Abstract
Chronic lung allograft dysfunction (CLAD) is the major factor limiting long-term success of lung transplantation. Polymorphisms of surfactant protein D (SP-D), an important molecule within lung innate immunity, have been associated with various lung diseases. We investigated the association between donor lung SP-D polymorphisms and posttransplant CLAD and survival in 191 lung transplant recipients consecutively transplanted. Recipients were prospectively followed with routine pulmonary function tests. Donor DNA was assayed by pyrosequencing for SP-D polymorphisms of two single-nucleotide variations altering amino acids in the mature protein N-terminal domain codon 11 (Met11Thr), and in codon 160 (Ala160Thr) of the C-terminal domain. CLAD was diagnosed in 88/191 patients, and 60/191 patients have died. Recipients of allografts that expressed the homozygous Met11Met variant of aa11 had significantly greater freedom from CLAD development and better survival compared to those with the homozygous Thr11Th variant of aa11. No significant association was noted for SP-D variants of aa160. Lung allografts with the SP-D polymorphic variant Thr11Th of aa11 are associated with development of CLAD and reduced survival. The observed genetic differences of the donor lung, potentially with their effects on innate immunity, may influence the clinical outcomes after lung transplantation.
Keywords: Lung allograft immunity, lung transplantation, pulmonary surfactant proteins, polymorphisms
Introduction
SP-D is a hydrophilic polymer, a member of the family of collectins and is produced in the lung primarily by type II pneumocytes (1–3). It is composed of homotrimeric subunits with each polypeptide in the subunit consisting of an N-terminal domain with two conserved cysteines that are critical in cross-linking within and between subunits, a collagen-like region, an α-helical neck region and a C-type lectin carbohydrate recognition domain (4). The SP-D molecule is found as single subunits or as oligomers (5,6). SP-D subunits bind to carbohydrate and lipid ligands by the carbohydrate recognition domain, but high affinity interactions require oligomeric assembly mediated by the N-terminal (7). SP-D, as a collectin, binds to the surfaces of bacteria, viruses and fungi, enhancing phagocytosis and intracellular killing, and has been shown to directly inhibit growth of bacteria and fungi by increasing membrane permeability (1,2,8–10).
SP-D orchestrates lung immunity by regulating cytokine production from macrophages and neutrophils and by providing direct or indirect modulation of lymphocyte activity and proliferation (1,2,8,11). SP-D modulates dichotomously, depending on the binding orientation of the molecule, to suppress or enhance lung inflammation by interacting with macrophage cell surface molecules like signal inhibitory regulatory protein-α, CD91 and CD14 (12–15). Mice deficient in SP-D accumulate surfactant phospholipids, exhibit enhanced acute inflammatory responses in the lung to a variety of stimuli (16), and develop chronic inflammation, emphysema and fibrosis (17).
Three coding single nucleotide polymorphism (SNPs) of the SP-D gene (SFTPD, chromosome 10q22.2–23.1) have been described for codons corresponding to amino acid residue 11 (Met11Thr), 160 (Ala160Thr) and 270 (Ser270Thr) in the mature protein, latter one being very infrequent (18,19). Genetic regulation of the quality and/or quantity of the SP-D protein has been suggested as certain SP-D variants have been found to have an association with a varying susceptibility to lung disease. SP-D gene single nucleotide polymorphic variants have been associated with respiratory syncytial virus infection, broncho-pulmonary dysplasia, COPD and risk for tuberculosis infection (18–24).
We hypothesized that SP-D polymorphisms are associated with clinical outcomes after lung transplantation, which represents a complex context of ongoing stimuli of the innate immunity. The goal of our study was to investigate the relationship of the two most frequent polymorphic variants, residue 11 (Met11Thr) and residue 160 (Ala160Thr) (19,23) with the development of chronic lung allograft dysfunction (CLAD) (25) and survival following lung transplantation.
Methods
The study was approved by the Institutional Review Board of the Columbia University Medical Center. Informed consent in adherence to the principles set forth in the Helsinki Declaration was obtained from each patient for the collection of blood samples from the donor of the lung allograft prior to implantation. We performed a retrospective analysis of prospectively banked samples. All samples were prospectively collected specifically to test the hypothesisofgeneandproteinexpressionassociatedwithpostlungtransplant outcomes. This study included 191 patients consecutively transplanted between 2004 and 2010. Post lung transplantation patient underwent pulmonary function testing in the lung transplant clinic weekly for the first 3 months. Beyond 3 months, patients were seen monthly for the first year and on alternating months for the second year. Follow-up frequency was extended to every 3 months beyond the second year after surgery.
Recipient data were prospectively collected with regards to the development of CLAD as determined by the permanent drop of the FEV1 by more than 20% from their posttransplant baseline and survival. CLAD development was monitored using routine pulmonary function tests. The interval of time from transplant to development of CLAD and survival after transplant was monitored.
