Abstract
Objective
To evaluate serovar and antimicrobial resistance patterns of Salmonella spp isolated from healthy, diseased and necropsied cows and calves in this observational study.
Methods
Nineteen isolates recovered from feces and tissues of salmonellosis-affected animals of two commercial farms in north-east of Iran. In second part of the study, the two farms were sampled 4 times with an interval of 2 month. The samples included calves' feces, adult cows' feces, feeds, water, milk filters, and milk fed to calves. Five Salmonella were isolated from 332 fecal samples collected from calves and peri-parturient cows. No Salmonella was recovered from water, feed, milk filers and milk fed to calves.
Results
Salmonella Typhimurium was the most frequently isolate among all sero-groups. S. Dublin was only accounted for 8% (two out of 24) of isolates. Isolated Salmonella strains were used for the ERIC PCR DNA fingerprinting assay. Our results grouped Salmonella isolates into 3 clusters, suggesting that specific genotypes were responsible for each sero-group of Salmonella. The results also revealed diversity among Salmonella isolates in cluster III (sero-group B). Eighteen out of 19 Salmonella spp. were resistant to oxytetracycline. Five isolates out of 19 showed more than one drug resistance. Multi-drug resistance was seen only among Salmonella Typhimurium isolates. Enrofloxacin was the most susceptible antibiotic against all isolates in this study.
Conclusion
The emergence of multiple antibiotic-resistant strains of Salmonella Typhimurium should be of great concern to the public. No correlation between ERIC fingerprinting and resistance patterns of Salmonella isolates was found, which indicates resistance to antimicrobial agents was not related to specific genetic background.
Keywords: Dairy cattle, Salmonella Typhimurium, Antibiotic resistance, ERIC PCR
1. Introduction
Salmonellosis is a common disease of livestock animals; manifestations include diarrhea, dehydration, abortion, depressed mentation, pneumonia, septic arthritis, meningitis, gangrene of distal extremities, and sudden death. The effects of infection can range from subclinical to endotoxemia and death. Salmonella outbreaks can be detrimental to dairy producers due to increased mortality and treatment costs in clinically infected cows[1].
Salmonellosis is one of the most frequently reported bacterial foodborne diseases and is a major economic and public health issue worldwide. In the United States, Salmonella serotypes cause an estimated 1.4 million cases of foodborne disease[2] and 400 deaths annually[3]. European data show that Salmonella is the second most predominant bacterial pathogen, causing around 132 000 human cases in 2008[4]. Ninety-five percent of human cases estimated to be foodborne origin. Humans can be infected with Salmonella from animal sources by many routs. More cases of bovine- associated salmonellosis in humans might result from direct contact with cattle, and ingestion of foods of bovine origin (milk and uncooked beef meat)[5]. Antimicrobial resistance in Salmonella inhibits the ability of physicians and veterinarians to treat severe infections, and results in increases in health care costs and mortality in human patients[6].
The present study designed to provide an on-farm view of the prevalence of Salmonella spp. among healthy, diseased and necropsied cows and calves, to determine the serotypic diversity of Salmonella isolates, and to monitor antimicrobial drug susceptibility of isolates in two large scale dairy farms of north-east of Iran.
2. Materials and methods
2.1. On-farm study
2.1.1. Study population
Two commercial dairy farms belonged to the same company with the same management in north-east of Iran in the province of Khorasan were sampled. The farms were chosen due to previous history of salmonellosis. Both herds milked more than 800 cows. The farms were close herds and did not receive animals from other herds. Cows were housed in open shed with sand and straw for bedding. Non-lactating cows and heifers were housed in separate pens. The calves were housed in individual cement hutches. Calves were typically fed un-pasteurized milk. Non-salable milk (i.e., milk not allowed for human consumption) was fed to calves. Milk was fed with buckets.
2.1.2. Sampling procedure
Farms sampled 4 times with an interval of 2 month. Types and number of samples collected at each visit were summarized in Table 1. The samples included calves' feces, adult cows' feces, feeds, water, milk filters, and milk fed to calves. Fresh fecal samples were collected directly from healthy animals by rectal grab. Milk filters were washed with peptone water into a sterile bottle. All samples were collected between July 5, 2009 and March 8, 2010.
