Abstract
Enzymes provide an exquisitely tailored chiral environment to foster high catalytic activities and selectivities, but their native structures are optimized for very specific biochemical transformations. Designing a protein to accommodate a non-native transition metal complex can broaden the scope of enzymatic transformations while raising the activity and selectivity of small molecule catalysis. Herein, we report the creation of a bifunctional artificial metalloenzyme in which a glutamic acid or aspartic acid residue engineered into streptavidin acts in concert with a docked biotinylated rhodium(III) complex to enable catalytic asymmetric C–H activation. The coupling of benzamides and alkenes to access dihydroisoquinolones proceeds with up to nearly a hundredfold rate acceleration compared to the activity of the isolated Rh complex and enantiomeric ratios as high as 93: 7.
Thanks to the advent of genetic engineering, enzymes are attracting increasing attention as versatile synthetic tools, even displacing established organometal-catalyzed industrial processes. (1) However, creating an enzyme for an abiotic reaction from a non-catalytic scaffold remains a major challenge. (2,3,4,5) One class of strategies has relied on the incorporation of non-natural metal cofactors within a protein scaffold to afford artificial metalloenzymes. (6,7,8,9) The main focus in the area has been improving the selectivity of the hybrid catalysts, rather than reaction rates, which are by-and-large dictated by the first coordination sphere interactions around the metal. (10,11,12) Among the various cofactor localization strategies, (13,14) the biotin-(strept)avidin technology has proven versatile: the geometry of the biotin-binding pocket is ideally suited to accommodate organometallic moieties, leaving enough room for substrate binding and activation. (15,16,17,18,19)
In recent years, [Cp*RhCl2], where Cp* is pentamethylcyclopentadienyl, has emerged as a versatile catalyst for electrophilic aromatic C–H activation reactions. (20) Elegant work by the groups of Fagnou and Glorius showed that pivaloyl-protected benzhydroxamic acids may be efficiently coupled with alkenes to access dihydroisoquinolones in good yield at room temperature. (21, 22) An exogeneous base is required for the orthometallation step. (23) Computations suggest that the C–H activation process occurs via a concerted metalation-deprotonation (CMD) mechanism. (24) The presence of a base significantly lowers the activation energy of this step. As three coordination sites are required around the {Cp*Rh}-moiety for catalysis (25), it has been difficult to introduce an asymmetric ligand, and no enantioselective version of this attractive benzannulation reaction has been reported thus far. In a biomimetic spirit, we hypothesized that incorporation of a biotinylated {Cp*RhX2} combined with an engineered aspartate or glutamate residue might yield an asymmetric catalyst for the production of enantioenriched dihydroisoquinolones (Fig. 1).
Fig. 1.
a. Synergistic action of a basic residue introduced by site-directed mutagenesis and a biotinylated [RhCp*biotinCl2] moiety acting as catalyst for an abiotic reaction. b. Benzannulation reaction catalyzed by the artificial metalloenzyme for the synthesis of enantioenriched dihydroisoquinolones. c. Postulated transition state for the C–H activation step. d. Auto-Dock model of biotinylated [RhCpbiotin(OAc)2] complex anchored in the proposed active site of the streptavidin tetramer with key residues highlighted (adjacent complex in Sav monomer B omitted for clarity).