Biological samples from lung allograft
Blood samples from the donor of the lung allograft were collected at the time of lung procurement. Blood samples were stored at −80°C for subsequent analysis. Total DNA was extracted from blood with a DNA extraction kit (Qiagen, USA), according to the manufacturer’s instructions. Donor demographics and clinical data were collected by Organ Procurement Organization personnel and recorded in the donor medical record.
Lung allograft SP-D variants
The SP-D gene polymorphisms for codons corresponding to amino acid residue 11 (Met11Thr), residue 160 (Ala160Thr) were assessed. Assessment was performed blinded to the clinical data. The SP-D SNPs assessment was performed by pyrosequencing. The PCR-based RFLP genotype method for SP-D (26,27) provided the basis for the pyrosequencing protocol, which is a primer based DNA sequencing method (20). A sequencing primer hybridizes to the single stranded template containing a given SNP and initiates nucleotide incorporation by DNA polymerase. Following nucleotide incorporation, a pyrophosphate group is released and converted to ATP, which then drives the conversion of luciferin to oxyluciferin producing a light, which in turn generates a pyrogram proportional to the number of nucleotides incorporated into the DNA strand. Pyrograms are scored by pattern-recognition software that compares the predicted SNPs pattern (histogram) to the observed pattern (pyrogram; Pyrosequencing AB, Uppsala, Sweden).
Statistical analysis
We analyzed categorical data using the Chi Square and Fisher’s Exact tests. We constructed stratified Cox proportional hazards models to examine associations between genotype and time to events of interest (CLAD and death) with strata for donor race/ethnicity and with adjustment for a priori purposefully selected recipient variables known to affect outcome after lung transplantation (i.e. precision variables). For time-to-death analyses, we censored follow-up time at the end of the study. For the CLAD analyses, we censored follow-up time at death and at end of study. Alleles were assumed to have additive effects. We also generated predicted survival curves adjusted for these covariates using these models. A competing risk analysis using Gray’s model was performed to confirm our findings (28). There were no missing covariate data. Adjusted survival curves were created based on Aalen’s additive model (29). Since this approach does not allow for a stratified Cox model, race was included as fixed effects in the model. Differences were considered significant when the p value was less than 0.05. Continuous variables are expressed as medians and 25th to 75th percentile range. Statistical analysis was performed using SAS 9.2 software (SAS Institute Inc., Cary, NC, USA).
Results
The overall characteristics of the 191 lung transplant recipients and of the donors are shown in Table 1. The overall median follow-up time was 1290 days (823–1843). To date 60/191 patients have died, 4 within 30 days from the lung transplantation and 56 with a median survival of 719 days (274–1269). The 131 patients currently alive have a median follow-up of 1595 days (1091–2054). CLAD was diagnosed in 88 patients, 38 of whom have died with median survival of 902 days (575–1422) and 50 of whom are currently alive with a median follow-up of 1841 days (1450–2296). Patients alive and free of CLAD are 81 with a median follow-up of 1303 days (1006–1823).
Table 1.
Lung transplant recipient and donor characteristics
| Recipients | 191 | Lung donor | |
|---|---|---|---|
| Males | 90 (47%) | Males | 99 (52%) |
| Ethnicity | Ethnicity | ||
| Caucasian | 155 (51%) | Caucasian | 99 (52%) |
| African American | 19 (28%) | African American | 41 (21.5%) |
| Hispanic | 13 (19%) | Hispanic | 38 (20%) |
| Asian Indian | 3 (1.5%) | Asian | 6 (3%) |
| Arabic | 1 (0.5) | Filipino | 2 (1%) |
| Age | 57 (43–62) | Hawaii | 3 (1.5%) |
| Bilateral | 141 (74%) | Asian Indian | 2 (1%) |
| Disease | Age | 35 (23–47) | |
| COPD | 56 (29%) | Last pO2 | 457 (402–506) |
| ILD | 76 (40%) | Smoking | 63 (33%) |
| CF | 37 (20%) | ||
| PPH | 6 (3%) | ||
| Bronchiectasis | 6 (3%) | ||
| Sarcoidosis | 7 (4%) | ||
| Scleroderma | 3 (2%) | ||
| CMV mismatch | 52 (27%) | ||
| PGD-3 at 72 h | 16 (8%) |
The SP-D polymorphic variations for aa11 and aa160 their frequency and the frequency combination between variants of the aa11 and aa160 alleles for all 191 patients are shown in Tables 2 and 3.
Table 2.
SP-D genotype frequency
| Met11Met | 59 (31%) |
| Met11Thr | 104 (54%) |
| Thr11Thr | 28 (15%) |
|
| |
| Ala160Ala | 90 (47%) |
| Ala160Thr | 83 (44%) |
| Thr160Thr | 18 (9%) |
In brackets number of patients.
Table 3.