Table 1. Types and numbers of samples collected on visits to 2 dairy farms during farm study.
| Feces |
Raw milk fed to calves | Milk filters | Water | Feeds | |||||||
| Calves<2 weeks | Calves 2-4 weeks | Calves 1-4 month | Calves 4-6 month | Close-up cows and heifers | Fresh cows and heifers | ||||||
| Farm A(Z) | 1st Sampling | 8 | 5 | 10 | 6 | 10 | 5 | 1 | 1 | 1 | 1 |
| 2nd Sampling | 5 | 8 | 5 | 5 | 10 | 14 | 1 | 1 | 1 | 2 | |
| 3rd Sampling | 5 | 5 | 5 | 5 | 10 | 9 | 1 | 1 | 1 | 1 | |
| 4th Sampling | 5 | 5 | 5 | 5 | 10 | 10 | 1 | 1 | 1 | 1 | |
| Farm B(G) | 1st Sampling | 5 | 5 | 5 | 5 | 10 | 10 | 1 | 1 | 1 | 2 |
| 2nd Sampling | 5 | 5 | 7 | 5 | 10 | 11 | 1 | 1 | 1 | 1 | |
| 3rd Sampling | 5 | 5 | 5 | 5 | 10 | 10 | 1 | 1 | 1 | 2 | |
| 4th Sampling | 5 | 5 | 4 | 5 | 10 | 10 | 1 | 1 | 1 | 1 | |
| Total | 43 | 43 | 46 | 41 | 80 | 79 | 8 | 8 | 8 | 11 | |
2.2. Diagnostic study on clinical cases
From June 2007 to March 2010, samples of diseased and necropsied animals from farms A and B were referred to the Diagnostic Laboratory of Center of Excellence in Ruminant Abortion and Neonatal Mortality, Ferdowsi University of Mashhad. Six Salmonella were isolated from feces of diarrheic and 13 isolates from tissues of necropsied animals. These isolates were used for sero-typing and antimicrobial susceptibility testing. No other pathogen was isolated.
The isolation methods of Salmonella spp. are based on Cobbold et al. (2006)[7]. Samples transported to the laboratory besides ice bags. Five grams of each fecal sample added to 45 mL of tetrathionate broth and enriched for 48 h at 37 °C. Liquid samples such as milk and water (60-80 mL) were combined with equal volume of double concentration selenite-F broth and enriched for 24 h at 37 °C. Solid samples such as feeds at the amount of 25 g was added to 225 mL of buffered peptone water, mixed thoroughly, and pre-enriched for 24 h before being added to tetrathionate broth and re-enriched for 24 h at 37 °C.
All enrichments were streaked for isolation on McConkey agar plates and incubated at 37 °C overnight. Lactose negative colonies from each plate were confirmed as Salmonella on the basis of growth pattern (alkaline/acid+H2S and urea negative) on triple-sugar iron and urea agar slants. Isolates were stored at nutrient broth with 15% glycerol at -20 °C for future reference.
2.3. Serogrouping
Salmonella isolates were grouped by use of a commercial slide agglutination method (Kooshafar Biotechnology Research Institute, Karaj, Iran).
2.4. PCR
2.4.1. Oligonucleotide primers
Three sets of primer pairs were used: invA, specific for the invA gene from Salmonella spp.; fliC, specific for the fliC gene from Salmonella Typhimurium; H:eh, specific for the H:eh gene found in Salmonella Newport; and SopE, specific for SopE gene from Salmonella Dublin. Primer sequences are shown in Table 2.
Table 2. Primers used for the detection of Salmonella spp.
| Primer | Nucleotides Sequences (5′-3′) | Use for identify of | Amplification product (bp) | Reference |
| invA | F: GTG AAA TTA TCG CCA CGT TCG GGC AA | Salmonella spp. | 284 | Oliveira et al. (2002)[8] |
| R: TCA TCG CAC CGT CAA AGG AAC C | ||||
| fliC | F: CGG TGT TGC CCA GGT TGG TAA T | Salmonella Typhimurium | 620 | Oliveira et al. (2002)[8] |
| R: ACT GGT AAA GAT GGC T | ||||
| H:eh | F: GCA GAT CAA CTC TCA GAC CCT GGG | Salmonella Newport | 200 | Herrera-Leon et al. (2004)[9] |
| R: AAC GAA AGC GTA GCA GAC AAG | ||||
| sopE | F: ACA CAC TTT CAC CGA GGA AGC G | Salmonella Dublin | 398 | Rahman et al. (2004)[10] |
| R: GGA GCC TTC TGA TGT TGA CTG G |
2.4.2. DNA extraction
The procedure was the same for both pure cultures grown in nutrient agar and field samples enriched in tetrathionate broth. In both the cases DNA extracted by using AccuPrep® Genomic DNA Extraction Kit (Bioneer, Korea).