We initially examined the viability of the [RhCp*Cl2]2-catalyzed reaction between pivaloyl-protected benzhydroxamic acid (1a) and methyl acrylate (2a) to dihydroisoquinolone (3a) under aqueous conditions. Although this reaction is typically performed in MeOH or EtOH, (22, 23) we were pleased to find that the reaction proceeds to completion in a 4:1 mixture of H2O/MeOH under basic conditions (200 mol % CsOAc), despite the sparing solubility of the substrates in this solvent mixture. Next, we designed a biotinylated analog [RhCp*biotinCl2]2 (26) for incorporation within streptavidin (Sav hereafter, Fig. 1). Two equivalents of [RhCp*biotinCl2]2 were required to displace weakly-bound 2-(4-hydroxyphenylazo)benzoic acid (HABA) in tetrameric streptavidin. (27) This suggests that the dimeric catalyst precursor dissociates in aqueous solution to [RhCp*biotinCl2(H2O)] and that the four biotin-binding sites of Sav can be loaded with the monomeric biotinylated catalyst precursor.(28)
When we combined benzhydroxamic acid 1a with 1.1 equivalents of methyl acrylate 2a in a 4:1 mixture of H2O:MeOH in the presence of tetrameric wild-type Sav and [RhCp*biotinCl2]2, only a trace amount of product was observed after 36h at room temperature (Table 1, entry 3). To increase conversion, we introduced a basic residue in the proximity of the rhodium moiety. As highlighted in the docking study,(29) residues S112A and K121B (of the adjacent Sav monomer B) lie closest to the metal center upon incorporation within WT Sav (Figure 1d). We thus introduced by site-directed mutagenesis a basic residue at either of these positions. The presence of a glutamate residue at position 112 (S112E) has a marginal effect on the activity of the catalyst (Table 1, entry 5). Introduction of a glutamate residue at position 121 (i.e. K121E) again gives low conversion (Table 1, entry 7). Gratifyingly, mutation to an aspartate at position 121 (i.e. K121D) improved the conversion to 89% after 72 h (Table 1, entry 8). To confirm that this increase in activity was indeed caused by the presence of a carboxylate residue, we introduced an asparagine residue (i.e. K121N). Asparagine is sterically and electronically similar to aspartic acid, but lacks the ability to facilitate the critical C–H activation step. As anticipated, K121N gave low conversion after 36 h (Table 1, entry 9). The data thus suggest that the reaction is critically dependent on the precise localization of a carboxylate residue provided by Sav We speculated that if the position of the carboxylate residue at position 121 could be further finetuned, increased conversions might result. This was realized upon combining a glutamate at position 121 with a lysine at position 118 (i.e. N118K-K121E). In addition to increased activity, this catalyst also gave enhanced levels of regioselectivity for alkene insertion (15:1) by comparison to the reaction in the absence of protein (4:1) (compare Table 1, entry 1 vs entry 10). With a mutant exhibiting superior activity in hand, we were eager to determine if the transformation could be rendered asymmetric thanks to the chiral nature of the active site. A survey of our arsenal of Sav mutants revealed that the mutant with superior activity was also reasonably enantioselective (Table 1, entry 10). Because position 121 was identified as the best location for the carboxylate residue, we focused on position 112 to modify the chiral environment. A screen of nine mutants at position 112 using an acetate buffer revealed that aromatic residues gave superior reactivity by comparison to non-aromatic residues (30-50% yield) as well as enhanced enantioselectivity (Table 1, entries 12-20). Prolonged reaction times were required to achieve enhanced conversion. The best mutant under our conditions proved to be S112Y with 30% yield, 12:1 regioselectivity and 88:12 enantiomeric ratio (er) (Table 1, entry 19). We hypothesized that if the superior activity imparted by the carboxylate mutation at position 121 could be combined in a synergistic way with the improved selectivity provided by S112Y, a highly active and selective artificial metalloenzyme might result.(30) The use of S112Y-K121D gave fair yield, but only a modest increase in er (Table 1, entry 21). Substituting the aspartic acid residue with glutamic acid resulted in a superior mutant that gave 80% yield with 20:1 regiomeric ratio, and 90:10 enantiomeric ratio (Table 1, entry 22). The yield and er could be further improved by exchanging H2O with 3-(N-morpholino)propanesulfonic acid buffer to yield the desired product in 95% yield, 19:1 rr, and 91:9 er (Table 1, entry 23).
Table 1.
Optimization of the performance of the artificial benzannulase.

| entry | Sav Mutant | Solvent | yield (%)* | regioisomeric ratio (rr) |
enantiomeric ratio (er) |
|---|---|---|---|---|---|
| 1 | - | Acetate Buffer | 80 | 4:1 | 51.5:48.5 |
| 2 | - | H2O | < 5% | - | - |
| 3 | WT† | H2O | < 5% | - | - |
| 4 | WT† | Acetate Buffer | 46 | 9:1 | 75:25 |
| 5 | S112E† | H2O | 10 | 15:1 | 78:22 |
| 6 | S112D† | H2O | < 5% | - | - |
| 7 | K121E† | H2O | 7 | 15:1 | 78:22 |
| 8 | K121D | H2O | 89 | 15:1 | 78:22 |
| 9 | K121N† | H2O | < 5% | - | - |
| 10 | N118K-K121E | H2O | 99 | 15:1 | 82:18 |
| 11 | N118K-K121E†,‡ | H2O | 8 | 6:1 | 52:48 |
| 12 | S112A | Acetate Buffer | 12 | 6:1 | 75:25 |
| 13 | S112C | Acetate Buffer | 6 | 10:1 | 71:29 |
| 14 | S112F | Acetate Buffer | 50 | 6:1 | 86:14 |
| 15 | S112K | Acetate Buffer | 6 | 6:1 | 76:24 |
| 16 | S112M | Acetate Buffer | 1 | 6:1 | 81:19 |
| 17 | S112T | Acetate Buffer | 35 | 6:1 | 79:21 |
| 18 | S112V | Acetate Buffer | 4 | 5:1 | 81:19 |
| 19 | S112Y | Acetate Buffer | 30 | 12:1 | 88:12 |
| 20 | S112W | Acetate Buffer | 32 | 12:1 | 86:14 |
| 21 | S112Y-K121D | H2O | 30 | 20:1 | 90:10 |
| 22 | S112Y-K121E | H2O | 80 | 20:1 | 90:10 |
| 23 | S112Y-K121E | MOPS Buffer | 95 | 19:1 | 91:9 |
Yield determined by gas chromatography integration
Reaction conducted for 36 h.