Observed frequency for aa11 and aa160
| Ala160Ala | Ala160Thr | Thr160Thr | Total | |
|---|---|---|---|---|
| Met11Met | 18 (9.4%) | 23 (12%) | 18 (9.4%) | 59 (31%) |
| Met11Thr | 46 (24%) | 58 (30.2%) | 0 | 104 (54%) |
| Thr11Thr | 26 (14%) | 2 (1%) | 0 | 28 (15%) |
| Total | 90 (47%) | 83 (43.6%) | 18 (9.4%) | 191 (100%) |
Fisher’s exact test: χ2 = 68.7; p < 0.0001.
The characteristics of the patient grouped according to the SP-D single nucleotide polymorphic variants for aa11 and aa160 are shown in Tables 4 and 5, including the ethnicity of the donor, recipient age, end stage pulmonary disease, type of transplant, cytomegalovirus (CMV) mismatch.
Table 6 describes non-adjusted survival models as well as the survival models stratified on donor ethnicity, and with adjustment for donor polymorphic variations in aa11 and aa160, recipient age, disease, procedure type and CMV status. The homozygous Thr11Thr variant of aa11 among donors was associated with a 70% increased rate of CLAD (95% CI, 1.1–2.6), a 40% increased mortality rate (95% CI, 0.9–2.2) with very similar point estimates for the non-adjusted and the adjusted multivariate model. Ad hoc competing risk analysis was also done showing similar results (data not shown). Multivariable-adjusted predicted survival curves are shown in Figures 1 and 2. Variations in the aa160 allele were not associated with the rate of CLAD or death (Table 6). Table 7 shows the cause of death according to the donor aa11 variants.
Table 6.
Nonadjusted survival models
| HR
|
95% CI
|
p-value
|
||
|---|---|---|---|---|
| CLAD | AA11 | 1.6 | 1.1–2.3 | 0.02 |
| AA160 | 1.5 | 1.0–2.1 | 0.047 | |
|
| ||||
| HR
|
95% CI
|
p
|
||
| DEATH | AA11 | 1.4 | 0.89–2.1 | 0.16 |
| AA160 | 0.9 | 0.5–1.4 | 0.5 | |
| Adjusted Survival Models stratified for donor ethnicity
| ||||
|---|---|---|---|---|
| HR
|
95% CI
|
p-value
|
||
| CLAD | AA11 | 1.7 | 1.1–2.6 | 0.01 |
| AA160 | 1.402 | 0.9–2.1 | 0.11 | |
| Age at Transplant | 0.955 | 0.97–1.0 | 0.68 | |
| Disease: ILD | Ref | Ref | Ref | |
| Disease: CF | 0.643 | 0.3–1.6 | 0.33 | |
| Disease: COPD | 1.342 | 0.8–2.3 | 0.29 | |
| Disease: Other | 1.543 | 0.7–3.4 | 0.28 | |
| Bilateral Lung-Tx | 0.759 | 0.4–1.3 | 0.33 | |
| CMV | 1.240 | 0.7–2.1 | 0.41 | |
|
| ||||
| HR
|
95% CI
|
p-value
|
||
| Death | AA11 | 1.4 | 0.9–2.2 | 0.14 |
| AA160 | 0.8 | 0.5–1.4 | 0.43 | |
| Age at Transplant | 1 | 0.97–1.0 | 0.94 | |
| Disease: ILD | Ref | Ref | Ref | |
| Disease: CF | 0.4 | 0.1–1.3 | 0.12 | |
| Disease: COPD | 0.8 | 0.4–1.6 | 0.53 | |
| Disease: Other | 1.0 | 0.5–2.4 | 0.92 | |
| Bilateral Lung-Tx | 1.4 | 0.7–2.9 | 0.31 | |
| CMV | 1.5 | 0.9–2.7 | 0.16 | |
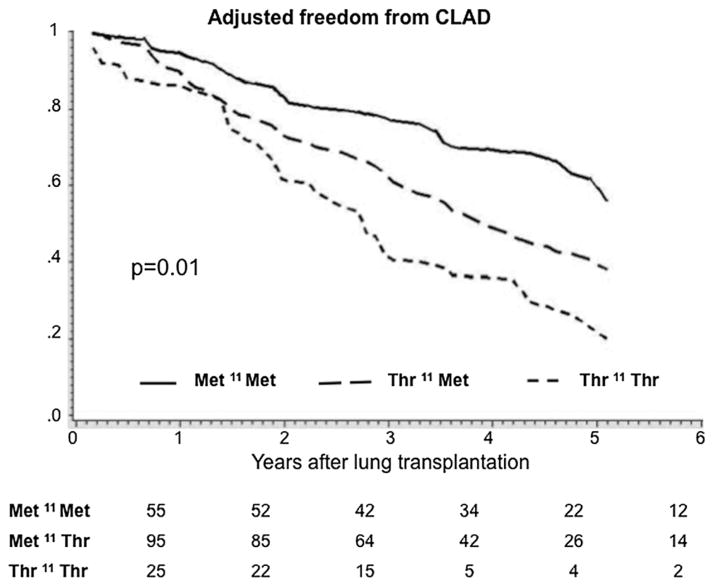
Figure 1. Multivariable adjusted freedom from CLAD curves for recipients grouped according to the lung allograft donor SP-D polymorphic variations for aa11 (Met11Thr).