2.4.3. DNA amplification
PCRs were performed with 5 µL of DNA sample and 25 pmol of each primer by using PCR Premix vials (Bioneer, Korea), containing Taq DNA polymerase 1 U, dNTP mix 250 µmol/L, Tris-HCl 10 mmol/L, KCl 30 mmol/L, MgCl2 1.5 mmol/L and Stabilizer and tracking dye in a final volume of 20 µL. For independent reactions were made for each DNA sample each with one set of primers.
Amplifications were carried out using following conditions: initial denaturation was at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 1.5 min, with a final extension at 72 °C for 10 min.
Electrophoresis of amplification products was on 1.5% agarose gel containing 5 µg/mL ethidium bromide with a 100 bp ladder as molecular weight marker.
2.5. ERIC PCR
Twenty-four Salmonella strains were used for the ERIC PCR DNA fingerprinting assay. And 6 µL of the DNA extract from each isolate was used in a 24 µL of premix PCR tubes (Bioneer Company, Korea). One µl of ERIC primer (5′ AAG TAA GTG ACT GGG GTG AGC G 3′) was added. The PCR amplification included a 2 min hot start at 94 °C, denaturation at 94 °C for 30 seconds, annealing at 60 °C for 1 min, and extension at 72 °C for 4.5 min and after 35 cycles, a final extension for 1 min at 72 °C. Amplified PCR product were visualized by electrophoresis in 1.5% agarose gel containing 0.2 µg/mL ethidium bromide. Gel was run at 100 V for 45 min, and visualized by using Gel Doc.
2.6. Antimicrobial susceptibility test
Antimicrobial susceptibility of Salmonella isolates was performed with the disc diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2008) in Mueller-Hinton agar with 9 antimicrobial agents (amoxicillin, oxytetracycline, enrofloxacin, trimetoprim-sulfametoxazole, gentamycin, cefalexin, kanamycin, chloramphenicol and nalidixic acid) at given concentrations (Mast Diagnostic, UK) as shown in Table 3. The antimicrobials were selected due to commonly using against Salmonella in dairy farms.
Table 3. Frequency and percentage of Salmonella isolates by level of resistance to various antimicrobials.
| Antimicrobial agent | Disc Content (µg) | Susceptible | Intermediate | Resistant |
| Amoxicillin | 10 | 9(47%) | 10(53%) | 0 |
| Oxytetracycline | 30 | 0 | 1(5%) | 18(95%) |
| Enrofloxacin | 5 | 19(100%) | 0 | 0 |
| Trimetoprim-Sulfametoxazole | 25 | 8(44%) | 11(56%) | 0 |
| Gentamycin | 10 | 12(63%) | 4(21%) | 3(16%) |
| Cefalexin | 30 | 2(11%) | 17(89%) | 0 |
| Kanamycin | 30 | 8(42%) | 10(53%) | 1(5%) |
| Chloramphenicol | 30 | 18(95%) | 0 | 1(5%) |
| Nalidixic Acid | 30 | 10(53%) | 7(37%) | 2(5%) |
Salmonella isolates were transferred to BHI broth and incubated at 37 °C until it achieved the turbidity of the 0.5 McFarland standards. The suspension was inoculated uniformly to Mueller-Hinton agar with a sterile cotton swab. Then antibiotic discs were placed onto the plate and incubated at 37 °C for 18 h. According to the sizes of the inhibition zones, interpretation of the strains as susceptible, intermediate, or resistant as made according to the CLSI.
3. Results
Salmonella isolates were recovered from 5 of 332 (1.5%) fecal samples collected from calves and peri-parturient cows. One isolate from 170 and 4 (0.6%) isolates from 162 (2.5%) fecal samples recovered from farm A and B, respectively. Three Salmonella were isolated from calves less than 6 month of age (isolation prevalence, 1.7%). In adults, two isolates were recovered from feces of a fresh cow in farm B and a close-up dry cow in farm A. No Salmonella was recovered from water, feed, milk filers and milk fed to calves.
Nineteen isolates were recovered from clinical cases, which referred to the Laboratory (Table 3).
Isolates of Salmonella belonged to group B (n=19), group D (n=2), and group C (n=1). The sero-groups of two isolates were undermined. The predominant sero-group was B. Thirteen out of 19 sero-group B isolates (68.4%) were identified as Salmonella Typhimurium. In addition, Salmonella Typhimurium was the most frequently isolate among all sero-groups isolates, and accounting for 54% of the all isolates. Other six isolates of sero-group B were undetermined. Salmonella Dublin was only accounted for 8% (two out of 24) of isolates (Table 4).
