Abstract
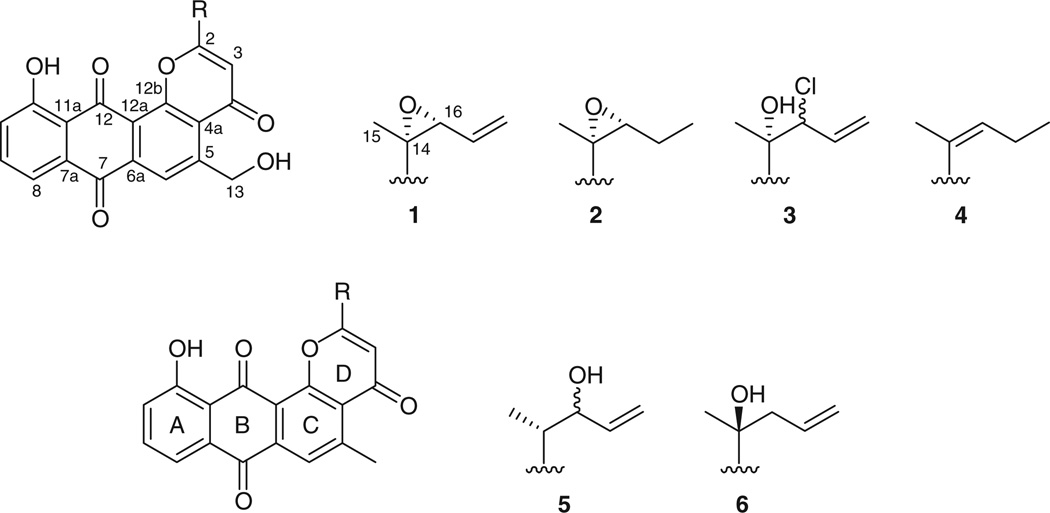
Six new anthraquinone-γ-pyrones, saliniquinones A–F (1–6), which are related to metabolites of the pluramycin/altromycin class, were isolated from a fermentation broth of the marine actinomycete Salinispora arenicola (strain CNS-325). Their structures were determined by analysis of one- and two-dimensional NMR spectroscopic and high-resolution mass spectrometric data. The relative and absolute configurations of compounds 1–6 were determined by analysis of NOESY NMR spectroscopic data and by comparison of circular dichroism and optical rotation data with model compounds found in the literature. Saliniquinone A (1) exhibited potent inhibition of the human colon adenocarcinoma cell line (HCT-116) with an IC50 of 9.9 × 10−9 M. In the context of the biosynthetic diversity of S. arenicola, compounds 1–6 represent secondary metabolites that appear to be strain specific and thus occur outside of the core group of compounds commonly observed from this species.
Introduction
Actinomycetes have been an enduring source of bioactive secondary metabolites for over 50 years,[1] although it is becoming ever more difficult to isolate novel bioactive secondary metabolites from terrestrial strains. As part of an effort to isolate new agents of potential use in the treatment of cancer, we have focussed our efforts toward actinomycete bacteria found in deep ocean sediments. As part of these studies, we have previously described the first obligate marine actinomycete genus, Salinispora,[2] which has proven to be a prolific producer of structurally unique bioactive molecules, including salinosporamide A,[3] a potent proteasome inhibitor currently completing Phase I clinical trials for the treatment of various cancers. Previous studies of Salinispora species have revealed that certain secondary metabolites are produced in species-specific patterns while others appear to be population or strain-specific.[4] As part of our current investigation into the relationships between actinomycete diversity and secondary metabolite production, strain CNS-325 was isolated from a marine sediment sample collected from Palau and identified as Salinispora arenicola by 16S rRNA gene sequence analysis. Fractions of the crude extract displayed potent antiproliferative activity againstHCT-116 colon carcinoma cells in vitro. Herein, we describe the isolation and characterization of six new anthraquinone-γ-pyrones, saliniquinones A–F (1–6) (Fig. 1). The isolation of new members of this potently cytotoxic skeletal class from S. arenicola expands the scope of secondary metabolite production observed from this species.
Fig. 1.
Structures of saliniquinones A–F (1–6).
Results
Saliniquinone A (1) was obtained as a yellow amorphous solid. Positive-ion high-resolution electrospray ionization time-of- flight mass spectrometry (HRESI-TOF MS) analysis gave a pseudomolecular ion at m/z 405.0970, which suggested the molecular formula C23H16O7. The 13C and g-HSQC NMR spectra of 1 in CDCl3 showed three α,β-unsaturated ketone carbonyls (δC 179.7 (C-4), 181.4 (C-7), 187.0 (C-12)), three sp2-oxygenated carbons (δC 168.9 (C-2), 162.8 (C-110), 156.6 (C-12b)), and an additional 13 sp2 carbons to be present in 1 (see Table 1). There was also evidence of three sp3-oxygenated carbons (δC 65.2 (C-13), 59.6 (C-14), 66.1 (C-16)) and one sp3 methyl carbon (δC 14.0 (C-15)). Select 1H NMR resonances from 1 in CDCl3 exhibited the characteristic pattern of an aromatic ABC spin system (δH 7.84 (dd, J 7.5, 1.5, H-8), 7.71 (t, J 7.5, H-9), 7.39 (dd, J 7.5, 1.5, H-10)). In addition, resonances for two aromatic methine hydrogens (δH 6.63 (s, H-3), 8.34 (s, H-6)), one terminal olefin (δH 5.91 (ddd, J 7.0, 11, 17, H-17), 5.59 (d, J 11, Hα-18), 5.66 (d, J 17, Hβ-18)), an oxymethylene (δH 5.08 (d, J 6.0, H2-13)), an oxymethine (δH 3.88 (d, J 7.0, H-16)), and a methyl group (δH 1.87 (s, C-15)) were observed.