Adjusted survival estimates are stratified for donor ethnicity and adjusted for recipient covariates (age at the time of transplant, diagnosis, laterality and CMV mismatch). From Table 7 adjusted p = 0.01 for trend. The numbers at the bottom indicate the numbers of lung transplant recipients free of CLAD at each year. The X-axis represents the cumulative freedom from CLAD. The Y-axis represents the time in years after transplant.
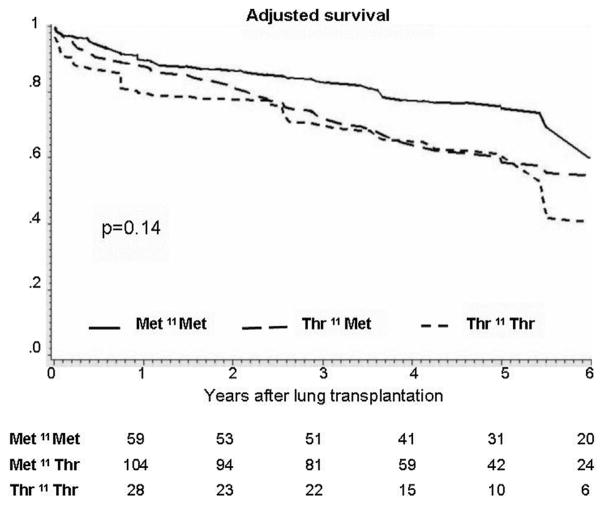
Figure 2. Multivariable adjusted survival curves for recipients grouped according to the lung allograft donor SP-D polymorphic variations for aa11 (Met11Thr).
Adjusted survival estimates are stratified for donor ethnicity and adjusted for recipient covariates (age at the time of transplant, diagnosis, laterality and CMV mismatch). From Table 7 adjusted p = 0.14 for trend. The numbers at the bottom indicate the numbers of lung transplant recipients alive at each year. The X-axis represents the cumulative survival. The Y-axis represents the time in years after transplant.
Table 7.
Cause of death according to donor aa11 variants
| Met11Met | Met11Thr | Thr11Thr | Total | |
|---|---|---|---|---|
| CLAD | 5 (36%) | 4 (12%) | 5 (42%) | 14 |
| CLAD-MOF | 0 | 2 (6%) | 0 | 2 |
| CLAD-PNA | 3 (21%) | 16 (47%) | 3 (25%) | 22 |
| Lung Cancer | 0 | 1 (3%) | 1 (8%) | 2 |
| MI | 0 | 2 (6%) | 0 | 2 |
| MOF | 0 | 1 (3%) | 0 | 1 |
| PGD-MOF | 1 (7%) | 1 (3%) | 2 (17%) | 4 |
| PNA | 5 (36%) | 5 (15%) | 1 (8%) | 10 |
| Sepsis | 0 | 2 (5%) | 0 | 2 |
| Total | 14 (100%) | 34 (100%) | 12 (100%) | 60 |
MOF, multiorgan failure; PNA, pneumonia; MI, myocardial infract.
Discussion
In this study we report a novel finding regarding the role of SP-D gene polymorphisms on clinical outcomes in lung transplantation. Lung allograft SP-D polymorphic variants may serve as a predictor of earlier posttransplant CLAD development and consequently survival. The SP-D gene polymorphisms of two single-nucleotide variations altering amino acids in the mature protein N-terminal domain codon 11 (Met11Thr), and in codon 160 (Ala160Thr) of the C-terminal domain were investigated. We set precedence by delineating a significant association between the donor lung single nucleotide polymorphic variations for aa11 (Met11Thr) with post lung transplant recipient clinical outcome as per CLAD and survival. Patients that received lung allograft from donors homozygous for allele variant Met11Met for aa11 showed a consistently better outcome with greater freedom from development of CLAD and a trend for improved survival. In contrast lung transplant recipients of allografts from donors with homozygous allele variant Thr11Thr for aa11 showed significantly earlier development of CLAD and reduced survival. No association was noted for the single nucleotide polymorphic variations for aa160 (Ala160Thr).