Table 4. Prevalence of Salmonella isolates sero-group and serotypes from infected, diseased and necropsied animals in two farms during 2007-2010.
| Farm | Animal | Condition of animal | Sample | Serogroup B |
Serogroup C | Serogroup D |
Undertermined | |
| Typhimurium | undertermined | Dublin | ||||||
| A | dry cow | infected | feces | 0 | 0 | 0 | 0 | 1 |
| B | calf | infected | feces | 1 | 0 | 0 | 0 | 0 |
| calf | infected | feces | 0 | 0 | 1 | 0 | 0 | |
| calf | infected | feces | 0 | 0 | 0 | 0 | 1 | |
| Fresh cow | infected | feces | 1 | 0 | 0 | 0 | 0 | |
| A | calf | necropsied | liver | 0 | 0 | 0 | 1 | 0 |
| B | fetus | aborted | liver, lung, kidney & abomasum contents | 0 | 1 | 0 | 0 | 0 |
| calf | necropsied | liver | 0 | 0 | 0 | 1 | 0 | |
| calf | necropsied | bile | 2 | 0 | 0 | 0 | 0 | |
| calf | necropsied | Lmyph node | 3 | 0 | 0 | 0 | 0 | |
| calf | necropsied | Lymph node | 0 | 1 | 0 | 0 | 0 | |
| calf | diseased | feces | 3 | 0 | 0 | 0 | 0 | |
| calf | diseased | feces | 0 | 3 | 0 | 0 | 0 | |
| cow | diseased | feces | 3 | 0 | 0 | 0 | 0 | |
| cow | diseased | feces | 0 | 1 | 0 | 0 | 0 | |
| Total | 13 | 6 | 1 | 2 | 2 | |||
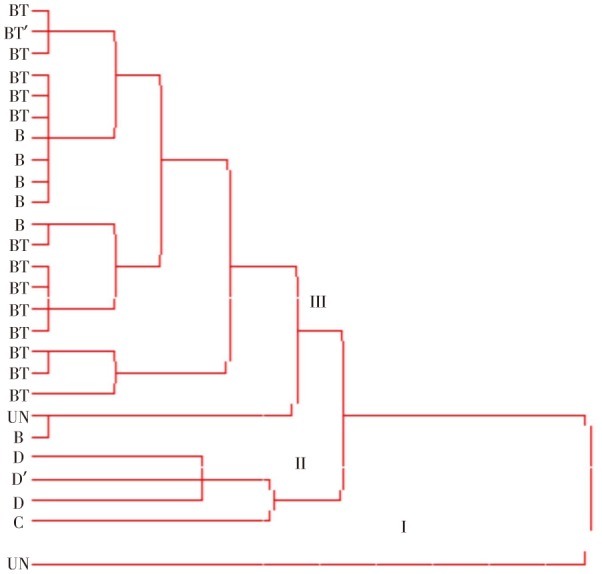
A total of 13 DNA bands were identified for all Salmonella isolates which ranging from 200 to 1 400 bp. The ERIC PCR profiles allowed the differentiation of the 24 isolates into 9 ERIC types (Figure 1), which were grouped into three main clusters (I, II and III). Cluster I comprised of one isolate which was undetermined sero-group. Cluster II comprised of sero-group D and C isolates. Isolates belong to sero-group B and all of the Salmonella Typhimurium were located into cluster III (Figure 2). There was no evidence of association between ERIC DNA fingerprinting and susceptibility to antimicrobial agents.
Figure 1. Detection of invA, sopE and fliC genes by PCR in Salmonella spp. Lane 1 and 3: product of 284 bp for invA gene (Salmonella genes), Lane 2: product of 398 bp for sopE gene (Samonella Dublin), Lane 4: product of 620 bp for fliC gene (Salmonella Typhimurium), Lane 5: 1 Kb DNA ladder.
