 24hai (Fig. 2B).
At 24hai, prf3 had the lowest numbers of proteins involved in pathogen related biological processes (Fig. 2B); however, more proteins were identified as being involved in response to abiotic stimulus when compared to prf3 4hai (Fig. 2B). Interestingly, no proteins were identified in signaling, protein complex biogenesis, cellular membrane organization, or cell death; in contrast, proteins were identified in those biological processes in PtoR 24hai, PtoR 4hai, and prf3 4hai (Fig. 2B). When comparing early and late time points in prf3, we identified glutamate dehydrogenase I (TC217332_2) in prf3 24hai (Fig. 3). This protein had an increase in expression and was annotated as being involved in response to abiotic stimulus (Fig. 2, Supporting Information Table S3). Glutamate dehydrogenase is involved in nitrogen metabolism and has been identified as having an increase in tomato plants inoculated with Pseudomonas syringae pv syringae[44]. This protein is also considered a leaf senescence marker[44] which indicates that leaf senescence is occurring in prf3 24hai in tomato when infected with Pst. There were several unique decreased proteins identified in prf3 24hai (Fig. 3, Supporting Information Table S3). One such protein, an abscisic acid, stress, and ripening protein 4 (ASR4) (TC218527_2), has previously been characterized as being involved in HR in Arabidopsis[45] (Fig. 3). Decreased proteins involved in oxidation reduction mechanisms include a chloroplast Cu Zn superoxide dismutase (TC231685_3) (Fig. 3). Superoxide dismutases are involved with reducing peroxide levels in the chloroplast and in consequence produces H2O2 [46], and has been associated with resistance response[47]. Decreased expression of this protein by prf3 24hai suggests a decrease in ROS regulation at this time point and suppression of a protein capable of contributing to resistance. Although the number of unique proteins with changes in expression identified in prf3 24hai was much smaller when compared to prf3 4hai and PtoR 24hai, the number of proteins with a decrease was higher in prf3 24hai when compared to prf3 4hai (Table 1, Supporting Information Table S3). Information derived from examining the unique proteins identified in prf3 24hai suggests reduced capability in pathogen defense in contrast to PtoR 24hai.
3.6 Biological processes of shared proteins involved in the response to pathogen
For each comparison, we also identified shared proteins with the same and different trends in expression (Fig. 2 and 3). These proteins suggest potential regulatory mechanisms between genotypes and time points. Interestingly, we identified thioredoxin y (TC220190_1) in both PtoR 24hai and prf3 4hai (Fig. 3, Supporting Information Table S3). Thioredoxins play an important role in cellular redox regulation, and thioredoxin y has been shown to be involved in NADP-malate dehydrogenase activation and is a substrate for peroxiredoxin Q[48, 49]. To date, thioredoxin y has not been characterized as being involved in pathogen response. Another protein, glyoxalase I (TC225421_1), identified as having a decrease in PtoR 4hai and an increase in PtoR 24hai (Fig. 3), is involved in detoxification of methylglyoxal, a cytotoxic byproduct of glycolysis, to S-lactoylglutathione (SLG) using reduced glutathione[50]. Increase of this protein can potentially enhance plant stress tolerance[5]. In PtoR 24hai, an increase in this protein suggests a role of this protein in pathogen defense process. Another protein of interest identified was an uncharacterized RNA-binding protein (TC222794_3); it had a decrease in expression in PtoR 4hai (Fig. 3) in this study as well transcript changes in previous studies[8]. This protein was also observed to have a decrease in expression in PtoR 24hai (Fig. 3). Changes in expression in this protein suggest a change in potential translational control, RNA splicing, and editing in response to pathogen infection in PtoR. Identification of this protein in PtoR at both time points with the same trend in expression suggests down regulation of this protein may be necessary for a resistance response against Pst. We also identified a peroxidase (TC225077_2) in the time point comparison in prf3 and between genotypes 4hai (Fig. 3). When time points were compared, the protein had a decrease in expression in prf3 4hai and an increase in prf3 24hai; when genotypes were compared, the protein was identified in prf3 4hai with a decrease in expression (Fig. 3, Supporting Information Table S3). The change in expression of this protein suggests an effort to increase H2O2 in a PAMPs response and decrease H2O2 in prf3 24hai [52].
