Abstract
Kidney stones are a global health problem, incurring massive health costs annually. Why stones recur in many patients remains unknown but likely involves environmental, physiological, and genetic factors. The solute linked carrier (SLC) 26A1 gene has previously been linked to kidney stones in mice. SLC26A1 encodes the sulfate anion transporter 1 (SAT1) protein, and its loss in mice leads to hyperoxaluria and calcium oxalate renal stones. To investigate the possible involvement of SAT1 in human urolithiasis, we screened the SLC26A1 gene in a cohort of 13 individuals with recurrent calcium oxalate urolithiasis, which is the commonest type. DNA sequence analyses showed missense mutations in seven patients: one individual was heterozygous R372H; 4 individuals were heterozygous Q556R; one patient was homozygous Q556R; and one patient with severe nephrocalcinosis (requiring nephrectomy) was homozygous Q556R and heterozygous M132T. The M132 amino acid in human SAT1 is conserved with 15 other species and is located within the third transmembrane domain of the predicted SAT1 protein structure, suggesting that this amino acid may be important for SAT1 function. These initial findings demonstrate genetic variants in SLC26A1 of recurrent stone formers and warrant wider independent studies of SLC26A1 in humans with recurrent calcium oxalate stones.
1. Introduction
The formation of insoluble calcium stones in the kidney and/or urinary tract is a major health problem worldwide with ≈6–9% of men and ≈3-4% of women affected [1]. This leads to significant cost in the form of expensive surgical procedures (>$2 billions annually in USA) and morbidity [2]. Most of these stones are composed of calcium and oxalate [3], the latter being a metabolic end product derived from the liver and diet [4], with high urinary oxalate levels (hyperoxaluria) being a major contributor to renal calcium oxalate deposition [5, 6]. We have previously shown normal levels of endogenous oxalate synthesis in recurrent stone formers [7] and individuals are generally compliant with dietary advice to reduce their oxalate intake, suggesting that most stones cannot be attributed to excess oxalate from the liver or diet. There is a genetic predisposition to renal stone formation with ≈15% of patients having an affected family member and ≈50% of patients form a second stone over the subsequent 10 years [8]. However, in most cases the aetiology of renal stone deposition is unknown [9].
Calcium oxalate stone formation is a multifactorial disease involving environmental, physiological, and genetic factors [12]. Numerous genetic studies have identified possible links between calcium oxalate stones and polymorphisms in genes that regulate calcium homeostasis [12–15], mediate inflammatory responses [16, 17], and maintain the structure and function of renal tubules [18–21]. Hyperoxaluria is the most frequent metabolic abnormality found in calcium oxalate stone formers and its basis can be idiopathic (most common), secondary (e.g., enteric hyperoxaluria due to intestinal dysfunction or resection), or due to primary hyperoxaluria (PH) [12]. The primary hyperoxalurias (PH 1 and PH 2) are rare forms of severe hyperoxaluria and calcium oxalate stones and are caused by mutations in the AGXT and GRHPR genes, respectively [22]. Monozygotic twin studies indicate that there is a genetic predisposition to idiopathic hyperoxaluria [12, 23], which may be relevant to the SAT1 sulfate anion transporter (protein is SAT1; gene is SLC26A1) that has been linked to calcium oxalate stone formation in mice [24].
SAT1 is expressed in the intestine, kidney, and liver where it exchanges sulfate (SO4 2−) with structurally similar anions including oxalate (C2O4 2−) [26–28]. This exchange process is proposed to be important for the removal of oxalate from the blood (via the intestine), as well as for maintaining circulating (via the kidneys) and hepatic levels of nutrient sulfate [4, 6, 24, 29–31]. Loss of SAT1 in mice led to disturbances in sulfate homeostasis (hyposulfataemia and hypersulfaturia), as well as hyperoxalaemia and hyperoxaluria [24]. The finding of calcium oxalate stones in SAT1 null mice in this landmark paper was remarkable because rodents are extremely refractive to forming stones, even when mild-moderate hyperoxaluria occurs [32].