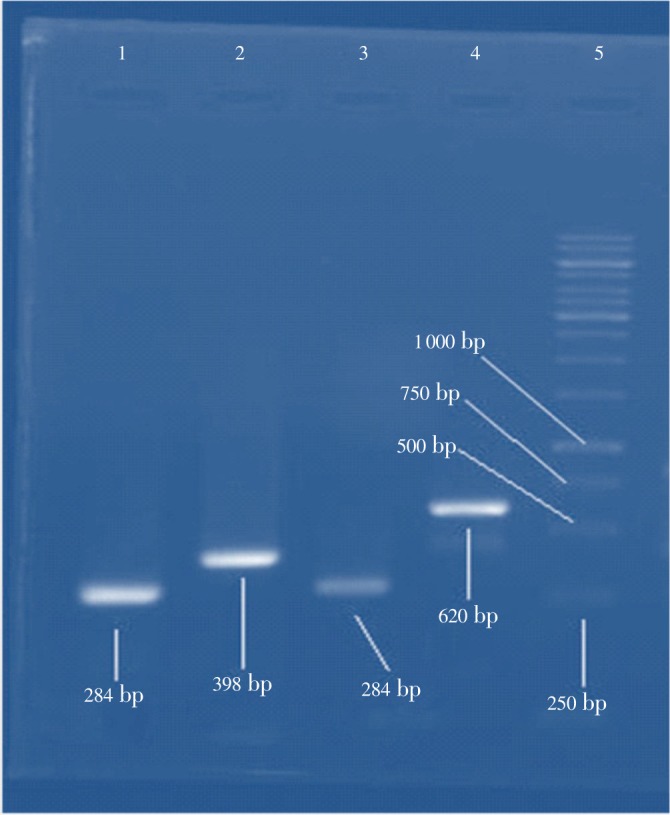
Figure 2. Phylogenetic diversity of 24 Salmonella isolates and 2 controls (Salmonella Typhimurium and Salmonella Dublin) identified by ERIC PCR. The isolates were grouped into three main clusters (I, II and III). UN: undetermined sero-group; C: serogroup C; D: serogroup D (Salmonella Dublin); B: serogroup B; BT: serogroup B (Salmonella Typhimurium); BT': control (Salmonella Typhimurium); D': control (Salmonella Dublin).
Salmonella isolates exhibited resistance to a number of antimicrobial agents. Eighteen out of 19 Salmonella spp. were resistant to oxytetracycline. Only one isolate had intermediate susceptibility to oxytetracycline (Table 4). Five isolates out of 19 (26% of all isolates) showed multi-drug resistance. More than one drug resistance was seen only among Salmonella Typhimurium isolates. Enrofloxacin was the most susceptible antibiotic against all isolates in this study. In addition, susceptibility to chloramphenicol was most common (95% of all isolates). Resistance patterns of Salmonella Typhimurium are described in Table 5.
Table 5. Resistance patterns of Salmonella Typhimurium against antimicrobial agents isolated from animals in farm B.
| Resistance Pattern | No. of Salmonella Typhimurium |
| Oxytetracycline | 8 |
| Oxytetracycline, Gentamycin | 1 |
| Oxytetracycline, Gentamycin, Kanamycin | 1 |
| Oxytetracycline, Gentamycin, Nalidixic acid | 1 |
| Oxytetracycline, Nalidixic acid | 1 |
| Oxytetracycline, Chloramphenicol | 1 |
4. Discussion
At least 3 potentially interacting elements are proposed to be necessary for long-term persistence of a Salmonella strain on a given dairy farm: carrier animals, chain infections and persistence of the organism in the environment[7]. The finding of this study that all isolates of on-farm study were recovered from feces of animals indicates the major role of carrier animals in persistence of infection in the farms. However, earlier studies revealed that prolonged maintenance of Salmonella within cattle herds is often associated with persistent mammary gland infections rather than fecal shedding[11].
Peri-parturient cows appear to be the adult group most susceptible to Salmonella infection, a finding that may be related to the influence of immunosuppression[12]. Two isolates were recovered out of 159 fecal samples (isolation prevalence, 1.25%) in peri-parturient cows in this study. This isolation prevalence in this susceptible group in farms with the history of clinical salmonellosis is relatively low. It is conceivable that the incidence of salmonellosis among dairy herds was underestimated. Furthermore, fecal culture does not have perfect sensitivity for detecting the presence of Salmonella[13]. Many animals that are shedding low numbers of Salmonella organisms are not actively infected and may not be detectable but this low-level shedding reflects environmental contamination[14].
The isolation prevalence of Salmonella (1.7%) in calves is close to a study that was focused on pre-weaned heifers, which 2.1% of calves were, infected[15]. Very few studies compare Salmonella prevalence among age groups of cattle, with mixed results regarding whether calves or cows have a higher prevalence of fecal shedding[13],[16].