Sav preloaded with excess biotin for 10 min and then treated with [RhCp*biotinCl2]2.
This transformation proved applicable to a variety of substrates (Table 2). Ethyl vinyl ketone was highly reactive under the reaction conditions, resulting in benzannulated product in good yield, high regioselectivity, but poor enantioselectivity. When methyl acrylate was replaced with benzyl acrylate, yield and enantioselectivity are diminished. Substitution on the benzamide is better tolerated under the reaction conditions. Bromo-substituted and naphthyl benzamides deliver product in good yield, with a modest erosion of er in comparison to the parent system. The enantioselectivity increased with para-substituted nitrobenzamides, albeit at the expense of lower yield. The remaining mass balance is represented by unreacted starting material.
Table 2.
Substrate Scope

|
The data presented above argue for the synergistic action of both the carboxylate side chain and the chiral cavity inside the metalloenzyme for optimal reactivity and selectivity. We sought further support for the role of the critical basic aminoacid residue by conducting mechanistic studies. Using the monodeuterated benzamide d1-1a, we observed a kinetic isotope effect (kH/kD, KIE) value of 3.8 for the reaction with [RhCp*biotinCl2]2 under buffered conditions (1:4 MeOH:Acetate Buffer (pH = 5.9, 0.69 M) – for the internal competition KIE study, see Fig. S1-S3). In the presence of WT Sav under identical conditions, a KIE value of 2.8 was obtained. With the most active mutant, N118K-K121E, a KIE value of 4.8 was found under acetate-free conditions (4:1 H2O:MeOH). These results suggest a primary kinetic isotope effect in all cases. Subtle differences in the geometry of the CMD mechanism (See Fig. 1c) is a likely source of the slightly different KIE values.
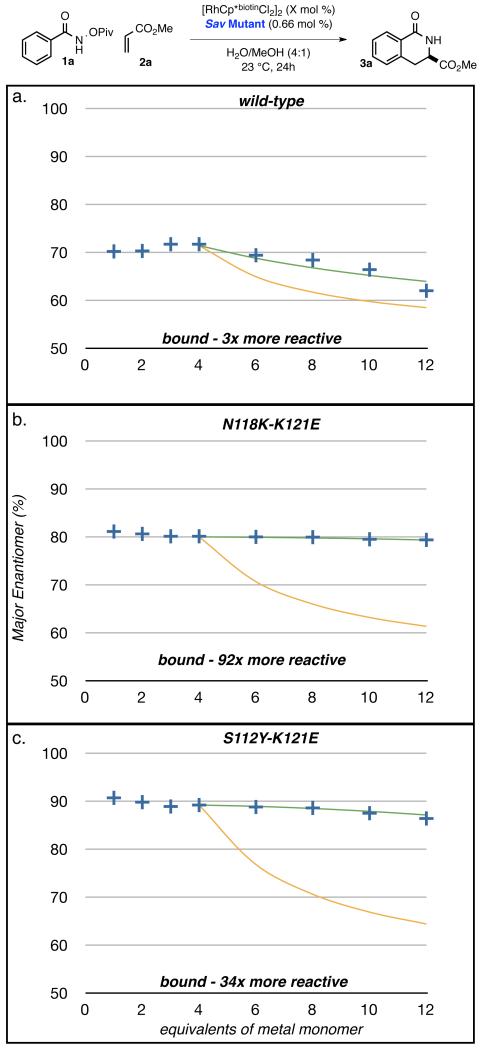
To provide further support for the critical role of the carboxylate residue within the enzyme’s active site, we conducted competition experiments between protein-bound and free [RhCp*biotinCl2]2 catalyst precursors. The very limited solubility of the starting material precludes the use of classical Michaelis-Menten kinetic experiments. Since the free biotinylated catalyst leads to nearly racemic product (51.5:48.5 er: Table 1, entry 1) whereas the Sav-bound catalyst leads to enantioenriched product, a comparison of the er as a function of the number of equivalents of [RhCp*biotinCl2(H2O)] vs. tetrameric Sav provides an estimate of the relative rates of the protein-bound (kbound) and the free biotinylated catalyst (kfree).(31) These experiments were performed with WT Sav, N118K-K121E and S112Y-K121E as host protein (Fig. 2 a, b and c respectively). As the er with up to four equivalents [RhCp*biotinCl2(H2O)] remains essentially constant, we conclude that the four Sav-bound catalysts operate independently (i.e. no cooperative effect) and induce the same level of enantioselectivity. Past four equivalents and if the relative rates kbound and kfree are identical, the er is expected to sharply and asymptotically decrease, (orange lines in Fig. 2). The rate acceleration provided by the protein-environment can be estimated by eq. 1:
| (1) |
Fig 2.