Compared to PtoR 4hai, the increase in the number of proteins with altered expression (Table 1) as well as the number of biological processes related to the HR process in PtoR 24hai (Figure 2B) highlights the importance of proteomic changes in pathogen defense. In this study we also observed an increase in the number of functions associated with PAMP response in both PtoR 4hai and prf3 4hai (Figure 2B). PAMP associated proteins and functions are expected to be observed since basal responses should be shared between the genotypes. This suggests PAMP response occurs at an earlier time point as suggested in Arabidopsis [53]. This work has provided information as to the differences in proteomic changes occurring between the two genotypes at 4hai and 24hai and constitutes another line of evidence for the ETI regulation of PTI responses[6]. The comparison between time points in prf3 supports past findings on this compatible interaction[22] and provides new information at the protein level regarding pathways that are changing due to successful pathogen infection. At 4hai, the increase in proteins associated with PAMPs response indicates active basal defense. However, the decreased proteins at 24hai were observed in more processes associated with pathogen response; this could be a result of either bacterial control or a response to increasing bacterial growth and the beginning signs of death. When we compared the two genotypes 24hai, we observed an increase not only in the number of proteins in PtoR 24hai, but also an increase in the number of biological processes with differentially regulated protein expression. Shared processes were identified between the two genotypes 24hai; however, those unique processes/changes identified in PtoR 24hai may determine the resistance response.
3.7 Correlation of protein fold changes with transcripts
Similar experimental design has allowed for us to compare microarray information obtained from experiments in which PtoR and prf3 underwent treatment with Pst[8, 22] to the proteins identified in this study. However, the microarray data do not cover many proteins that we have identified. Q-RT PCR was performed to examine the transcript levels for a survey of proteins we identified (Table 2, Supporting Information Table S2). We decide to choose proteins involved in a variety of processes within the plant and most of them were not identified in previous microarray analysis. The calculated Pearson correlation value was 0.674 (p-value <0.0001). We observed mixed results with three sets of information. One set includes seven proteins and transcripts with a similar trend in expression in the particular genotype at 4hai or 24hai (Table 2). They include 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (TC217800_2), Kunitz-type protease inhibitor precursor (TC226966_3), leucine-amino peptidase (TC217231_1), lipoxygenase (TC234372_3), polyphenol oxidase precursor (TC219183_1), pathogenesis-related protein 1 (TC240418_3), and prosystemin (TC218153_3) (Table 2). The second set we gathered includes eight additional proteins that had been observed in the proteomics data as having a significant fold change in expression and showed no significant change between the inoculated and control transcript levels (Table 2, Supporting Information Table S2). These proteins are cystatin (TC217539_3), cyanate hydratase (TC228351_1), elongation factor-1α (TC229158_2), isocitrate dehydrogenase (NADP+) (TC236100_2), methionine synthase (TC220343_2), ribulose-bisphosphate carboxylase oxygenanse large subunit (TC217295_3), and a poly (A)-binding protein (TC223115_1) (Table 2). This information provides insight into differential regulation between transcripts and proteins. The third set observations made was that of proteins with correlated transcript and protein levels at one genotype and time point but uncorrelated transcript and protein level at a different genotype and time point (Table 2, Supporting Information Table S2). A luminal binding protein (TC217353_3) was observed to have an increase in fold change at transcription and protein levels in PtoR 24hai (Table 2). But in prf3 24hai, this protein showed no significant changes at the transcript level, although changes at the protein level were observed. Another protein, transketolase (TC220395_2) had a significant change in transcript levels in PtoR 4hai, but had a different trend in expression when compared to protein fold change. In prf3 24hai, transcript and protein levels were both decreased. These observations indicate different regulatory mechanisms at transcriptional and posttranscriptional levels in different genotypes in the course of bacterial infection, and highlight the importance of protein level characterization towards understanding defense/disease mechanisms and phenotypes.
Table 2.
Observation of transcript and protein level correlation.