It is not unexpected that no Salmonella was isolated from water, feed, milk filers and milk fed to calves. It seems eight samples are not sufficient for Salmonella isolation by conventional culture techniques. In a study of New York dairy herds, Salmonella were isolated from 1.5 percent of 404 milk filters[17]. In another study conducted on 12 dairy farms from Minnesota, Michigan, New York and Wisconsin, it was found that only 1.1% of bulk tank milk (n=91) were positive for Salmonella spp.[18]. It seems conventional culture techniques are not sensitive method to detect low number of the bacteria in milk, feed and water. In the study of Karns et al. (2005), 101 samples (11.8%) were shown to contain the bacteria using the real time PCR assay, whereas conventional culture techniques detected the pathogen in only 22 (2.6%) of the samples[19].
Feeds, water and particularly milk can contain many other organisms that may compete with Salmonella in the enrichment medium, keeping the total number of Salmonella lower than our detection limit on plates[19]. However, generalizations that can be drawn from results of the present study are limited by the low number of farms studied.
Isolates of Salmonella belonged to groups B (n=19), D (n=2), and C (n=1). These sero-groups have been identified as important causes of diseases in calves, dairy cows, and humans[20]. A major finding from our study was the prevalence of Salmonella Typhimurium in these two herds accounting for 54% of the isolates. There is a great diversity in Salmonella serotypes shed by dairy cows. Salmonella Montevido was the most common serovar detected in several studies in United States[14],[21]. In healthy cows, Salmonella Montevido was the prevalent isolate and Salmonella Typhimurium was not among 10 most common isolates[22]. However, Salmonella Newport and Salmonella Typhimurium were the prominent serotypes in salmonellosis affected herds[13]. In agreement with the previous cited study, Salmonella Typhimurium was most isolated from salmonellosis affected animals in the present study. This finding is noteworthy in regard to public health. Salmonella Typhimurium was the second most common Salmonella serotypes isolated from people with laboratory -confirmed food-borne infection in United States[23]. The low isolates of Salmonella Dublin were another striking feature apparent from the present study. Salmonella Dublin is host-adapted to cattle. Host-adapted serotypes are found most often in their host species, where true long-term carriers exist. In contrast, non-host-adapted serotypes rarely achieve carrier status and usually infect an animal for a period of 3 to 16 weeks[24]. In general, the specific serotype of Salmonella to be found on dairy farms is impossible to predict. However, all Salmonella serotypes are pathogens of humans or animals[21].
ERIC PCR was used to assess phylogenetic diversity and source typing of Salmonella isolates. Our results grouped Salmonella isolates into 3 clusters, suggesting that specific genotypes were responsible for each sero-group of Salmonella. The results also revealed diversity among Salmonella isolates in cluster III (sero-group B). This polymorphism could be due to chromosomal recombination or mutation, which underlies certain individual isolates, or due to unrelated isolates with different sources. In a study by Campioni et al (2012), the genotypic diversity was assessed by ERIC-PCR and PFGE. The ERIC-PCR results revealed that 112 strains exhibited a similarity of >85.4% and the PFGE that 96 strains exhibited a similarity of >80.0%[25]. However, in agreement to our results, ERIC-PCR was found to be valuable in the analysis of Salmonella Typhimurium, Salmonella Virchow, Salmonella Enteritidis, Salmonella Abortusequi, Salmonella Choleraesuis, Salmonella Bareilly and Salmonella Dublin[26]. ERIC fingerprinting may be useful in epidemiological studies and the identification of new strains with unknown origins.
Another notable finding was the antibiotic resistance of Salmonella Typhimurium strains in affected and healthy calves and cows. Resistant of Salmonella Typhimurium against oxytetracycline and other antimicrobial agents were remarkable. It seems widespread and inappropriate usage of oxytetracycline in dairy operations is the major cause of Salmonella resistance against this antibiotic. However, resistance to nalidixic acid and chloramphenicol which occurred in three isolates is also concerning. Nalidixic acid and cholamphenicol are prescribed to treat human typhoid fever.
The emergence of multiple antibiotic-resistant strains of Salmonella Typhimurium should be of great concern to the public. An outbreak following the handling of sick calves and consumption of raw milk that contained Salmonella Typhimurium received national attention in United States. In the process of molecular epidemiological investigation of bovine salmonellosis caused by extended-spectrum cephalosporins resistant Salmonella Typhimurium isolates, we observed multiple anti- microbial resistance patterns were observed among the isolates detected in a beef cattle farm in Japan[27].
We found no correlation between ERIC fingerprinting and resistance patterns of Salmonella isolates, which indicates resistance to antimicrobial agents was not related to specific genetic background.