Determination of the relative catalytic rates of free- and Sav-bound catalysts using a) WT Sav, b) N118K-K121E, and c) S112Y-K121E. + experimentally determined er (blue crosses); - predicted er in the case of no protein rate acceleration (i.e. krel = (kbound)/(kfree) = 1) (orange line); - fitted er allowing the determination of krel (green line).
where ksav and k are the rate constants for the Sav-bound and the free catalyst respectively. The parameters μbound and μfree are the number of Sav-bound and free catalysts (i.e. at eight equivalents, μbound = μfree = 4); %(R)bound and %(R)free are the % (R) for the Sav-bound and the free catalyst (i.e. b = 0.515, Table 1 entry 1. Using equation (1) and performing a least square minimization on the calculated and experimentally determined %(R) (green line and blue crosses respectively, Fig. 2, see Supplementary text, Table S1) we can determine the relative rates krel = (kSav)/(k). For WT Sav, we compute a 3 fold rate acceleration compared to the protein-free catalyst. This phenomenon of protein acceleration is significantly enhanced in the presence of carboxylate-bearing Sav isoforms: in pure water and for N118K-K121E and S112Y-K121E, we compute rate accelerations of 92 and 34 respectively. The substantially increased rate is diagnostic of the key role of the active site carboxylate in the turnover limiting step, which we suggest is the C–H activation event. This confirms the hypothesis that the engineered carboxylate residue within the active site is key to generating a highly active and selective artificial benzannulase.
Supplementary Material
Acknowledgments
We thank NIGMS (GM80442), the Swiss National Science Foundation (Grant 200020-126366), the NCCR Nano, the Marie Curie Training Network (FP7-ITN-238434), Amgen and Roche, for support. We thank Johnson Matthey for a loan of rhodium salts and Prof. C. R. Cantor for the Sav gene. Malcolm Jeremy Zimbron is thanked for providing an initial sample of [RhCp*biotinCl2]2 and for stimulating discussions.