Table 1.
13C NMR spectroscopic data for saliniquinones A–F (1–6, in CDCl3)A
| Position | Compound δC | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 2 | 168.9 | 169.9 | 169.6 | 165.6 | 170.1 | 171.6 |
| 3 | 109.8 | 109.0 | 110.7 | 108.5 | 113.0 | 109.7 |
| 4 | 179.7 | 179.8 | 179.7 | 180.3 | 178.7 | 179.4 |
| 4a | 126.6 | 126.6 | 126.3 | 126.4 | 126.5 | 126.3 |
| 5 | 150.2 | 150.0 | 150.2 | 149.6 | 150.3 | 150.4 |
| 6 | 124.0 | 124.0 | 124.0 | 123.5 | 126.0 | 126.0 |
| 6a | 136.9 | NRB | NRB | NRB | 135.9 | 136.0 |
| 7 | 181.4 | 181.4 | 181.1 | 181.5 | 181.7 | 181.9 |
| 7a | 132.3 | 132.1 | 132.0 | 132.1 | 132.3 | 132.1 |
| 8 | 119.9 | 119.8 | 119.8 | 119.5 | 119.6 | 119.5 |
| 9 | 137.3 | 136.9 | 137.1 | 136.6 | 136.8 | 136.6 |
| 10 | 125.8 | 125.7 | 125.6 | 125.6 | 125.5 | 125.4 |
| 11 | 162.8 | 162.8 | 162.8 | 162.7 | 162.6 | 162.6 |
| 11a | 116.8 | 116.7 | 116.4 | 116.6 | 116.6 | 116.7 |
| 12 | 187.0 | 187.1 | 187.1 | 187.2 | 187.6 | 187.4 |
| 12a | 121.6 | 121.5 | 121.0 | 121.4 | 119.5 | 119.6 |
| 12b | 156.6 | 156.6 | 155.9 | 156.4 | 156.5 | 156.1 |
| 13 | 65.2 | 65.2 | 64.9 | 65.1 | 24.4 | 24.1 |
| 14 | 59.6 | 57.9 | 75.7 | 125.7 | 44.7 | 72.9 |
| 15 | 14.0 | 13.7 | 25.0 | 12.3 | 15.8 | 26.3 |
| 16 | 66.1 | 67.7 | 68.7 | 142.9 | 74.5 | 45.2 |
| 17 | 130.9 | 22.0 | 133.2 | 22.7 | 138.5 | 131.8 |
| 18 | 123.1 | 10.4 | 120.3 | 13.3 | 116.4 | 120.8 |
Chemical shifts (δ) in ppm.
Resonances were not observed.
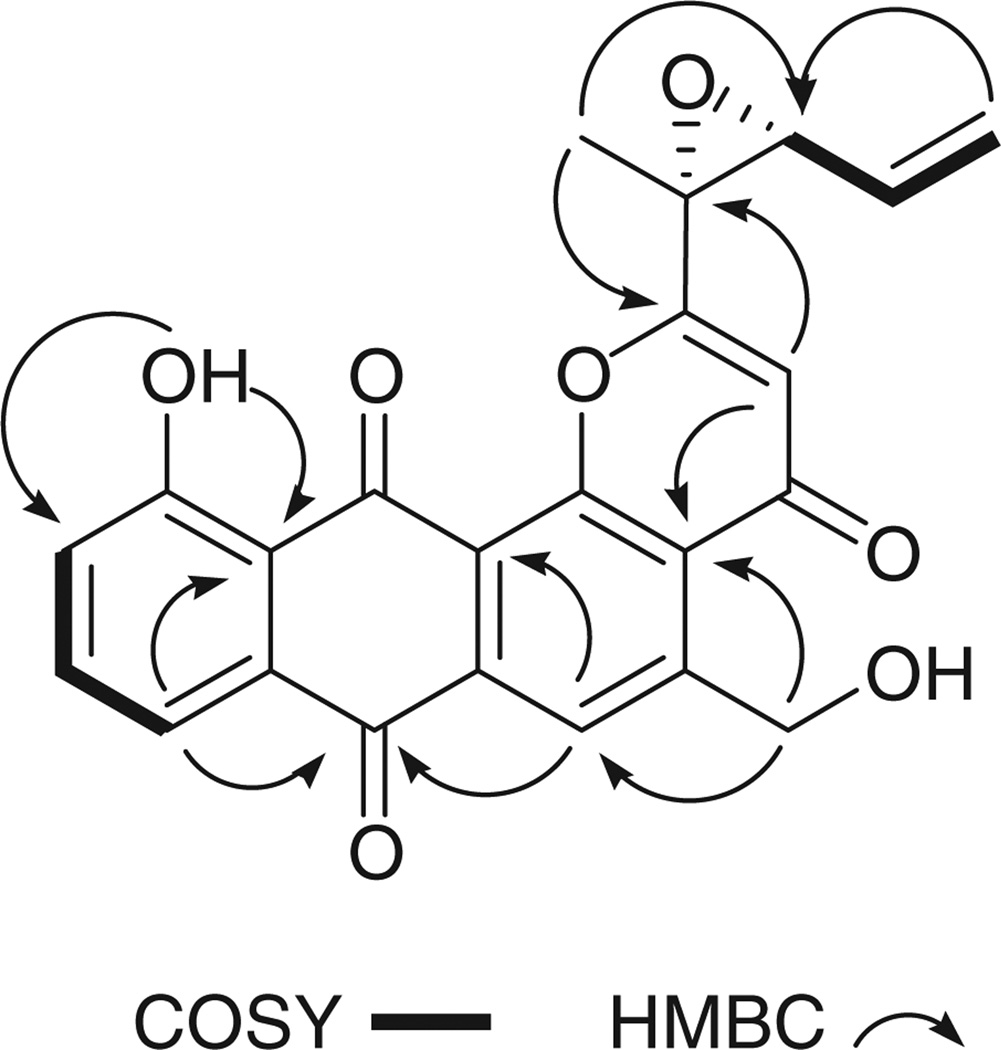
Analysis of combined g-COSY and g-HMBC NMR spectra allowed the assembly of rings A–D (Fig. 2). Ring A was constructed from interpretation of COSY correlations from H-8 to H-9, and H-9 to H-10. HMBC correlations of a deshielded peri-hydroxy proton (δH 12.73 (s, OH-11)) to C-10, C-11a, and C-12 (J4) allowed the hydroxy group to be placed at C-11 and also linked ring A to the quinone ring B. The presence of an anthraquinone moiety was evident from long-range correlations of H-8 and H-6 to the quinone carbonyl at C-7. Proton H-6 also exhibited long-range correlations to C-4a, C-5, and C-12a. Protons H2-13 correlated with C-6 and C-4a, effectively placing an oxymethylene carbon at position-5. Several key J4-HMBC correlations were also used. A correlation from H-6 and H2-13 to C-12b solidified the C ring assignment of the anthraquinone, while H-6 and H2-13 also correlated with the carbonyl at C-4. These, in addition to the long-range coupling of H-3 to C-12b, served to establish a γ-pyrone moiety fused to ring C. Supporting this were additional HMBC correlations of H-3 to C-2, and C-4.