Lung transplantation in the past three decades has evolved into a widely applied therapy for end stage lung disease. Early and long-term graft and patient survival continue to be challenged by both primary graft dysfunction, and CLAD or bronchiolitis obliterans syndrome. Primary graft failure accounts for the majority of early mortality, while CLAD accounts for more than 30% of deaths occurring after the third postoperative year, with a 5-year survival after its onset of only 30–40% (30–32). Liver, kidney and heart transplantation have a recipient and graft 5-year survival of over 70% while lung and intestine linger around 50%. Interestingly both latter organs suffer the disadvantage of having continuous exposure to the environment. Thus they rely on a more active organ-specific innate immunity to provide for the first line of protection from the ongoing environmental noxious agents (33–36). Interactions between innate and adaptive immune responses in these organs in the setting of transplantation are likely a major contributor to the increased graft dysfunction that is observed. Lung allografts are continuously exposed to noxious insults starting with donor brain death, cold ischemic preservation, reperfusion injury, acute rejection, immunosuppression, acute and chronic infections and other injuries such as gastro-esophageal reflux and aspiration (30–32,37,38).
Pulmonary surfactant and the surfactant-related proteins serve as one of the first host defense mechanisms mounted against the various insults. Surfactant phospholipids, in addition to their role in lowering the alveolar surface tension, also serve as part of the physical mucosal barrier to noxious agents (39,40). Surfactant protein D plays different and/or specific roles with respect to surface tension lowering function, phospholipid homeostasis and innate and adaptive immunity (1,2,8,11,39–43). Of the described coding SNPs of the SP-D gene, one results in an alteration of the codon corresponding to amino acid 11 in the mature protein where a methionine (Met) is exchanged for a threonine (Thr) (18–19). The aa11 variants have proved particularly interesting given that studies on patients with tuberculosis have indicated that the presence of the Thr variant may increase the susceptibility to tuberculosis (22), while presence of the Met variant has been associated with increased susceptibility to respiratory syncytial virus infection (18).
The Thr11Thr variant has previously been associated with low serum levels of SP-D in three studies from two independent groups (19,42,44), and was also demonstrated to inhibit the oligomerized state of SP-D and thus reduce binding of bacterial ligands (19).
The SP-D variant frequencies seen in our study patient population (Tables 2 and 3) reflect that of other patient populations described in the literature (19). In fact the population homozygous for allele variant in aa11 and aa160 represent 37% of the total population. This is very consistent with what was previously observed in similar size cohort (206 pts) of Danish Caucasian blood donors. Similarly, linkage disequilibrium was noted between alleles at locus determining aa11 and aa160 as shown in Table 3 (19).
In our study the adjusted survival models and curves that we have presented consistently show better post lung transplantation clinical outcomes in recipients of lung allografts from donors showing homozygous Met11Met variant for the aa11 SNPs. Poor clinical outcomes have demonstrated a consistent association with the homozygous Thr11Thr variant for the aa11 with a significantly increased rate for CLAD or death posttransplantation as shown in Table 6. The aa11 SNPs pertains to the N-terminal domain of the mature protein while no association was noted with the SNPs for the aa160, which is in the C-terminal domain. Of note the N-terminal domain mediates the oligomerization required for the high affinity interactions facilitating simultaneous engagement of individual SP-D molecule with multiple sites on membranes and microbial surfaces (7).
In this study we elucidated a novel association between the SP-D SNPs of the aa11 with the development of CLAD and survival after lung transplantation. The study follows the principles central to Mendelian randomization and should be noted that the multivariate model confirms the point estimate from the unadjusted analysis and the fact that the point estimates are so similar greatly strengthens the likelihood of a true signal. However, limitations are the retrospective nature of the study and the lack of a mechanistic link between the putative changes in multimeric structure due to the SNP’s and increase in CLAD and death. CLAD includes different chronic lung pathology associated with permanent drop of the pulmonary lung function post lung transplantation including bronchiolitis obliterans syndrome, the clinical correlate of chronic rejection (25,45). Moreover, although there are studies from independent groups supporting the relationship between the genotype of interest and the phenotype as per levels of SP-D protein expression (19,42,44), this has not been investigated in the current study. Monitoring for levels of SP-D protein within the broncho-alveolar lavage and/or in serum stratified by SP-D genotype and in relation to clinical and biologic markers of infection, inflammation and rejection may identify whether the association is driven by an increase in infection and/or if there is an immunological effect. These points should be addressed in a prospective investigation to validate the current observations and further study the role of SP-D polymorphisms in post lung transplant outcome.
Lung transplantation may be considered the ultimate “test bench” to study the lung and its responses to stress. In fact, no other model contextualizes the lung with a series of sequential acute and chronic noxious events: brain death, mechanical ventilation, cold ischemic preservation, ischemia reperfusion injury, rejection, chronic immunosuppression, viral, bacterial and fungal infections and chronic silent aspiration from gastro-esophageal reflux. It is indeed quite conceivable that the capability of the lung allograft to withstand the various transplant related insults is driven by its genetic background. The organ-specific innate immunity provided by the surfactant and its associated proteins plays a large part in the first defense mechanisms that the lung exhibits in its protective strategy against the various insults. Further studying of the contribution of genetic variability of the lung allograft’s innate immunity molecules may provide insights into the mechanism of injury occurring post-transplantation.