Footnotes
References
- 1.Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 2.Seelig B, Szostak JW. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park HS, Nam SH, Lee JK, Yoon CN, Mannervik B, Benkovic SJ, Kim HS. Science. 2006;311:535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- 4.Khare SD, Kipnis Y, Greisen PJ, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Nature Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinisch T, Ward TR. Curr. Opin. Chem. Biol. 2010;14:184–199. doi: 10.1016/j.cbpa.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 8.Roelfes G. ChemCatChem. 2011;3:647. [Google Scholar]
- 9.Mugford PF, Wagner UG, Jiang Y, Faber K, Kazlauskas RJ. Angew. Chem., Int. Ed. 2008;47:8782–8793. doi: 10.1002/anie.200705159. [DOI] [PubMed] [Google Scholar]
- 10.Yeung N, Lin Y-W,, Gao Y-G, Zhao X, Russell BS, Lei L, Miner KD, Robinson H, Lu Y. Nature. 2009;462:1079–1082. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y-W, Yeung N, Gao Y-G, Miner KD, Tian S, Robinson H, Lu Y. Proc. Nat. Acad. Sci. USA. 2010;107:8581–8586. doi: 10.1073/pnas.1000526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y-W, Yeung N, Gao Y-G, Miner KD, Lei L, Robinson H, Lu Y. J. Am. Chem. Soc. 2010;132:9970–9972. doi: 10.1021/ja103516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deuss PJ, Heeten R. d., Laan W, Kamer PCJ. Chem. Eur. J. 2011;17:4680–4698. doi: 10.1002/chem.201003646. [DOI] [PubMed] [Google Scholar]
- 14.Boersma AJ, Megens RP, Feringa BL, Roelfes G. Chem. Soc. Rev. 2010;39:2083–2092. doi: 10.1039/b811349c. [DOI] [PubMed] [Google Scholar]
- 15.Wilson ME, Whitesides GM. J. Am. Chem. Soc. 1978;100:306–307. [Google Scholar]
- 16.Lin C-C, Lin C-W, Chan ASC. Tetrahedron: Asymmetry. 1999;10:1887–1893. [Google Scholar]
- 17.Reetz MT, Peyralans JJP, Maichele A, Fu Y, Maywald M. Chem. Commun. 2006:4318–4320. doi: 10.1039/b610461d. [DOI] [PubMed] [Google Scholar]
- 18.Skander M, Humbert N, Collot J, Gradinaru J, Klein G, Loosli A, Sauser J, Zocchi A, Gilardoni F, Ward TR. J. Am. Chem. Soc. 2004;126:14411–14418. doi: 10.1021/ja0476718. [DOI] [PubMed] [Google Scholar]
- 19.Ward TR. Acc. Chem. Rev. 2011;44:47–57. doi: 10.1021/ar100099u. [DOI] [PubMed] [Google Scholar]
- 20.Satoh T, Miura M. Chem. Eur. J. 2010;16:11212–11222. doi: 10.1002/chem.201001363. [DOI] [PubMed] [Google Scholar]
- 21.Guimond N, Gorelsky SI, Fagnou K. J. Am. Chem. Soc. 2011;133:6449–6457. doi: 10.1021/ja201143v. [DOI] [PubMed] [Google Scholar]
- 22.Rakshit S, Grohmann C, Besset T, Glorius F. J. Am. Chem. Soc. 2011;133:2350–2353. doi: 10.1021/ja109676d. [DOI] [PubMed] [Google Scholar]
- 23.Hyster TK, Rovis T. J. Am. Chem. Soc. 2010;132:10565–10569. doi: 10.1021/ja103776u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D Lapointe, Fagnou K. Chem. Lett. 2010;39:1118–1126. [Google Scholar]
- 25.Xu L, Zhu Q, Huang G, Cheng B, Xia Y. J. Org. Chem. 2012;77:3017–3024. doi: 10.1021/jo202431q. [DOI] [PubMed] [Google Scholar]
- 26.Reiner T, Jantke D, Raba A, Marziale AN, Eppinger J. J. Organomet. Chem. 2009;694:1934–1937. [Google Scholar]
- 27.Green NM. Methods Enzymol. 1970;18A:418–424. [Google Scholar]
- 28.After optimization, the ideal conditions with WT Sav were found to be aqueous conditions (1:4 MeOH:Acetate Buffer (0.67M, pH=5.9)) with a 1.3:1 ratio of Sav binding sites to biotinylated rhodium monomer, delivering product in 23% yield with 9:1 regioselectivity after 36 h. Identical product distribution was obtained upon using [Cp*biotinRh(H2O)3]2+.
- 29.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reetz MT. Angew. Chem. Int. Ed. 2011;50:138–174. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 31.Collot J, Humbert N, Skander M, Klein G, Ward TR. J. Organomet. Chem. 2004;689:4868–4871. [Google Scholar]
- 32.Zheng L, Baumann U, Reymond J-L. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janin YL, Roulland E, Beurdeley-Thomas A, Decaudin D, Monneret C, Poupon M-F. J. Chem. Soc., Perkin Trans. 1. 2002;529 [Google Scholar]
- 34.Vicente J, Saura-Llamas I, García-López J-A, Calmuschi-Cula B. Organometallics. 2007;26:2768. [Google Scholar]
- 35.Vicente J, Saura-Llamas I, Garcia-Lopez J-A. Organometallics. 2009;28:448. [Google Scholar]
- 36.Humbert N, Zocchi A, Ward TR. Electrophoresis. 2005;26:47. doi: 10.1002/elps.200406148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.