Fig. 2.
Key COSY and HMBC NMR correlations for saliniquinone A (1).
Interpretation of NMR data also showed that a five-carbon allylic, epoxide-bearing side chain was located at C-2 of the γ-pyrone. COSY correlations from H2-18 to H-17, and H-17 to H-16, in addition to long-range coupling of H3-15 to C-14, C-16, and C-2 effectively finalized the planar structure assignment of 1.The assignment of the absolute configuration of 1 was determined on the basis of a comparison of optical rotation data with that of the aglycone of altromycin, a compound that contains a nearly identical carbon skeleton as the saliniquinones.[5] The optical rotation of the altromycin aglycone, which possesses 14S, 16S configurations, was measured as [α]D −114 (c 0.3, CHCl3), while 1 exhibited [α]D +66 (c 0.20, CHCl3). Although not a perfect inverse relationship, the absolute configuration of 1 is proposed as 14R, 16R.
Saliniquinone B (2) was also obtained as an amorphous yellow solid. High-resolution electron impact mass spectrometry (HREI MS) analysis gave a molecular ion at m/z 406.1053 ([M]+), which suggested the molecular formula C23H18O7. Compound 2 exhibited nearly identical 1H and 13C NMR resonances to those of 1, and comparison of their g-COSY and g-HMBC spectra further confirmed that both compounds shared an identical anthraquinone skeleton. The distinction between the two sets of spectra was demonstrated by the addition of two aliphatic resonances in the upfield region of the 1H NMR spectrum (δH 1.81 (m, H2-17), 1.17 (t, J 7.5, H3-18)) in place of three olefinic resonances that appear in the spectrum of 1 (H-17, Hα-18, Hβ-18). COSY correlations of H3-18 to H2-17, and H2-17 to H-16 (δH 3.26 (t, J 6.5)) affirmed that the terminal olefin in 1 had been saturated to yield 2. This was supported by the additional two mass units observed in the mass spectrum. The absolute configuration of 2 was also assigned by analysis of the circular dichroism (CD) spectrum and optical rotation data. Since compound 2 displayed a similar sign of rotation ([α]D +35; c 0.10, CHCl3), in addition to a nearly superimposable CD pattern with that of 1, the absolute configuration could also be assigned as 14R, 16R.