| Gene | Accession | Genotype & treatment |
log2proteina | p-value | log2transcriptb | p-value |
|---|---|---|---|---|---|---|
| Correlated transcript changes and protein changes | ||||||
| Transketolase | TC220395_2 | prf3 24hai | −0.375 | 0.015 | −5.128 | 0.014 |
| Luminal binding protein | TC217353_3 | PtoR 24hai | 0.837 | 0.000 | 1.377 | 0.001 |
| 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase | TC217800_2 | PtoR 24hai | 1.550 | 0.001 | 2.564 | 0.014 |
| prf3 24hai | 0.983 | 0.001 | 2.116 | 0.001 | ||
| Glutathione-S-transferase | TC218071_3 | PtoR 4hai | 0.592 | 0.000 | 1.711 | 0.004 |
| Peroxidase | TC225077_2 | PtoR 24hai | 1.368 | 0.004 | 1.115 | 0.044 |
| Kunitz-type protease precursor | TC226966_3 | PtoR 24hai | 2.300 | 0.000 | 11.369 | 0.001 |
| prf3 24hai | 2.319 | 0.000 | 10.600 | 0.005 | ||
| Leucine-amino peptidase | TC217231_1 | PtoR 24hai | 1.937 | 0.000 | 4.481 | 0.004 |
| prf3 24hai | 1.820 | 0.000 | 4.373 | 0.000 | ||
| Lipoxygenase | TC234372_3 | PtoR 24hai | 1.470 | 0.000 | 1.994 | 0.007 |
| prf3 24hai | 0.566 | 0.026 | 1.432 | 0.014 | ||
| Monodehydroascorbate reductase | TC217592_1 | PtoR 24hai | 0.979 | 0.000 | 0.756 | 0.029 |
| prf3 4hai | 0.384 | 0.000 | 2.391 | 0.025 | ||
| Polyphenol oxidase | TC219183_1 | PtoR 24hai | 2.375 | 0.000 | 4.224 | 0.025 |
| prf3 24hai | 1.507 | 0.000 | 4.831 | 0.018 | ||
| Pathogenesis-related protein 1 | TC240418_3 | PtoR 24hai | 1.868 | 0.000 | 2.194 | 0.002 |
| Prosystemin | TC218153_3 | PtoR 24hai | 1.066 | 0.003 | 1.176 | 0.034 |
| prf3 24hai | 0.629 | 0.033 | 1.582 | 0.014 | ||
| Uncorrelated transcript changes and protein changes | ||||||
| Transketolase | TC220395_2 | PtoR 4hai | 0.437 | 0.000 | −1.723 | 0.001 |
| No transcript change | ||||||
| Monodehydroascorbate reductase | TC217592_1 | PtoR 4hai | 0.649 | 0.000 | 0.401 | 0.171 |
| prf3 24hai | 0.354 | 0.002 | 0.049 | 0.414 | ||
| Luminal binding protein | TC217353_3 | prf3 24hai | 0.529 | 0.027 | 0.732 | 0.065 |
| Peroxidase | TC225077_2 | prf3 4hai | −0.450 | 0.035 | 0.313 | 0.896 |
| prf3 24hai | 0.832 | 0.029 | −0.388 | 0.337 | ||
| Glutathione-S-transferase | TC218071_3 | PtoR 24hai | 0.904 | 0.000 | 0.118 | 0.598 |
| prf3 4hai | 0.284 | 0.000 | 0.046 | 0.915 | ||
| prf3 24hai | 0.485 | 0.000 | −1.038 | 0.163 | ||
| Cysteine protease inhibitor | TC217554_2 | PtoR 24hai | 2.157 | 0.000 | 0.679 | 0.495 |
| prf3 24hai | 2.955 | 0.000 | 0.617 | 0.231 | ||
| Cystatin | TC217539_3 | PtoR 4hai | 0.696 | 0.032 | 0.243 | 0.461 |
| PtoR 24hai | 0.755 | 0.002 | 0.631 | 0.087 | ||
| Cyanate hydratase | TC228351_1 | PtoR 24hai | 0.915 | 0.000 | 0.421 | 0.351 |
| Elongation factor 1α | TC229158_2 | PtoR 4hai | 0.545 | 0.000 | −0.068 | 0.742 |
| prf3 4hai | 0.408 | 0.000 | −0.368 | 0.414 | ||
| Isocitrate dehydrogenase (NADP+) | TC236100_2 | PtoR 4hai | 0.308 | 0.017 | 0.079 | 0.717 |
| PtoR 24hai | 0.695 | 0.002 | −0.039 | 0.803 | ||
| prf3 4hai | 0.351 | 0.000 | 0.655 | 0.740 | ||
| prf3 24hai | 0.576 | 0.000 | −0.083 | 0.400 | ||
| Methionine synthase | TC220343_2 | PtoR 4hai | 0.537 | 0.000 | −0.155 | 0.239 |
| PtoR 24hai | 0.504 | 0.001 | −0.422 | 0.199 | ||
| prf3 4hai | 0.304 | 0.001 | 0.155 | 0.893 | ||
| prf3 24hai | 0.407 | 0.000 | −0.785 | 0.583 | ||
| Poly (A)-binding protein | TC223115_1 | PtoR 24hai | 0.877 | 0.011 | −0.175 | 0.282 |
| prf3 4hai | 0.322 | 0.000 | −0.218 | 0.583 | ||
| Ribulose-bisphosphate carboxylase oxygenase large subunit | TC217295_3 | PtoR 4hai | 0.514 | 0.000 | −0.413 | 0.159 |
| PtoR 24hai | −0.800 | 0.000 | −0.699 | 0.211 | ||
proteomic expression difference obtained from iTRAQ.