To date, the contribution of human SAT1 to calcium oxalate stones is unknown. As an initial approach, we compared the SLC26A1 gene sequence of 13 male patients with recurrent calcium oxalate stones to NCBI reference SLC26A1 sequences for human and 15 other species. Our findings of three nonsynonymous gene variants in SLC26A1, including a missense variant at a highly conserved residue in one patient with severe nephrocalcinosis, warrant wider independent studies of SLC26A1 in recurrent calcium oxalate stone formers.
2. Materials and Methods
2.1. Patient Cohort
The research protocol was approved by the Mater Human Research Ethics Committee. All persons gave their informed consent prior to their inclusion in the study. The eligibility criteria for our study were adult patients with recurrent calcium oxalate stones, based on laboratory stone analysis (calcium oxalate >75% of the stone), for whom the aetiology of calcium oxalate stones was unknown, and if the diagnosis of primary hyperoxaluria or enteric hyperoxaluria was able to be excluded by their treating clinician. Buccal swab samples for DNA analysis were collected but individual patient characteristics were not.
2.2. Genomic DNA Isolation and PCR and DNA Sequencing
Genomic DNA was prepared from buccal swabs according to the manufacturer's protocol (Isohelix). Five overlapping fragments of SLC26A1 (Figure 1(a)) were PCR-amplified using 200 nM forward and reverse primers (Table 1) in a C1000 thermal cycler (BioRad). The thermal cycling protocol was 94°C for 1 min; 35 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 2 min, followed by 1 cycle 72°C for 5 min. DNA sequencing of SLC26A1 was performed using 50 ng PCR product (purified with NucleoSpin Extract II, Macherey-Nagel) and 9.6 pmol forward and reverse primers (Table 1), ABI Prism BigDye Terminator Sequence chemistry and an AB3730x/96-capillary sequencer (Applied Biosystems). DNA sequence analyses included the SLC26A1 coding region (2 exons of 2103 bp total), exon-intron junctions, 5′-(378 bp) and 3′-(1189 bp) UTRs and ≈300 bp 5′-flanking region containing the minimal (135 bp) SLC26A1 promoter region [33]. Nucleotide changes in SLC26A1 were verified by DNA sequence analysis of 2 independent PCR-amplified products for both sense and antisense strands.
Figure 1.