Table 4.
Donor and recipient characteristics according to donor aa11 variants
| Met11Met | Met11Thr | Thr11Thr | |
|---|---|---|---|
| No. 191 | 59 (31%) | 104 (54%) | 28 (15%) |
| Donor | |||
|
| |||
| Age | 40 (25–47) | 33 (21–47) | 42 (27–49) |
| Ethnicity | |||
| Caucasian | 30 (51%) | 55 (53%) | 14 (50%) |
| African American | 14 (28%) | 21 (20%) | 6 (21%) |
| Hispanic | 11 (19%) | 24 (23%) | 3 (11%) |
| Other | 4 (7%) | 4 (4%) | 5 (18%) |
| Smoking | 18 (31%) | 34 (33%) | 11 (39%) |
| Last pO2 | 453 (411–508) | 460 (411–507) | 411 (388–479) |
|
| |||
| Recipient | |||
|
| |||
| Age | 56 (43–61) | 57 (44–62) | 60 (39–64) |
| Disease | |||
| COPD | 21 (36%) | 28 (27%) | 7 (25%) |
| ILD | 19 (32%) | 44 (42%) | 13 (46%) |
| CF | 13 (22%) | 18 (17%) | 6 (21%) |
| Other | 6 (10%) | 14 (13%) | 2 (7%) |
| Bilateral lung-Tx | 53 (90%) | 69 (66%) | 19 (68%) |
| CMV mismatch | 19 (32%) | 21 (20%) | 12 (43%) |
| PGD-3 at 72 h | 6 (10%) | 8 (8%) | 2 (7%) |
PGD, primary graft dysfunction score 3. Numbers in brackets show 25th to 75th percentile range.
Table 5.
Donor and recipient characteristics according to donor aa160 variants
| Ala160Ala | Ala160Thr | Thr160Thr | |
|---|---|---|---|
| No. 191 | 90 (47%) | 83 (44%) | 18 (9%) |
| Donor | |||
|
| |||
| Age | 36 (21–47) | 35 (25–48) | 40 (24–44) |
| Ethnicity | |||
| Caucasian | 32 (36%) | 54 (65%) | 13 (72%) |
| African American | 30 (33%) | 11 (13%) | 0 (0%) |
| Hispanic | 18 (20%) | 16 (19%) | 4 (22%) |
| Other | 10 (11%) | 2 (2%) | 1 (6%) |
| Smoking | 22 (24%) | 32 (39%) | 8 (44%) |
| Last pO2 | 453 (398–490) | 460 (141–512) | 415 (389–504) |
|
| |||
| Recipient | |||
|
| |||
| Age | 58 (43–62) | 57 (43–62) | 53 (36–60) |
| Disease | |||
| COPD | 24 (27%) | 24 (29%) | 8 (44%) |
| ILD | 38 (42%) | 34 (41%) | 4 (22%) |
| CF | 16 (18%) | 15 (18%) | 6 (33%) |
| Other | 12 (13%) | 10 (12%) | 0 (0%) |
| Bilateral lung-Tx | 66 (73%) | 59 (71%) | 16 (89%) |
| CMV mismatch | 19 (32%) | 21 (20%) | 12 (43%) |
| PGD-3 at 72 h | 8 (9%) | 6 (7%) | 2 (11%) |
PGD, primary graft dysfunction score. Numbers in brackets show 25th to 75th percentile range.
Acknowledgments
The study is supported by the National Institutes of Health, NHLBI, R21HL092478; and NIH HL34788.
Dr. Beatrice Aramini, Ph.D. student supervised by Dr. Frank D’Ovidio and supported by the “Scuola di Dottorato in Scienze Mediche e Chirurgiche Cliniche” of the University of Bologna, Italy.