Log2((inoculated – control)/control).
4. Conclusion
Provided in this work is the proteomic analysis of four responses (two genotypes and two time points) of tomato (S. lycopersicum) to Pst DC3000 infection. This information is the next piece of the puzzle that has been built around microarray data and the initial proteomics work done in Arabidopsis. Our analysis of proteome changes and the processes involved has complemented microarray analysis of processes involved in pathogen response during a compatible and incompatible interaction. We have also added another dimension to the knowledge base by including not just the two genotypes but also two time points during the infection process. Through extensive comparisons between the time points and genotypes, we have corroborated known proteins and functions in plant response to pathogen infection and added a new list of proteins with potential functions in plant defense. It is interesting to note that the resistance genotype shows more protein/function changes at 24hai, while the susceptible genotype exhibits more protein/function changes at 4hai. Although both genotypes appear to share PTI, our data have demonstrated the ETI regulation of PTI and revealed potential resistance proteins, many of which are targets for future investigation. We have also examined the relationship between identified proteins and their transcripts. A set of proteins chosen for their coverage of processes supports the notion that protein levels and transcript levels are differentially regulated. The results from this study are valuable on both a basic and applied level as it forms a foundation for future plant defense research as well as for protection of crops against pathogen infection.
Supplementary Material
Acknowledgements
We thank Professor Greg Martin and Ms. Diane Dunham from Boyce Thompson Institute for Plant Research and Dr. Zhonglin Mou at University of Florida for providing the experimental materials and invaluable advice. We acknowledge the Proteomics Division of the University of Florida’s Interdisciplinary Center for Biotechnology Research (ICBR) and Dr. Bruce Stanley from Pennsylvania State University for assistance in LC-MS/MS analysis. This work was funded by University of Florida and the National Institute of Health (1S10RR025418-01) to S Chen.
Abbreviations
- Pst
Pseudomonas
- hai
hour after inoculation
- RG
Rio Grande
- PAMP
pathogen associated molecular patterns
- PTI
PAMP triggered immunity
- ETI
effector triggered immunity
- ROS
reactive oxidative species
- HR
hypersensitive response
- PR
pathogen response
- DAB
3’-3’ diaminobenzidine
Footnotes
The authors have declared no conflict of interest.
Reference
- 1.Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuppels DA. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Applied and Environmental Microbiology. 1986;51:323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobek JL, Smith JA, Lindgren PB. Suppression of bean defense responses by Pseudomonas syringae. Plant Cell. 1993;5:57–63. doi: 10.1105/tpc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin NC, Martin GB. Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse pseudomonas syringae pathovars to infect tomato. Mol. Plant Microbe Interact. 2007;20:806–815. doi: 10.1094/MPMI-20-7-0806. [DOI] [PubMed] [Google Scholar]
- 8.Mysore KS, Crasta OR, Tuori RP, Folkerts O, et al. Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32:299–315. doi: 10.1046/j.1365-313x.2002.01424.x. [DOI] [PubMed] [Google Scholar]
- 9.Jones J, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.O'Donnell PJ, Schmelz E, Block A, Miersch O, et al. Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 2003;133:1181–1189. doi: 10.1104/pp.103.030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wi SJ, Ji NR, Park KY. Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic Phytophthora parasitica in tobacco plants. Plant Physiol. 2012;159:251–265. doi: 10.1104/pp.112.194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem. J. 1997;322(Pt 3):681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramovitch RB, Martin GB. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Thomma BP, Nurnberger T, Joosten MH. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell. 2011;23:4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiology. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scofield SR, Tobias CM, Rathjen JP, Chang JH, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 17.Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 18.Block A, Alfano JR. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011;14:39–46. doi: 10.1016/j.mib.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh CS, Martin GB. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011;16:132–140. doi: 10.1016/j.tplants.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrov VD, Van Breusegem F. Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants. 2012;2012:pls014. doi: 10.1093/aobpla/pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohn JR, Martin GB. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 2005;44:139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- 23.Jones AM, Bennett MH, Mansfield JW, Grant M. Analysis of the defense phosphoproteome of Arabidopsis thaliana using differential mass tagging. Proteomics. 2006;6:4155–4165. doi: 10.1002/pmic.200500172. [DOI] [PubMed] [Google Scholar]