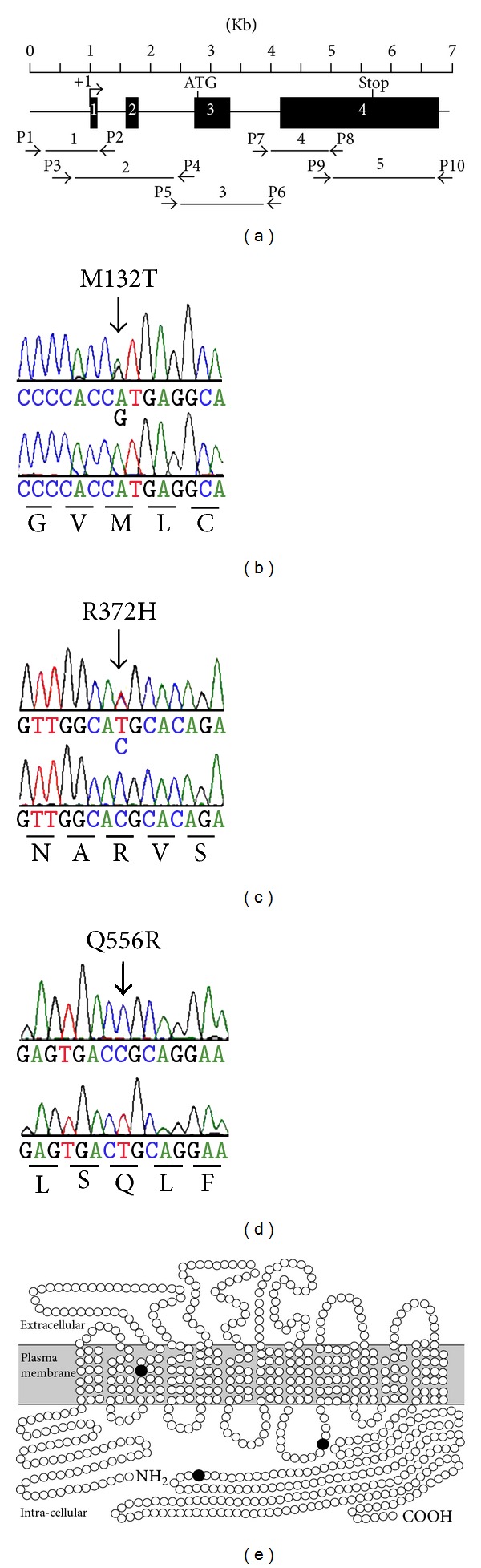
Identification of variants in the human SLC26A1 gene and predicted amino acid substitutions. (a) Schematic of SLC26A1 showing 4 exons (boxes), transcription initiation site (+1, arrow), and the relative location of PCR primers (P1–P10) and PCR amplicon fragments (1–5) used for DNA sequence analysis. ((b)–(d)) Representative DNA sequence chromatographs showing variant (top panels) and control (bottom panels) sequences that predict (b) heterozygous M132T, (c) heterozygous R372H, and (d) homozygous Q556R. Reverse complement nucleotide sequences are shown. (e) Position of amino acid substitutions (•) in the predicted secondary structure model of the human SAT1 protein [25]. M132T within the third transmembrane domain; R372H in the fourth intracellular loop; and Q556R in the intracellular carboxy-terminal domain.
Table 1.
Primers used for PCR and DNA sequencing.
| Primer | aDirection | Sequence (5′ to 3′) | bPCR fragment |
|---|---|---|---|
| Primers used for PCR | |||
| P1 | F | GCTGAGGCAGGAGAATGGCGTGAAC | 1 |
| P2 | R | CTGGGGTCCAGGTGTGTGGAATGG | 1 |
| P3 | F | CAGCTTGGACCAGGCTGGCTCCTTG | 2 |
| P4 | R | GGATTGGTGCTGGGCCTTCTCCT | 2 |
| P5 | F | GTAATCCCATCTCACCTCACGATG | 3 |
| P6 | R | CATCGAGCAGTGGCTGTGAGAGGTAG | 3 |
| P7 | F | GATGCTCACGTGGATGTCACAGTTG | 4 |
| P8 | R | CGAACTCTGTGGCATCCTCGTAGAAG | 4 |
| P9 | F | CACCTGTATGCTGGTCAGCACAGAG | 5 |
| P10 | R | TGTCCCATTCCTTCCACCTAGAG | 5 |
| Primers used for sequencing | |||
| P11 | F | CCAGGAGGCAGAGCTTCCAG | 1 |
| P12 | F | GCATATGCACGCACCGGCAGCCTTG | 1 |
| P13 | R | AGCCTGAGGGTGCCCCTAAGAAACC | 1 |
| P14 | R | CAAGGAGCCAGCCTGGTCCAAGCTG | 1 |
| P15 | F | CTGGCTCCTTG CAGGACCAG | 2 |
| P16 | F | CCATTCCACACACCTGGACCCCAG | 2 |
| P17 | R | CTTCTCAGCGACGTTAGGCAAAGAC | 2 |
| P18 | R | CATCGTGAGGTGAGATGGGATTAC | 2 |
| P19 | F | AGGAGAAGGCCCAGCACCAATCC | 3 |
| P20 | F | CAAGCACAGGGTTGGCAGAGGAGGTG | 3 |
| P21 | R | CCTCACCAGCACCTGGCATGG | 3 |
| P22 | R | TGCCCAAGCCTTGCTGTCTTG | 3 |
| P23 | F | TTGTCAGGTCCTGTGCCGTGAC | 4 |
| P24 | R | CTCTGTGCTGACCAGCATACAGGTG | 4 |
| P25 | F | CTTCTACGAGGATGCCACAGAGTTCG | 5 |
| P25 | F | ATCACACGCAGGACCCAAACACTCAG | 5 |
| P26 | R | ATTCCTTCCACCTAGAGCTGAGG | 5 |
| P27 | R | CTGAGTGTTTGGGTCCTGCGTGTGAT | 5 |
aF: forward primer; R: reverse primer. bPCR fragment denoted in Figure 1(a).