Saliniquinone C (3) was obtained as an amorphous yellow solid. HRESI-TOF MS analysis gave a pseudomolecular ion at m/z 441.0739 ([M+H]+), which analyzed for the molecular formula C23H17O7Cl. Compound 3 exhibited nearly identical 1H and 13C NMR resonances to those of 1, and comparison of the g-COSY and g-HMBC spectra further confirmed that both compounds shared the identical carbon skeleton. The observed mass spectrometry fragmentation pattern was typical of a chlorinated molecule, and since the major difference between 3 and 1 in the 1H NMR spectrum was a deshielding effect on H-16 (δH 5.14 (d, J 9.5)), it was apparent that chlorine was positioned on the side chain forming a vicinal halohydrin. A COSY correlation between H-16 and H-17 (δH 5.99 (dt, J 17, 9.5)) and a HMBC correlation from H-16 to the terminal olefinic carbon (δC 120.3 (C-18)) further confirmed the planar structure with chlorine positioned at C-16.
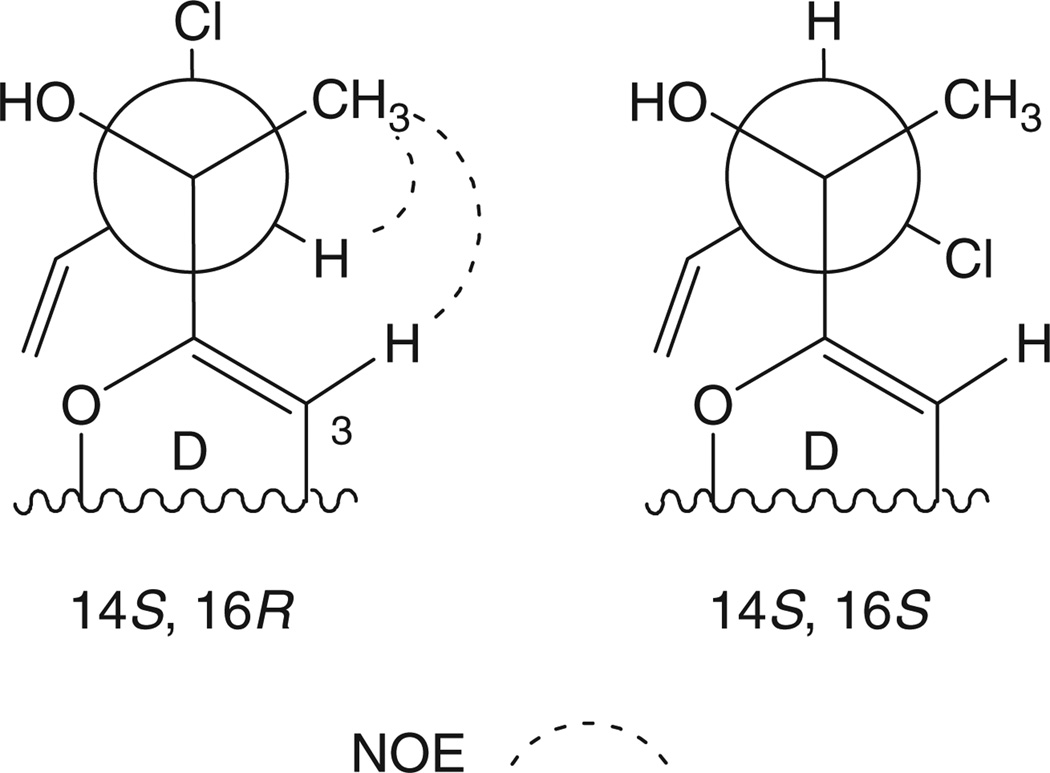
The CD spectrum of 3 displayed similar Cotton effects to those of 1, suggesting that the hydroxy stereocentre in the side chain was of identical configuration. In addition, the CD spectrum of 3 displayed Cotton effects opposite to those of 6 (see Accessory Publication). Furthermore, comparison of optical rotation data of compounds 3 and 6 in chloroform allowed us to conclude that the two molecules have opposite configurations at C-14, primarily since C-14 in 6 is the only stereocenter in the molecule ([α]D +10, c 0.10, CHCl3, 14S, 3; [α]D −10, c 0.10, CHCl3, 14S, 6). Unfortunately, data was not conclusive enough to determine the absolute configuration at C-16. Data from a two-dimensional NOESY experiment allowed a probable conformation at C-16 to be proposed. A correlation was observed between the methyl group H3-15 and methine protons H-16 and H-3. Analysis of Newman projections ruled out any conformation in which C-15 and H-16 were anti, thus narrowing down the list of possibilities to two conformers (Fig. 3). Some evidence indicated that the absolute configuration of C-16 was R. First, an NOE interaction was not observed between H-16 and OH-14, and second, a more energetically favourable confirmation exists when the chlorine at position 16 is anti to the bulky pyrone moiety. Despite these observations, we do not feel confident in definitively stating the configuration at C-16.