Abbreviations
- AA
amino acid
- Ala
alanine
- BOS
bronchiolitis obliterans syndrome
- CD
cluster differentiation
- CF
cystic fibrosis
- CLAD
chronic lung allograft dysfunction
- CMV
cytomegalovirus
- COPD
chronic obstructive pulmonary disease
- DNA
deoxyribonucleic acid
- FEV1
forced expiratory volume in 1 second
- ILD
interstitial lung disease
- Met
methionine
- PCR
polymerize chain reaction
- PGD
primary graft dysfunction
- RFLP
restriction fragment length polymorphism
- Ser
serine
- SFTPD
surfactant protein D
- SNP
single nucleotide polymorphism
- SP-D
surfactant protein D
- Thr
threonine
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Holmskov U, Thiel S, Jensenius JC. Collectins and Ficolins: Humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 2.McCormack FX, Whitsett JA. The pulmonary collectins SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest. 2002;109:707–712. doi: 10.1172/JCI15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner MW. The role of mannose-binding lectin in health and disease. Mol Immunol. 2003;40:423. doi: 10.1016/s0161-5890(03)00155-x. [DOI] [PubMed] [Google Scholar]
- 4.Hakansson K, Lim NK, Hoppe HJ, Reid KB. Crystal structure of the trimeric α-helical coiled-coil and the three lectin domains of human lung surfactant protein D. Structure Fold Des. 1999;7:255. doi: 10.1016/s0969-2126(99)80036-7. [DOI] [PubMed] [Google Scholar]
- 5.Hartshorn K, Chang D, Rust K, White M, Heuser J, Crouch E. Interactions of recombinant human pulmonary surfactant protein D and SP-D multimers with influenza A. Am J Physiol. 1996;271:L753. doi: 10.1152/ajplung.1996.271.5.L753. [DOI] [PubMed] [Google Scholar]
- 6.Crouch E, Chang D, Rust K, Persson A, Heuser J. Recombinant pulmonary surfactant protein D: Post-translational modification and molecular assembly. J Biol Chem. 1994;269:15808. [PubMed] [Google Scholar]
- 7.Palaniyar N, Zhang L, Kuzmenko A, et al. The role of pulmonary collectin N-terminal domains in surfactant structure, function, and homeostasis in vivo. J Biol Chem. 2002;277:26971. doi: 10.1074/jbc.M110080200. [DOI] [PubMed] [Google Scholar]
- 8.Crouch E, Hartshorn K, Ofek I. Collectins and pulmonary innate immunity. Immunol Rev. 2000;173:52–565. doi: 10.1034/j.1600-065x.2000.917311.x. [DOI] [PubMed] [Google Scholar]
- 9.McCormack FX, Gibbons R, Ward SR, Kuzmenko A, Wu H, Deepe GS., Jr Macrophage-independent fungicidal action of the pulmonary collectins. J Biol Chem. 2003;278:36250. doi: 10.1074/jbc.M303086200. [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Kuzmenko A, Wan S, et al. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. 2003;111:1589. doi: 10.1172/JCI16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phelps DS. Surfactant regulation of host defense function in the lung: A question of balance. Pediatric Path Mol Med. 2001;20:269–292. [PubMed] [Google Scholar]
- 12.Gardai SJ, Xiao YQ, Dickinson M, et al. By binding SIRP or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 13.Sano H, Chiba H, Iwaki D, Sohma H, Voelker DR, Kuroki Y. Surfactant proteins A and D bind CD14 by different mechanisms. J Biol Chem. 2000;5:22442–22451. doi: 10.1074/jbc.M001107200. [DOI] [PubMed] [Google Scholar]
- 14.Vandivier RW, Ogden CA, Fadok VA, et al. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: Calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169:3978–3986. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- 15.Gardai SJ, Xiao XY, Dickinson M, et al. By binding SIRPα or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 16.LeVine AM, Whitsett JA, Gwozdz JA, et al. Distinct effects of surfactant protein A or D deficiency during bacterial infection on the lung. J Immunol. 2000;165:3934. doi: 10.4049/jimmunol.165.7.3934. [DOI] [PubMed] [Google Scholar]
- 17.Wert SE, Yoshida M, LeVine AM, et al. Increased metallo-proteinase activity, oxidant production, and emphysema in surfactant protein D gene inactivated mice. Proc Natl Acad Sci USA. 2000;97:5972. doi: 10.1073/pnas.100448997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahti M, Lofgren J, Marttila R, et al. Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr Res. 2002;51:696–699. doi: 10.1203/00006450-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Leth-Larsen R, Garred P, Jensenius H, et al. A common polymorphism in the SFTPD gene influences assembly, function and concentration of surfactant protein D. J Immunol. 2005;174:1532–1538. doi: 10.4049/jimmunol.174.3.1532. [DOI] [PubMed] [Google Scholar]
- 20.Pavlovic J, Papagaroufalis C, Xanthou M, et al. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Disease Markers. 2006;22:277–299. doi: 10.1155/2006/817805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman MG, Kong X, DeMeo DL, et al. Polymorphisms in surfactant protein D are associated with chronic obstructive pulmonary disease. Am J Respis Cell Mol Biol. 2011;44:316–322. doi: 10.1165/rcmb.2009-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floros J, Lin HM, Garcia M, et al. Surfactant protein genetic marker alleles identify a subgrioup of tubercolosis in a Mexican population. J Infect Dis. 2000;182:1473–1478. doi: 10.1086/315866. [DOI] [PubMed] [Google Scholar]