2.3. Bioinformatic Analyses
Prediction of protein stability changes for amino acid substitutions was determined as previously described [10] using SAT1 amino acid sequence NP_998778.1. Alignment of SAT1 amino acid sequences between human (NP_998778.1) and multiple species was generated using the Clustal W program [11]. Putative transcription factor binding sites in the human SLC26A1 5′-flanking region were determined using MatInspector [34] and NCBI reference sequence NT_037622.5.
3. Results and Discussion
To date, two variants (G368S and Q556R) in the human SLC26A1 gene have been reported, based on information from the NCBI single nucleotide polymorphism (SNP) database [25]. Whilst SLC26A1 has yet to be associated with any human disease, the mouse Slc26a1 gene has been linked to hyperoxaluria and calcium oxalate kidney stones [24]. Since human SAT1 and mouse Sat1 share high homology (78% amino acid identity) [33] and a similar tissue distribution with strong expression in the kidneys [27, 28], we propose a potential role for human SLC26A1 in renal stone pathology.
In this study, we screened the SLC26A1 gene in a cohort of recurrent calcium oxalate kidney stone formers and identified single nucleotide variants in the coding region of eight individuals (Table 2). Three of these variants predict amino acid substitutions (M132T, R372H, and Q556R), and four nucleotide variants are silent changes (T225T, S306S, R439R, and A625A). In addition, one variant was detected in the 5′-untranslated region of SLC26A1 exon 1 for three individuals, and four variants were found within the first 210 nucleotides of the SLC26A1 5′-flanking region for eight individuals (Table 2). The allele frequencies of 5 variants are shown in Table 3 and Figure 2. As yet, we do not know the allelic frequencies of SLC26A1 variants in recurrent calcium oxalate stone formers but that will be the next phase of our work.
Table 2.
SLC26A1 sequence variants identified in a cohort of recurrent calcium oxalate stone formers.
| aIndividuals with recurrent calcium oxalate kidney stones | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence variant | A | B | C | bD | E | F | G | H | I | J | K | L | M |
| Coding nonsynonymous | |||||||||||||
| c.395T>C, p.M132T | − | − | − | + | − | − | − | − | − | − | − | − | − |
| c.1115G>A, p.R372H | − | − | − | − | − | − | − | − | − | − | − | + | − |
| c.1667A>G, p.Q556R | − | ++ | − | ++ | + | + | − | − | − | − | + | − | + |
| Coding synonymous | |||||||||||||
| c.675C>A, p.T225T | − | − | − | − | − | + | − | − | − | − | − | − | + |
| c.918G>A, p.S306S | − | + | − | − | + | + | + | − | − | − | + | − | + |
| c.1315C>A, p.R439R | − | − | − | + | − | − | − | − | − | − | − | − | − |
| c.1875C>T, p.A625A | − | + | − | − | + | + | + | − | − | − | + | − | + |
| 5′ Untranslated region c | |||||||||||||
| g.977205C>T | + | − | + | − | − | − | + | − | − | − | − | + | − |
| 5′ Flanking region c | |||||||||||||
| g.977276A>G | − | + | + | ++ | + | − | + | + | + | − | + | − | − |
| g.977343C>A | − | + | − | + | + | − | + | + | + | − | + | − | − |
| g.977391G>C | − | − | − | − | + | − | + | + | − | − | + | − | − |
| g.977420C[6] | − | − | − | − | − | − | + | − | − | − | − | − | − |
aThe genotypes of thirteen individuals (A–M) are indicated as + (heterozygous) or ++ (homozygous) for each sequence variant. bIndividual (D) with severe nephrocalcinosis requiring left nephrectomy. cGenomic DNA reference NT_037622.5.
Table 3.
Predicted effect of missense gene variants on SAT1 protein structure stability, and allelic frequencies of these variants in the NCBI SNP database.
| SLC26A1 variant | aStability prediction | bSVM score | Allelic frequency this study | Allelic frequency cNCBI database | cNCBI SNP accession |
|---|---|---|---|---|---|
| M132T | decrease | −0.9 | 0.038 | — | — |
| R372H | decrease | −1.0 | 0.038 | 0.0082 | rs73219719 |
| Q556R | increase | 0.2 | 0.307 | 0.3484 | rs3796622 |
aPrediction of protein stability and bSupport Vector Machine modelling scores (values less than zero are significant) [10]. cNCBI database accessed on October 30 2012.