Fig. 3.
NOE correlations and potential C-16 configurations of saliniquinone C (3).
Saliniquinone D (4) was obtained as a yellow non-crystalline solid. HRESI-TOF MS analysis gave a pseudomolecular ion at m/z 391.1176 ([M+H]+), which suggested the molecular formula for this derivative as C23H18O6. Analysis of the one and two-dimensional NMR data suggested the structure of 4 was nearly identical to that of 2, and that the difference resided in the side chain. A loss of 16 mass units suggested that the epoxide oxygen was replaced by a double bond at positions 14 and 16, a conclusion that was supported by the presence of olefinic resonances in the 1H and 13C NMR spectra (δH 7.48 (t, J 9.0, H-16) 142.9 (C-16), 125.7 (C-14)) and long-range HMBC correlations of H-16 to C-2, C-15, and C-18 (δC 165.6, 12.3, and 13.3, respectively). Finally, based on a NOESY correlation from H3- 15 to H-16, the geometry of the double bond at position 14 was determined to be Z.
Saliniquinone E (5) was obtained as a yellow solid. HRESI-TOF MS analysis gave a pseudomolecular ion at m/z 391.1168 ([M+H]+), which suggested a molecular formula of C23H18O6. The molecular weight was identical to that of 4, although we observed a few minor spectroscopic differences. The oxygenated methylene resonance observed in the 1H NMR spectrum of 4 (δH 5.07 (d, J 9.5, H2-13)) was not present in the spectrum of 5. Instead, a methyl resonance (δH 3.02 (s, C-13)) was found to show HMBC correlations to three aromatic carbons (δC 126.5 (C-4a), 150.3 (C-5), 126.0 (C-6)). This prompted us to position a methyl group at C-5. COSY connectivities between H3-15 (δH 1.50 (d, J 6.5)) and H-14 (δH 2.94 (m)), H-14 and H-16 (δH 4.52 (t, J 6.0)), H-16 and H-17 (δH 5.93 (ddd, J 17, 11, 6.0)), and H-17 and H2-18 (δH 5.38 (d, J 17, Hα-18), 5.19 (d, J 11, Hβ- 18)) allowed us to construct the entirety of the side chain, while HMBC correlations from H-14 to C-3 (δC 113.0), and both H3- 15 and H-16 to C-2 of the fused γ-pyrone (δC 170.1) allowed it to be positioned at C-2. Partial determination of the absolute configuration of the two stereocentres at C-14 and C-16 was based on optical rotation and CD data, and comparison to a similar pluramycin-derived molecule, AH-1763 IIa.[6, 7] Compound 5 displayed similar Cotton effects to 6, and opposite Cotton effects to 1–3. Given C-14 is the only stereocentre in 6, we suggest the configuration at C-14 be assigned as R. Compound 5 ([α]D +4.2, c 0.10) afforded a net positive rotation in chloroform, which agreed with the sign of the natural product (AH-1763 IIa, [α]D +6.6, c 0.10, CHCl3, 14R, 16S) and was opposite to that of the synthetic analogue (AH-1763 IIa, [α]D −28, c 0.10, CHCl3, 14S, 16R). Given the possible existence of the 14R, 16R diastereomer and unavailability of those optical rotation and CD data, we were unable to definitively conclude the configuration at C-16.
Saliniquinone F (6) was obtained as a yellow solid. HRESI-TOF MS analysis gave a pseudomolecular ion at m/z 391.1174 ([M+H]+), which suggested a molecular formula of C23H18O6. The molecular weight was identical to that of 4 and 5, although 1H and 13C NMR resonances for a C-5 substituted methyl group were observed. Thus, 6 shared an identical anthraquinone-γ- pyrone skeleton to that of 5. This was established by analysis of COSY and HMBC data. A COSY correlation from H-17 (δH 5.76 (m)) to an allylic methylene (δH 3.03 (dd, J 13, 6.6, Hα-16), 2.64 (dd, J 13, 8.4, Hβ-16)) in addition to HMBC correlations from H-17 and H-3 (δH 6.55 (s)) to an sp3 oxygenated quaternary carbon (δC 72.9 (C-14)), suggested that a hydroxy group was positioned at C-14. The absolute configuration of the stereocentre at C-14 was determined on the basis of comparison of CD data to metabolites 1 and 2, and optical rotation values to both the synthetic and natural anthraquinone-pyran metabolite, γ-indomycinone.[8] As described earlier, compound 6 displays Cotton effects nearly opposite to those of 1 and 2. In addition, 6 ([α]D −49, c 0.10, dimethyl sulfoxide (DMSO)) afforded a net negative rotation, which coincided with the sign of the natural product ([α]D −5.5, c 0.11, DMSO, 14S) and was opposite to that of the synthetic analogue ([α]D +4.0, c 0.10, DMSO, 14R). Thus, 6 was assigned the (14S)-saliniquinone F.