Acknowledgments
This work was supported by research fund of Ferdowsi University of Mashhad (grant number: 353- 88/4/20) and center of excellence in ruminant abortion and neonatal mortality (project number: 342-89/4/7). The authors thank Dr. Zahra Naseri and Mrs. Tahereh Gholamhosseini Moghadam for technical assistance.
Comments
Background
Salmonella enterica is the most frequent cause of foodborne-related deaths and hospitalizations in humans. Salmonella are symptomatically or asymptomatically excreted by dairy cattle and are present in the environment of many dairy farms. The serovars, resistance phenotypes, and genetic subtypes of Salmonella isolated from dairy cattle have considerable overlap with those that cause disease in humans. The epidemiology of salmonellosis in dairy farms and antimicrobial resistance of Salmonella spp. are important aspects of prevention against salmonellosis.
Research frontiers
The carrier animals of Salmonella spp. have major role in the distribution of the disease. In addition, emergence of multi drug resistance serovars of Salmonella in dairy farms has a great impact on public health.
Related reports
The finding of this study that all isolates of on-farm study were recovered from feces of animals is in contrast to earlier studies which shwed that prolonged maintenance of Salmonella within cattle herds is often associated with persistent mammary gland infections rather than fecal shedding.
Innovations and breakthroughs
Salmonella Typhimurium is the prevalent serovar in dairy farms of east-north of Iran. This study has showed that five isolates out of 19 showed more than one drug resistance.
Applications
It may be significant to know the distribution of antimicrobial resistant Salmonella in dairy farms. The results of the present study suggest that carrier animals may act as a significant reservoir of the infection.
Peer review
This is a well-done study on the epidemiology of Salmonella infection in dairy farms. The importance of the article lies on the high prevalence of Salmonella Typhimurium as the major isolated Salmonella spp. in the studied farms. The resistance of the isolates to several antimicrobial agents is remarkable of the public health viewpoint.
Footnotes
Foundation Project: supported by research fund of Ferdowsi University of Mashhad (grant number: 353- 88/4/20) and center of excellence in ruminant abortion and neonatal mortality (project number: 342-89/4/7).
Conflict of interest statement: Authors declare no conflict of interest.
References
- 1.Mohler VL, House J. Salmonellosis in ruminants. In: Anderson DE, Rings DM, editors. Current veterinary therapy-Food animal practice. 5th ed. St. Louise, Mi: Saunders Elseviere; 2009. pp. 106–111. [Google Scholar]
- 2.Bugarel M, Granier SA, Weill FX, Fach P, Brisabois A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC Microbiol. 2011;11:151. doi: 10.1186/1471-2180-11-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA Journal. 2010;1496:288. [Google Scholar]
- 5.Habing GG, Lombard JE, Kopral CA, Dargatz DA, Kaneene JB. Farm-level associations with the shedding of Salmonella and antimicrobial-resistant Salmonella in U.S. dairy cattle. Foodborne Pathog Dis. 2012;9:815–821. doi: 10.1089/fpd.2012.1149. [DOI] [PubMed] [Google Scholar]
- 6.Maragakis LL, Perencevich EN, Cosgrove SE. Clinical and economic burden of antimicrobial resistance. Expert Rev Anti Infect Ther. 2008;6:751–763. doi: 10.1586/14787210.6.5.751. [DOI] [PubMed] [Google Scholar]
- 7.Cobbold RN, Rice DH, Davis MA, Baser TE, Hancock DD. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J Am Vet Med Assoc. 2006;228:585–591. doi: 10.2460/javma.228.4.585. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira SD, Santos LR, Schuch DMT, Silva AB, Salle CTP, Canal CW. Detection and identification of salmonellas from poultry by PCR. Vet Microbiol. 2002;87:25–35. doi: 10.1016/s0378-1135(02)00028-7. [DOI] [PubMed] [Google Scholar]
- 9.Herrera-León S, McQuiston JR, Usera MA, Fields PI, Garaizar J, Echeita MA. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J Clin Microbiol. 2004;42:2581–2586. doi: 10.1128/JCM.42.6.2581-2586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman H, Streckel W, Prager R, Tschape H. Presence of sopE gene and its phenotypic expression among different serovars of Salmonella isolated from man and animals. Indian J Med Res. 2004;120:35–38. [PubMed] [Google Scholar]