Figure 2.
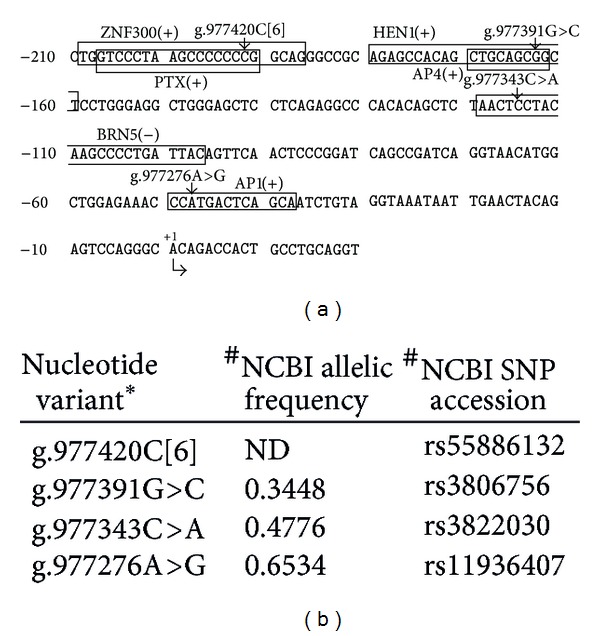
Sequence variants identified in this study and their location within putative transcription factor binding sites in the human SLC26A1 5′-flanking region. (a) Nucleotide sequence of the SLC26A1 5′-flanking region (NCBI accession number NT_037622.5) from −210 to +20 is shown. Position +1 denotes the putative transcription initiation site (NCBI accession number AK292747). Putative transcription factor binding motifs are boxed, and their position on the plus (+) or minus (−) strands is indicated: ZNF300: KRAB-containing zinc finger protein 300; PTX1: pituitary homeobox 1; HEN1: helix-loop-helix transcription factor; AP4: activator protein 4; BRN5: BM-5 POU domain factor; AP1: activator protein 1. (b) Allelic frequencies and accession numbers of nucleotide variants in the NCBI database: #accessed 7 November 2012. *Nucleotide numbering based on NCBI reference sequence NT_037622.
Destabilisation of protein structure is the most common mechanism by which an amino acid variant causes loss of protein function [35]. Using the Support Vector Machine program [10], we identified M132T and R372H to have a significant deleterious effect on the structural stability of human SAT1 protein (Table 3). In addition, the location of variants within membrane proteins can also be useful for predicting the functional consequences of amino acid changes. For example, nonconservative amino acid substitutions within the transmembrane domains (TMDs) of plasma membrane transporters frequently have a negative consequence on protein function [25]. The human SAT1 protein is predicted to contain 12 TMDs, with intracellular amino and carboxy-termini [28]. The nonconservative (hydrophobic to hydrophilic) M132T substitution is located within the third putative TMD of SAT1 (Figure 1(e)), suggesting that M132 may be important for maintaining the TMD structure of SAT1. R372H is located within the fourth intracellular loop, whereas Q556R is positioned in the intracellular carboxy-terminal sequence of the predicted secondary structure of SAT1 (Figure 1(e)), suggesting the possibility that these 2 amino acid substitutions may not disrupt the TMD structure of SAT1.