Discussion
Saliniquinones A–F are members of the pluramycin class of antitumor antibiotics, whose core structural feature consists of an anthraquinone ring system fused with a γ-pyrone moiety. Most members of this structural class also contain C-glycosidic amino sugars at the 5-, 8-, and 10-positions. Commonly isolated from strains of Streptomyces, the antitumour properties of pluramycins have been thoroughly studied; such bioactivity is the result of their ability to alkylate N7 of the guanine base in DNA by intercalation.[9–15] Derivatives that contain an epoxide on the C-2 side chain displayed especially potent bioactivity, as this was shown to be the site of DNA alkylation. As expected, saliniquinone A exhibited potent antiproliferative activity against the human colon adenocarcinoma cell line, HCT-116 (half maximum inhibitory concentration (IC50)=9.9 × 10−9 M).
Pluramycin-class antibiotics have also been shown to inhibit the growth of several Gram-positive bacteria and appear to be largely ineffective against Gram-negative bacteria. Minimum inhibitory concentrations (MIC) of sapurimycin,[16] the altromycins,[17] topopyrones A and C,[18] and antibiotic SF- 2330[19] range from (0.10 to 8.0) × 10−6M against various strains of Staphylococcus, Bacillus, Streptococcus, and other Gram-positive pathogens. Saliniquinone A was tested against methicillin-resistant S. aureus and exhibited a weak MIC of 9.9 × 10−6 M.
Although the bioactivity of this structural class of metabolites has been examined, there have been few studies focussed on the biosynthetic assembly of various pluramycin-type skeletons. In regard to the structures described in the current study, the closest structural relatives of the saliniquinones include the indomycinones,[20] and the aglycones of kidamycin and hedamycin.[5, 21, 22] Hedamycin shares an identical core skeleton with 5 and 6 but fashions a six carbon bis-epoxide side chain at C-2 and two C-glycosylated amino sugars at C-5 and C-8. In a cloning experiment, Bililign et al. were the first to describe the biosynthetic gene cluster responsible for the production of hedamycin in Streptomyces griseoruber.[23] In a rare collaborative assembly, the organism employs an iterative type I polyketide synthase (PKS) to assemble the C-2 side chain, and thus ‘primes’ the type II PKS with a starter unit. Subsequent chain elongation and cyclization/aromatization results in the assembly of the hedamycin aglycone.
Although it has not been confirmed if S. arenicola strain CNS-325 produces saliniquinones A–F by a pathway similar to hedamycin biosynthesis, it is known that the closely related S. arenicola strain CNS-205, for which a complete genome sequence is available, does not possess the genetic machinery required to produce this class of secondary metabolites.[24] This observation further emphasizes the metabolic diversity that can exist between strains of the same species, even when isolated from the same location (in this case both strains originated from Palau), despite evidence that some metabolites appear to be common to all strains.[4] In terms of structural novelty, the saliniquinones represent the first members of the pluramycin class of antibiotics to contain both a terminal olefin and five carbons in the C-2 side chain; this is a deviation from all other known analogues that contain the prototypic four and six carbon units. If the architecture of the CNS-325 biosynthetic gene cluster that is responsible for production of the saliniquinones is similar to the one previously discussed, this may indicate that one malonyl CoA and one propionyl CoA unit combine to form a five-carbon side chain, as opposed to three malonyl CoA units that compose the six-membered priming side chain found in hedamycin. Regardless of their biosynthetic origin, the saliniquinones represent yet another example of the metabolic diversity of the genus Salinispora.
Experimental
General Experimental Procedures
Optical rotations were measured on a JASCO P-2000 polarimeter. UV spectra were measured on a Beckman Coulter DU800 spectrophotometer. CD spectra were recorded on an AVIV model 215 spectrometer. IR spectra were recorded on a Perkin–Elmer 1600 FTIR spectrometer. NMR spectra were obtained on Varian Inova 500 and 300MHz spectrometers, and a Bruker 600MHz DRX-600 equipped with a 1.7 mm cryoprobe and Avance III console. Chemical shifts (δ) are given in ppm, and coupling constants (J) are reported in Hz. For 13Cand 1H NMR characterization data of compounds 1–6, see Tables 1 and 2. High-resolution mass spectra were obtained on an Agilent ESI-TOF at the Scripps Center for Mass Spectrometry. Low-resolution liquid chromatography mass spectrometry (LC/MS) data were acquired using a Hewlett–Packard series 1100 system equipped with a reversed-phase C18 column (Phenomenex Luna, 4.6 × 100 mm2, 5 µm) at a flow rate of 0.7 mL min−1. High-performance liquid chromatography (HPLC) separations were performed using a Waters 600E system controller and pumps with a Model 480 spectrophotometer. Separation was achieved using Phenomenex Luna semi-preparative C18 (250 × 10 mm2, 5 µm) and Varian Dynamax preparative silica (250 × 21.4 mm2, 8 µm) columns at flow rates of 2.0 and 10 mL min −1, respectively.
Table 2.