- 23.Guo X, Lin HM, Lin Z, et al. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. 2001;18:482–490. doi: 10.1183/09031936.01.00043401. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z, Pearson C, Chinchilli V, et al. Polymorphism of human SP-A, SP-B, and SP-D genes: Association of SP-B Thr131Ile with ARDS. Clin Genet. 2000;58:181–191. doi: 10.1034/j.1399-0004.2000.580305.x. [DOI] [PubMed] [Google Scholar]
- 25.Woodrow JP, Shlobin OA, Barnett SD, Burton N, Natha SD. Comparison of bronchiolitis obliterans syndrome to other forms of chronic lung allograft dysfunction after lung transplantation. J Heart Lung Transplant. 2010;29:1159–1164. doi: 10.1016/j.healun.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 26.DiAngelo S, Lin Z, Philips S, Ramet M, Floros J. Novel non radioactive, simple and multiplex PCR-cRLP methods for genotyping human SP-A and SP-D gene alleles. Dis Markers. 1999;15:269–281. doi: 10.1155/1999/961430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. 1998;1408:312–322. doi: 10.1016/s0925-4439(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 28.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 29.Zhang X, Akcin H. A SAS macro for direct adjusted survival curves based on Aalen’s additive model. Comput Methods Programs Biomed. 2012;108:310–317. doi: 10.1016/j.cmpb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Trulock EP, Edwards LB, Taylor DO, et al. Registry of the international society for heart and lung transplantation: Twenty second official adult lung and heart-lung transplant report-2005. J Heart Lung Transplant. 2005;24:956–967. doi: 10.1016/j.healun.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Boehler A, Kesten S, Weder W, Speich R. Bronchiolitis obliterans after lung transplantation: A review. Chest. 1998;114:1411–1426. doi: 10.1378/chest.114.5.1411. [DOI] [PubMed] [Google Scholar]
- 32.Valentine VG, Robbins RC, Berry GJ, et al. Actuarial survival of heart-lung and bilateral sequential lung transplant recipients with obliterative bronchiolitis. J Heart Lung Transplant. 1996;15:371–383. [PubMed] [Google Scholar]
- 33.Ghanekar A, Grant D. Small bowel transplantation. Curr Opin Critic Care. 2001;7:133–137. doi: 10.1097/00075198-200104000-00013. [DOI] [PubMed] [Google Scholar]
- 34.Brown RS, Rush SH, Rosen HR, et al. Liver and intestine transplantation. Am J Transplant. 2004;4(S9):81–92. doi: 10.1111/j.1600-6135.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- 35.Wynn JJ, Distant DA, Pirsch JD, et al. Kidney and pancreas transplantation. Am J Transplant. 2004;4(S9):72–80. doi: 10.1111/j.1600-6135.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 36.Pierson RN, Barr ML, McCullough KP, et al. Thoracic organ transplantation. Am J Transplant. 2004;4(S9):93–105. doi: 10.1111/j.1600-6135.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 37.Davis RD, Jr, Lau CL, Eubanks S, et al. Improved lung allograft function after fundoplication in patients with gastroesophageal reflux disease undergoing lung transplantation. J Thorac Cardio-vasc Surg. 2003;125(3 Pt 1):533–542. doi: 10.1067/mtc.2003.166. [DOI] [PubMed] [Google Scholar]
- 38.D’Ovidio F, Mura M, Tsang M, et al. Bile acid aspiration and the development of bronchiolitis obliterans after lung transplantation. J Thorac Cardiovasc Surg. 2005;129:1144–1152. doi: 10.1016/j.jtcvs.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Hills BA. Surface active phospholipids: A Pandora’s box of clinical applications. Part II. Barrier and lubricating properties. Internal Med J. 2002;32:242–251. doi: 10.1046/j.1445-5994.2002.00201.x. [DOI] [PubMed] [Google Scholar]
- 40.Goerke J. Pulmonary surfactant: Functions and molecular composition. Biochim Biophys Acta. 1998;1408:79–89. doi: 10.1016/s0925-4439(98)00060-x. [DOI] [PubMed] [Google Scholar]
- 41.Haagsman HP, Diemel RV. Sufactant associated proteins: Functions and structural variation. Comp Biochem Physiol A. 2001;129:91–108. doi: 10.1016/s1095-6433(01)00308-7. [DOI] [PubMed] [Google Scholar]
- 42.Heidinger K, Konig IR, Bohnert A, et al. Polymorphisms in the human surfactant protein-D (SFTPD) gene: Strong evidence that serum levels of surfactant protein-D (SP-D) are genetically influenced. Immunogenetics. 2005;57:1–7. doi: 10.1007/s00251-005-0775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci. 2012;17:407–409. doi: 10.2741/3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen GL, Hjelmborg JB, Kyvik KO, et al. Genetic and environmental influences of surfactant protein D serum levels. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1010–L1017. doi: 10.1152/ajplung.00487.2005. [DOI] [PubMed] [Google Scholar]
- 45.Estenne M, Maurer JR, Boehler A, et al. Bronchiolitis obliterans syndrome 2001: An update of the diagnostic criteria. J Heart Lung Transplant. 2002;21:297–310. doi: 10.1016/s1053-2498(02)00398-4. [DOI] [PubMed] [Google Scholar]