- 11.Warnick LD, Crofton LM, Pelzer KD, Hawkins MJ. Risk factors for clinical salmonellosis in Virginia, USA cattle herds. Prev Vet Med. 2001;49:259–279. doi: 10.1016/s0167-5877(01)00172-6. [DOI] [PubMed] [Google Scholar]
- 12.Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Bender JB, et al. et al. Herd-level factors associated with isolation of Salmonella. Prev Vet Med. 2005;70:279–291. doi: 10.1016/j.prevetmed.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Cummings KJ, Warnick LD, Alexander KA, Cripp CJ, Grohn YT, McDonough PL, et al. et al. The incidence of salmonellosis among dairy herds in the northeastern states. J Dairy Sci. 2009;92:3766–3774. doi: 10.3168/jds.2009-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berge AC, Moore DA, Sischo WM. Prevalence and antimicrobial resistance patterns of Salmonella enterica in preweaned calves from dairies and calf ranches. Am J Vet Res. 2006;67:1580–1588. doi: 10.2460/ajvr.67.9.1580. [DOI] [PubMed] [Google Scholar]
- 15.Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Eberly LE, et al. et al. Cattle and environmental sample-level factors associated with the presence of Salmonella in a multi-state study of conventional and organic dairy farms. Prev Vet Med. 2005;67:39–53. doi: 10.1016/j.prevetmed.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Kirchner M, McLaren I, Clifton-Hadley FA, Liebana E, Wales AD, Davies RH. A comparison between longitudinal shedding patterns of Salmonella typhimurium and Salmonella dublin on dairy farms. Vet Rec. 2012;171:194–201. doi: 10.1136/vr.100865. [DOI] [PubMed] [Google Scholar]
- 17.Hassan L, Mohammed HO, McDonough PL, Gonzalez RN. A cross-sectional study on the prevalence of Listeria monocytogenes and Salmonella in New York dairy herds. J Dairy Sci. 2000;83:2441–2447. doi: 10.3168/jds.S0022-0302(00)75135-6. [DOI] [PubMed] [Google Scholar]
- 18.Warnick LD, Kaneene JB, Ruegg PL, Wells SJ, Fossler C, Halbert L, et al. et al. Evaluation of herd sampling for Salmonella isolation on midwest and northeast US dairy farms. Prev Vet Med. 2003;60:195–206. doi: 10.1016/s0167-5877(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 19.Karns JS, Van Kessel JS, McCluskey BJ, Perdue ML. Prevalence of Salmonella eneterica in bulk tank milk from US dairies as determined by polymerase chain reaction. J Dairy Sci. 2005;88:3475–3479. doi: 10.3168/jds.S0022-0302(05)73031-9. [DOI] [PubMed] [Google Scholar]
- 20.Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, FitzPatrick ES. Veterinary microbiology and microbial disease. 2nd ed. London: Wiley Blackwell; 2011. Enterobacteriacaece; pp. 263–287. [Google Scholar]
- 21.Cummings KJ, Warnick LD, Davis MA, Eckmann K, Gröhn YT, Hoelzer K, et al. et al. Farm animal contact as risk factor for transmission of bovine-associated Salmonella subtypes. Emerg Infect Dis. 2012;19:1929–1936. doi: 10.3201/eid1812.110831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells SJ, Fedorka-Cray PJ, Dargatz DA, Ferris K, Green A. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J Food Prot. 2001;64:3–11. doi: 10.4315/0362-028x-64.1.3. [DOI] [PubMed] [Google Scholar]
- 23.Center for Disease Control and Prevention (CDC) Preliminary Foodnet data on the incidence of infection with pathogens transmitted commonly through food-10 states, 2007. Morb Mortal Wkly Rep. 2008;57:366–370. [PubMed] [Google Scholar]
- 24.Smith BP. Salmonellosis in ruminants. In: Smith BP, editor. Large animal internal medicine. 4th ed. St. Louise, Mi: Mosby Elseviere; 2009. pp. 877–881. [Google Scholar]
- 25.Campioni F, Moratto Bergamini AM, Falcão JP. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012;32:254–264. doi: 10.1016/j.fm.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Bennasar A, de Luna G, Cabrer B, Lalucat J. Rapid identification of Salmonella typhimurium, S. enteritidis and S. virchow isolates by polymerase chain reaction based fingerprinting methods. Int Microbiol. 2000;3:31–38. [PubMed] [Google Scholar]
- 27.Sugawara M, Shahada F, Izumiya H, Watanabe H, Uchida I, Tamamura Y, et al. et al. Change in antimicrobial resistance pattern in Salmonella enterica serovar typhimurium isolates detected in a beef cattle farm. J Vet Med Sci. 2012;74:93–97. doi: 10.1292/jvms.11-0240. [DOI] [PubMed] [Google Scholar]