Conservation of amino acid sequences between multiple species can be useful to identify those residues that are important for protein function [36]. In order to determine if the nonsynonymous variants M132T, R372H, and Q556R occur at conserved positions of the SAT1 protein, we aligned the human SAT1 sequence with homologues from 15 different species (Table 4). Human M132 is conserved between all species, suggesting that this amino acid may play an important role in the structure and or function of SAT1. The R372H variant is a conservative amino acid change (i.e., both arginine and histidine are positively charged amino acids). Whilst human R372 is conserved with 11 other species (Table 4), this residue aligns with cysteine in rhesus monkey, serine in both rat and mouse, and glutamic acid in the African clawed frog (Table 4), implying that R372 may not be essential for SAT1 structure or function. Human Q556 aligns with an arginine residue in 10 species, including chimpanzee, rhesus monkey, dog, rat, and mouse, demonstrating that SAT1 sequence containing Q556R is conserved with other species. This finding implies that Q556 is a more recent amino acid in humans, due to genetic drift or selection, to become the major allele (≈0.65, Table 3). In addition, the presence of R556 in 10 species, together with the relatively high allelic frequency (≈0.35) of R556 in the human population (Table 3), suggests that the Q556R variant may not be pathogenetic in humans.
Table 4.
Alignment of human SAT1 amino acid variants across species.
| Species, accession number | % Identity to human SAT1 | Human SAT1 variants | ||
|---|---|---|---|---|
| aM132T | R372H | Q556R | ||
| Human | 100 | M | R | Q |
| Chimpanzee, XM_003310211.1 |
98 | M | R | R |
| Pygmy chimpanzee, XM_003811300.1 |
98 | M | R | R |
| Sumatran orangutan, XM_002814495.1 |
96 | M | R | R |
| Northern white-cheeked gibbon, XM_003280551.1 | 96 | M | R | R |
| Olive baboon, XM_003890923.1 |
95 | M | R | R |
| Rhesus monkey, XM_001093208.2 |
94 | M | C | R |
| Bolivian squirrel monkey, XM_003934620.1 |
93 | M | R | R |
| Dog, XM_545984.4 |
80 | M | R | R |
| Cattle, XM_002688445.1 |
79 | M | R | Q |
| Rat, CH474079.2 |
78 | M | S | R |
| Mouse, BC025824.1 |
78 | M | S | R |
| African clawed frog, NM_001090973.1 |
51 | M | E | K |
| Rainbow trout, NM_001124486.1 |
50 | M | R | K |
| Japanese eel, AB111927.1 |
49 | M | R | K |
| Zebra fish, NM_001080667.1 |
48 | M | R | K |
Human SAT1 variants detected in this study are shown at the top, and the aligned amino acid for each species is shown below. Alignments were generated using the Clustal W program [11]. aIdentical amino acid across species.
Previous studies identified the first 135 bp of the human SLC26A1 5′-flanking region to be sufficient for basal promoter activity [28]. In the SLC26A1 5′-flanking region of eight individuals, we identified four nucleotide variants (Table 2) which are located within putative transcription factor binding motifs (Figure 2(a)). One variant, g.977276A>G, is located within an AP1 site, previously shown to be important for the transcriptional activity of human SLC26A1 [28]. However, this variant does not alter the essential core nucleotides of the AP1 binding motif sequence. Similarly, the other three variants found within the 5′-flanking region do not alter the essential core nucleotide sequences of the ZNF300, HEN1, or BRN5 transcription factor binding motifs, indicating that these variants may not be detrimental to the transcriptional activity of the SLC26A1 promoter. These findings, together with the high allelic frequencies (0.34 to 0.65, NCBI database) of g.977391G>C, g.977343C>A, and g.977276A>G in the general population (Figure 2(b)), imply that these variants may not be pathogenetic.
4. Conclusions
This study provides preliminary and as yet unreported evidence of sequence variants in the human SLC26A1 gene of a small number of individuals with recurrent calcium oxalate kidney stones. Of great interest is the finding of a nonconservative amino acid substitution (M132T) within a predicted transmembrane domain of SAT1, in one patient with severe nephrocalcinosis. The novelty of these findings warrants further studies of SLC26A1 in humans with unexplained recurrent kidney stones.
Conflict of Interests
The authors declare that they have no conflict of interests regarding the publication of this paper.
Acknowledgments
The authors thank S. Diaz-Guilas, S. Hancock, and the Mater Urology Unit for patient recruitment and sample collection. This study was supported by the Mater Medical Research Institute and the Mater Foundation.