- 24.Clarke JD. Phenotypic analysis of Arabidopsis mutants: diaminobenzidine stain for hydrogen peroxide. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot4981. pdb.prot4981. [DOI] [PubMed] [Google Scholar]
- 25.Shan L, He P, Zhou J, Tang X. A Cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Molecular Plant-Microbe Interactions. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 26.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. The Plant Journal. 1997;11:1187–1194. [Google Scholar]
- 27.Koh J, Chen S, Zhu N, Yu F, et al. Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol. 2012 doi: 10.1111/j.1469-8137.2012.04251.x. 10.1111/j.1469-8137.2012.04251.x. [DOI] [PubMed] [Google Scholar]
- 28.Law ME, Corsino PE, Jahn SC, Davis BJ, et al. Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene. 2012 doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu M, Dai S, Zhu N, Booy A, et al. Methyl jasmonate responsive proteins in Brassica napus guard cells revealed by iTRAQ-based quantitative proteomics. J. Proteome Res. 2012;11:3728–3742. doi: 10.1021/pr300213k. [DOI] [PubMed] [Google Scholar]
- 30.Shilov IV, Seymour SL, Patel AA, Loboda A, et al. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Molecular & Cellular Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 32.Aidemark M, Andersson C, Rasmusson AG, Widell S. Regulation of callose synthase activity in situ in alamethicin-permeabilized Arabidopsis and tobacco suspension cells. BMC Plant Biology. 2009;9:1–13. doi: 10.1186/1471-2229-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Kim J. Callose synthesis in higher plants. Plant Signaling & Behavior. 2009;4:489–492. doi: 10.4161/psb.4.6.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fangel JU, Ulvskov P, Knox JP, Mikkelsen MD, et al. Cell wall evolution and diversity. Frontiers in Plant Science. 2012;3:1–8. doi: 10.3389/fpls.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nomura K, Mecey C, Lee YN, Imboden LA, et al. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10774–10779. doi: 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura K, DebRoy S, Lee YH, Pumplin N, et al. A Bacterial Virulence Protein Suppresses Host Innate Immunity to Cause Plant Disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 37.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Dean JD, Goodwin PH, Hsiang T. Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J. Exp. Bot. 2005;56:1525–1533. doi: 10.1093/jxb/eri145. [DOI] [PubMed] [Google Scholar]
- 39.Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, et al. Map-Based Cloning of a Protein Kinase Gene Conferring Disease Resistance in Tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 40.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim. Biophys. Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Jarsch IK, Ott T. Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 2011;24:7–12. doi: 10.1094/MPMI-07-10-0166. [DOI] [PubMed] [Google Scholar]
- 42.Moreno JI, Martin R, Castresana C. Arabidopsis SHMT1, a serine hydroxymethyl-transferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2005;41:451–463. doi: 10.1111/j.1365-313X.2004.02311.x. [DOI] [PubMed] [Google Scholar]
- 43.Pageau K, Reisdorf-Cren M, Morot-Gaudry JF, Masclaux-Daubresse C. The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicotiana tabacum L. leaves. J. Exp. Bot. 2006;57:547–557. doi: 10.1093/jxb/erj035. [DOI] [PubMed] [Google Scholar]
- 44.Xu QF, Cheng WS, Li SS, Li W, et al. Identification of genes required for Cf-dependent hypersensitive cell death by combined proteomic and RNA interfering analyses. J. Exp. Bot. 2012;63:2421–2435. doi: 10.1093/jxb/err397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002;53:1331–1341. [PubMed] [Google Scholar]
- 46.Buonaurio R, Torre DG, Montalbini P. Soluble superoxide dismutase (SOD) in susceptible and resistant host-parasite complexes of Phaseolus vulgaris and Uromyces phaseoli. Physiological and Molecular Plant Pathology. 1987;31:173–184. [Google Scholar]
- 47.Vieira Dos Santos C, Rey P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006;11:329–334. doi: 10.1016/j.tplants.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, et al. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol. 2004;136:4088–4095. doi: 10.1104/pp.104.052233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thornalley PJ. Glyoxalase I-structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 50.Singla-Pareek SL, Reddy MK, Sopory SK. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noctor G, Foyer CH. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 52.Jones AM, Thomas V, Bennett MH, Mansfield J, Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006;142:1603–1620. doi: 10.1104/pp.106.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.