Abstract
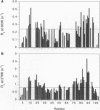
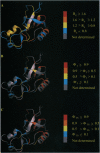
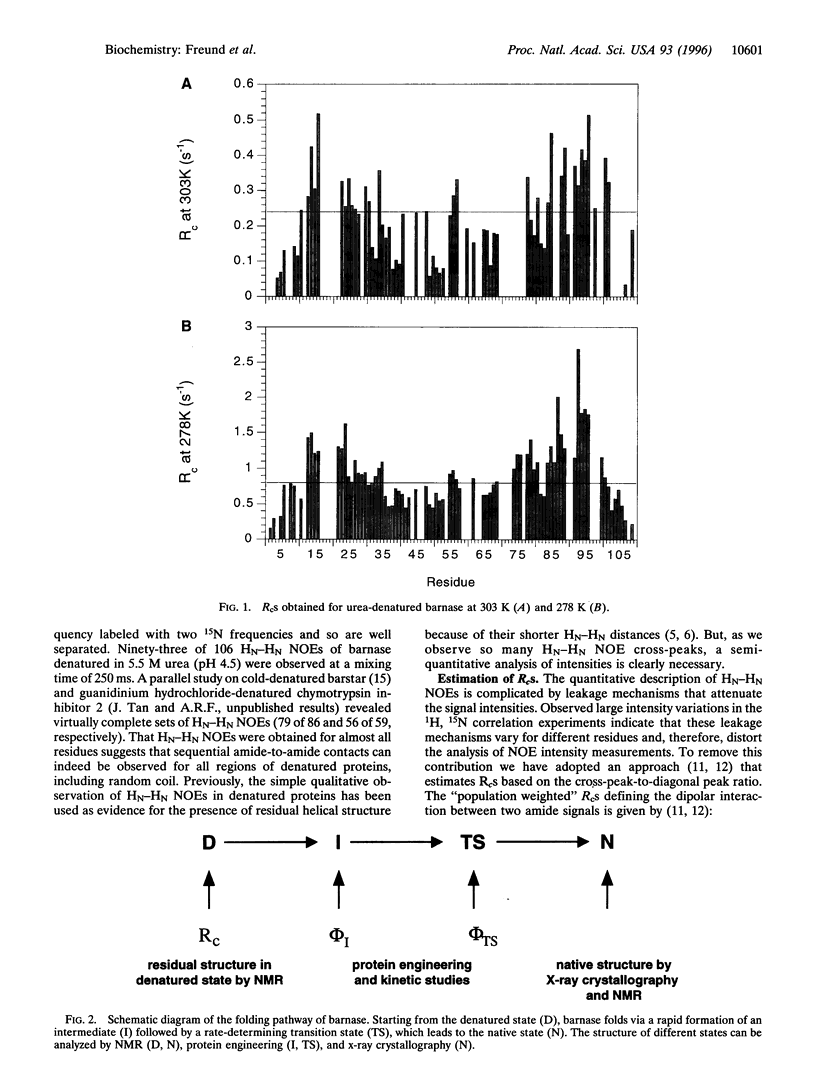
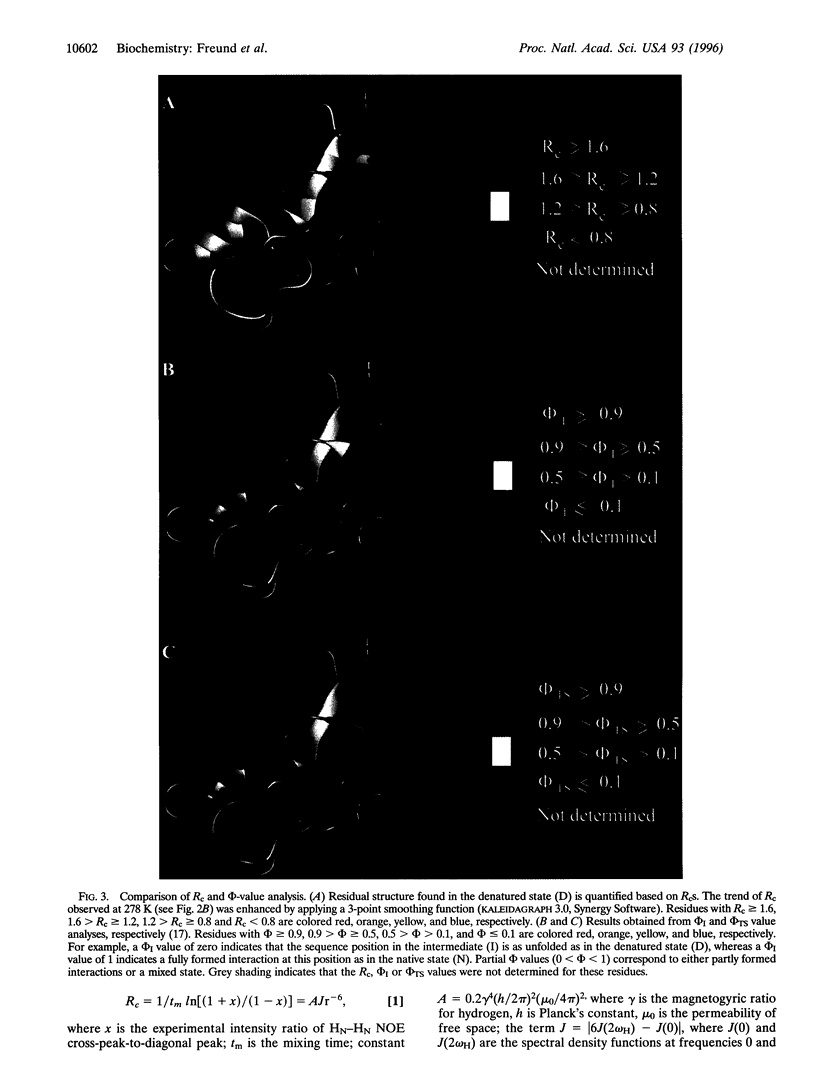
Detailed characterization of denatured states of proteins is necessary to understand the interactions that funnel the large number of possible conformations along fast routes for folding. Nuclear magnetic resonance experiments based on the nuclear Overhauser effect (NOE) detect hydrogen atoms close in space and provide information about local structure. Here we present an NMR procedure that detects almost all sequential NOEs between amide hydrogen atoms (HN-HN NOE), including those in random coil regions in a protein, barnase, in urea solutions. A semi-quantitative analysis of these HN-HN NOEs identified partly structured regions that are in remarkable agreement with those found to form early on the reaction pathway. Our results strongly suggest that the folding of barnase initiates at the first helix and the beta-turn between the third and the fourth strands. This strategy of defining residual structure has also worked for cold-denatured barstar and guanidinium hydrochloride-denatured chymotrypsin inhibitor 2 and so should be generally applicable.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcus V. L., Vuilleumier S., Freund S. M., Bycroft M., Fersht A. R. A comparison of the pH, urea, and temperature-denatured states of barnase by heteronuclear NMR: implications for the initiation of protein folding. J Mol Biol. 1995 Nov 24;254(2):305–321. doi: 10.1006/jmbi.1995.0618. [DOI] [PubMed] [Google Scholar]