References
- 1.Hughes P. Kidney stones epidemiology. Nephrology. 2007;12(1):S26–S30. doi: 10.1111/j.1440-1797.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 2.Lotan Y. Economics and cost of care of stone disease. Advances in Chronic Kidney Disease. 2009;16(1):5–10. doi: 10.1053/j.ackd.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Gault MH, Chafe L. Relationship of frequency, age, sex, stone weight and composition in 15,624 stones: comparison of results for 1980 to 1983 and 1995 to 1998. Journal of Urology. 2000;164(2):302–307. [PubMed] [Google Scholar]
- 4.Holmes RP, Goodman HO, Assimos DG. Contribution of dietary oxalate to urinary oxalate excretion. Kidney International. 2001;59(1):270–276. doi: 10.1046/j.1523-1755.2001.00488.x. [DOI] [PubMed] [Google Scholar]
- 5.Khan SR, Glenton PA, Byer KJ. Dietary oxalate and calcium oxalate nephrolithiasis. Journal of Urology. 2007;178(5):2191–2196. doi: 10.1016/j.juro.2007.06.046. [DOI] [PubMed] [Google Scholar]
- 6.Brown JM, Chalmers AH, Cowley DM, McWhinney BC. Enteric hyperoxaluria and urolithiasis. The New England Journal of Medicine. 1986;315(15):970–971. doi: 10.1056/NEJM198610093151516. [DOI] [PubMed] [Google Scholar]
- 7.McWhinney BC, Nagel SL, Cowley DM, Brown JM, Chalmers AH. Two-carbon oxalogenesis compared in recurrent calcium oxalate stone formers and normal subjects. Clinical Chemistry. 1987;33(7):1118–1120. [PubMed] [Google Scholar]
- 8.Rose GA. Urinary Stones: Clinical and Laboratory Aspects. Lancaster, UK: MTP; 1981. [Google Scholar]
- 9.Park S, Pearle MS. Pathophysiology and management of calcium stones. Urologic Clinics of North America. 2007;34(3):323–334. doi: 10.1016/j.ucl.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62(4):1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 11.Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Computer Applications in the Biosciences. 1992;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 12.Sayer JA. The genetics of nephrolithiasis. Nephron. 2008;110(2):e37–e43. doi: 10.1159/000151730. [DOI] [PubMed] [Google Scholar]
- 13.Reed BY, Gitomer WL, Heller HJ, et al. Identification and characterization of a gene with base substitutions associated with the absorptive hypercalciuria phenotype and low spinal bone density. Journal of Clinical Endocrinology and Metabolism. 2002;87(4):1476–1485. doi: 10.1210/jcem.87.4.8300. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y, Pasch A, Bonny O, Mohaupt MG, Hediger MA, Frey FJ. Gain-of-function haplotype in the epithelial calcium channel TRPV6 is a risk factor for renal calcium stone formation. Human Molecular Genetics. 2008;17(11):1613–1618. doi: 10.1093/hmg/ddn048. [DOI] [PubMed] [Google Scholar]
- 15.Vezzoli G, Terranegra A, Arcidiacono T, et al. R990G polymorphism of calcium-sensing receptor does produce a gain-of-function and predispose to primary hypercalciuria. Kidney International. 2007;71(11):1155–1162. doi: 10.1038/sj.ki.5002156. [DOI] [PubMed] [Google Scholar]
- 16.Mittal RD, Bid HK, Manchanda PK, Kapoor R. Association of interleukin-1β gene and receptor antagonist polymorphisms with calcium oxalate urolithiasis. Journal of Endourology. 2007;21(12):1565–1570. doi: 10.1089/end.2007.0071. [DOI] [PubMed] [Google Scholar]
- 17.Mittal RD, Bid HK, Manchanda PK. Genotype and haplotype determination of IL1B (g. − 511C > T and g. + 3954C > T) and (IL1RN) in pediatric nephrolithiasis. Clinica Chimica Acta. 2007;379(1-2):42–47. doi: 10.1016/j.cca.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Onaran M, YIlmaz A, Şen I, et al. Heparan sulfate gene polymorphism in calcium oxalate nephrolithiasis. Urological Research. 2009;37(1):47–50. doi: 10.1007/s00240-008-0167-z. [DOI] [PubMed] [Google Scholar]