1H NMR spectroscopic data for saliniquinones A–F (1–6, in CDCl3)A
| Position | Compound δH (multiplicity,B J [Hz]) | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 3 | 6.63 (s) | 6.58 (s) | 6.77 (s) | 6.52 (s) | 6.28 (s) | 6.55 (s) |
| 6 | 8.34 (s) | 8.33 (s) | 8.33 (s) | 8.28 (s) | 8.06 (s) | 8.08 (s) |
| 8 | 7.85 (dd, J 7.5, 1.5) | 7.85 (dd, J 7.5, 1.5) | 7.85 (d, J 8.0) | 7.84 (dd, J 9.0, 1.2) | 7.81 (dd, J 9.5, 1.5) | 7.84 (d, J 7.5) |
| 9 | 7.71 (t, J 7.5) | 7.71 (t, J 7.5) | 7.73 (t, J 8.0) | 7.71 (t, J 9.0) | 7.69 (t, J 9.5) | 7.70 (t, J 7.5) |
| 10 | 7.39 (dd, J 7.5, 1.5) | 7.39 (dd, J 7.5, 1.5) | 7.41 (d, J 8.0) | 7.48 (dd, J 9.0, 1.2) | 7.36 (dd, J 9.5, 1.5) | 7.38 (dd, J 7.5, 1.5) |
| 13 | 5.08 (d, J 6.0) | 5.07 (d, J 6.0) | 5.08 (d, J 5.5) | 5.07 (d, J 9.5) | 3.00 (s) | 3.03 (s) |
| 14 | – | – | – | – | 2.94 (m) | – |
| 15 | 1.87 (s) | 1.87 (s) | 1.84 (s) | 2.04 (s) | 1.50 (d, J 6.5) | 1.71 (s) |
| 16 | 3.87 (d, J 7.0) | 3.26 (t, J 6.0) | 5.14 (d, J 9.5) | 7.48 (t, J 9.0) | 4.52 (t, J 6.0) | 3.03 (dd, J 13, 6.6) |
| 2.64 (dd, J 13, 8.4) | ||||||
| 17 | 5.91 (ddd, J 17, 11, 7.0) | 1.81 (m) | 5.99 (dt, J 17, 9.5) | 2.43 (m) | 5.93 (ddd, J 17, 11, 6.0) | 5.76 (m) |
| 18 | 5.65 (d, J 17) | 1.17 t (7.5) | 5.28 (d, J 17) | 1.23 t (9.0) | 5.38 (d, J 17) | 5.23 (d, J 17) |
| 5.59 (d, J 11) | 5.15 (d, J 9.5) | 5.19 (d, J 11) | 5.21 (d, J 10) | |||
| 11-OH | 12.7 (s) | 12.7 (s) | 12.7 (s) | 12.8 (s) | 12.6 (s) | 12.9 (s) |
| 13-OH | 4.39 (br s) | Not observed | 4.42 (br s) | 4.72 t (9.5) | – | – |
| 14-OH | – | – | 3.13 (br s) | – | – | 2.90 (br s) |
| 16-OH | – | – | – | – | Not observed | – |
Chemical shifts (δ) in ppm. Data were acquired with a Bruker 600 MHz DRX-600 spectrometer equipped with a 1.7 mm cryoprobe.
br s: broad singlet; d: doublet; t: triplet; m: multiplet.
Antiproliferative Bioassay
Aliquot samples of HCT-116 human colon adenocarcinoma cells were transferred to 96-well plates and incubated overnight at 37 °C in 5% CO2/air. Test compounds were added to the plates in DMSO and serially diluted. The plates were then further incubated for another 72 h and at the end of this period a CellTiter 96 Aqueous non-radioactive cell proliferation assay (Promega) was used to assess cell viability. IC50 values were deduced from the bioreduction of (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)- 2-(4-sulfophenyl)-2H-tetrazolium/phenazine methosulfate) (MTS/PMS) by living cells into a formazan product. MTS/PMS was first applied to the sample wells, followed by incubation for 3 h. Etoposide (Sigma; IC50 =(1.5–4.9) × 10−6 M) and DMSO (solvent) were used as the positive and negative controls in this assay. The quantity of the formazan product (in proportion to the number of living cells) in each well was determined by the Molecular Devices Emax microplate reader set to 490 nm wavelength. IC50 values were calculated using the analysis program, SOFTMax.
Bacterial Isolation and Identification
Strain CNS-325 was isolated from a sediment sample collected in Palau at a depth of ~30 m in 2004 using previously described methods.[25] The identification of this strain was accomplished by 16S rRNA gene sequence analysis. The partial 16S rRNA gene sequence (843 base pairs) has been deposited in GenBank under accession number GU593973.
Fermentation and Extraction
Strain CNS-325 was cultured in 52 × 1 L Fernbach flasks that contained A1BFe+C medium for 7 days at 25–27 °C while shaking at 230 rpm. Sterilized Amberlite XAD-16 resin (20 g) was added to each Fernbach flask on day 7 to adsorb extracellular secondary metabolites. The resulting mixture was shaken for 4 h, filtered using cheesecloth to collect the resin, and washed with deionized water to remove salts. The resin, cell mass, and cheesecloth were then extracted overnight with acetone, concentrated under vacuum, partitioned between water and EtOAc, and the organic layer dried under vacuum to afford 3.3 g of crude extract.