- Arcus V. L., Vuilleumier S., Freund S. M., Bycroft M., Fersht A. R. Toward solving the folding pathway of barnase: the complete backbone 13C, 15N, and 1H NMR assignments of its pH-denatured state. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9412–9416. doi: 10.1073/pnas.91.20.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Bromberg S., Yue K., Fiebig K. M., Yee D. P., Thomas P. D., Chan H. S. Principles of protein folding--a perspective from simple exact models. Protein Sci. 1995 Apr;4(4):561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. Characterizing transition states in protein folding: an essential step in the puzzle. Curr Opin Struct Biol. 1995 Feb;5(1):79–84. doi: 10.1016/0959-440x(95)80012-p. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Optimization of rates of protein folding: the nucleation-condensation mechanism and its implications. Proc Natl Acad Sci U S A. 1995 Nov 21;92(24):10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. The sixth Datta Lecture. Protein folding and stability: the pathway of folding of barnase. FEBS Lett. 1993 Jun 28;325(1-2):5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- Karplus M., Sali A. Theoretical studies of protein folding and unfolding. Curr Opin Struct Biol. 1995 Feb;5(1):58–73. doi: 10.1016/0959-440x(95)80010-x. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I. Modeling protein folding: the beauty and power of simplicity. Fold Des. 1996;1(3):R50–R54. doi: 10.1016/s1359-0278(96)00027-2. [DOI] [PubMed] [Google Scholar]
- Shortle D. R. Structural analysis of non-native states of proteins by NMR methods. Curr Opin Struct Biol. 1996 Feb;6(1):24–30. doi: 10.1016/s0959-440x(96)80091-1. [DOI] [PubMed] [Google Scholar]
- Wolynes P. G., Onuchic J. N., Thirumalai D. Navigating the folding routes. Science. 1995 Mar 17;267(5204):1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- Wong K. B., Freund S. M., Fersht A. R. Cold denaturation of barstar: 1H, 15N and 13C NMR assignment and characterisation of residual structure. J Mol Biol. 1996 Jun 21;259(4):805–818. doi: 10.1006/jmbi.1996.0359. [DOI] [PubMed] [Google Scholar]