- 19.Onaran M, Yilmaz A, Şen I, et al. A HindIII polymorphism of fibronectin gene is associated with nephrolithiasis. Urology. 2009;74(5):1004–1007. doi: 10.1016/j.urology.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Ozturk M, Kordan Y, Cangul H, et al. Association of urokinase gene 3′-UTR T/C polymorphism with calcium oxalate urolithiasis in children. International Urology and Nephrology. 2008;40(3):563–568. doi: 10.1007/s11255-008-9335-x. [DOI] [PubMed] [Google Scholar]
- 21.Tasic V, Dervisov D, Koceva S, Weber S, Konrad M. Hypomagnesemia with hypercalciuria and nephrocalcinosis: case report and a family study. Pediatric Nephrology. 2005;20(7):1003–1006. doi: 10.1007/s00467-005-1853-5. [DOI] [PubMed] [Google Scholar]
- 22.Hoppe B, Beck BB, Milliner DS. The primary hyperoxalurias. Kidney International. 2009;75(12):1264–1271. doi: 10.1038/ki.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monga M, Macias B, Groppo E, Hargens A. Genetic heritability of urinary stone risk in identical twins. Journal of Urology. 2006;175(6):2125–2128. doi: 10.1016/S0022-5347(06)00272-2. [DOI] [PubMed] [Google Scholar]
- 24.Dawson PA, Russell CS, Lee S, et al. Urolithiasis and hepatotoxicity are linked to the anion transporter Sat1 in mice. The Journal of Clinical Investigation. 2010;120(3):706–712. doi: 10.1172/JCI31474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson PA, Markovich D. Genetic polymorphisms of human sulfate transporters. Current Pharmacogenomics. 2007;5(4):262–274. [Google Scholar]
- 26.Krick W, Schnedler N, Burckhardt G, Burckhardt BC. Ability of sat-1 to transport sulfate, bicarbonate, or oxalate under physiological conditions. American Journal of Physiology: Renal Physiology. 2009;297(1):F145–F154. doi: 10.1152/ajprenal.90401.2008. [DOI] [PubMed] [Google Scholar]
- 27.Lee A, Beck L, Markovich D. The mouse sulfate anion transporter gene sat1 (Slc26a1): cloning, tissue distribution, gene structure, functional characterization, and transcriptional regulation by thyroid hormone. DNA and Cell Biology. 2003;22(1):19–31. doi: 10.1089/104454903321112460. [DOI] [PubMed] [Google Scholar]
- 28.Regeer RR, Lee A, Markovich D. Characterization of the human sulfate anion transporter (hsat-1) protein and gene (SAT1; SLC26A1) DNA and Cell Biology. 2003;22(2):107–117. doi: 10.1089/104454903321515913. [DOI] [PubMed] [Google Scholar]
- 29.Brown JM, Stratmann G, Cowley DM. The variability and dietary dependence of urinary oxalate excretion in recurrent calcium stone formers. Annals of Clinical Biochemistry. 1987;24(4):385–390. doi: 10.1177/000456328702400407. [DOI] [PubMed] [Google Scholar]
- 30.Hatch M, Freel RW. Intestinal transport of an obdurate anion: oxalate. Urological Research. 2005;33(1):1–16. doi: 10.1007/s00240-004-0445-3. [DOI] [PubMed] [Google Scholar]
- 31.Marengo SR, Romani AMP. Oxalate in renal stone disease: the terminal metabolite that just won’t go away. Nature Clinical Practice Nephrology. 2008;4(7):368–377. doi: 10.1038/ncpneph0845. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, Asplin JR, Evan AP, et al. Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nature Genetics. 2006;38(4):474–478. doi: 10.1038/ng1762. [DOI] [PubMed] [Google Scholar]
- 33.Lee A, Dawson PA, Markovich D. NaSi-1 and Sat-1: structure, function and transcriptional regulation of two genes encoding renal proximal tubular sulfate transporters. International Journal of Biochemistry and Cell Biology. 2005;37(7):1350–1356. doi: 10.1016/j.biocel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Moult J. SNPs, protein structure, and disease. Human Mutation. 2001;17(4):263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]