Isolation of Saliniquinones A–F (1–6)
The crude extract was first fractionated using a silica flash column (33 g of silica gel) eluting with a step gradient of EtOAc in isooctane (increments of 20% EtOAc), to 20% MeOH in EtOAc. All fractions were analyzed by LC-MS (analytical RP-C18 column) and further isolation of the saliniquinones was facilitated by searching for their distinct UV pattern. The 40/60 isooctane/ EtOAc soluble fraction (171 mg)was fractionated using preparative silica HPLC(45/55, isooctane/EtOAc) to afford 17 fractions. Fraction 4 (tR 21.0 min, 5.0 mg) was further purified using R-PC18 semi-preparative HPLC (68/32, MeCN/H2O) to afford 1 (tR 14.0 min, 3.6 mg) and 2 (tR 16.0 min, 1.1 mg). Fraction 5 (tR 27.0 min, 2.4 mg) was further separated using RP-C18 semi-preparative HPLC (65/35, MeCN/H2O) to afford 3 (tR 12.0 min, 0.2 mg). The 60/40 isooctane/EtOAc soluble fraction (151 mg) was separated using preparative silica HPLC (80/20, isooctane/ EtOAc) to afford 28 fractions. Fraction 13 (tR 50.0 min, 3.6 mg) was further purified using RP-C18 semi-preparative HPLC (66/34, MeCN/H2O) to afford 6 (tR 14.0 min, 0.2 mg). Fraction 21 (tR 86.0 min, 13.4 mg) was further separated using RP-C18 semi-preparative HPLC (60/40, MeCN/H2O) to afford 5 (tR 15.0 min, 0.6 mg). Fraction 24 (tR 108 min, 12.5 mg) was further purified using RP-C18 semi-preparative HPLC (60/40, MeCN/H2O) to afford 4 (tR 19.0 min, 0.4 mg).
Saliniquinone A ( 1): yellow solid. [α]D +66 (c 0.20, CHCl3). λmax (MeOH)/nm (log ε) 237 (4.61), 259 (4.33), 268 (4.31), 414 (3.83). CD (MeOH, c 0.20) [θ]208 −12.2, [θ]222 26.0, [θ]262 10.3, [θ]316 5.1. νmax/cm−1 3737.4, 1649.2, 1455.9. m/z (HRESITOF MS)Anal. Calc. forC23H17O7: 405.0969. Found: 405.0970 [M+H]+.
Saliniquinone B ( 2): yellow solid. [α]D +35 (c 0.10, CHCl3). λmax (MeOH)/nm (log ε) 237 (4.61), 261 (4.33), 268 (4.32), 414 (3.83).CD(MeOH, c 0.10) [θ]208 −0.60, [θ]222 10.2, [θ]262 3.40, [θ]310 2.4. νmax/cm−1 3738.4, 1645.9, 1454.8. m/z (HREI MS) Anal. Calc. for C23H18O7: 406.1047. Found: 406.1053 [M]+.
Saliniquinone C ( 3): yellow solid. [α]D +10 (c 0.10, CHCl3). λmax (MeOH)/nm (log ε) 237 (4.38), 262 (4.09), 268 (4.08), 414 (3.59). CD (MeOH, c 0.10) [θ]208 4.2, [θ]218 9.1, [θ]266 2.8, [θ]314 0.30. νmax/cm−1 3445.2, 1646.9, 1457.3. m/z (HRESITOF MS) Anal. Calc. for C23H18O7Cl: 441.0736. Found: 441.0739 [M+H]+.
Saliniquinone D ( 4): yellow solid. λmax (MeOH)/nm (log ε) 237 (4.01), 263 (3.79), 268 (3.79), 414 (4.24). νmax/cm−1 1646.6, 1456.7. m/z (HRESI-TOF MS) Anal. Calc. for C23H19O6: 391.1176. Found: 391.1176 [M+H]+.
Saliniquinone E ( 5): yellow solid. [α]D +4.2 (c 0.10, CHCl3). λmax (MeOH)/nm (log ε) 237 (4.32), 266 (4.02), 414 (3.53). CD (MeOH, c 0.10) [θ]206 15.8, [θ]208 −6.9. νmax/cm−1 3524.7, 1644.1, 1458.2. m/z (HRESI-TOF MS) Anal. Calc. for C23H19O6: 391.1176. Found: 391.1168 [M+H]+.
Saliniquinone F ( 6): yellow solid. [α]D −10 (c 0.10, CHCl3). λmax (MeOH)/nm (log ε) 237 (4.33), 262 (4.04), 268 (4.02), 414 (4.54).CD(MeOH, c 0.10) [θ]208 −2.1, [θ]210 −11.6. νmax/cm−1 3363.8, 1644.1, 1454.1. m/z (HRESI-TOF MS) Anal. Calc. for C23H19O6: 391.1176. Found: 391.1174 [M+H]+.
Supplementary Material
Acknowledgements
This research was funded by the National Institutes of Health (NIH), National Cancer Institute (NCI) under grant R37 CA 044848 (to W.F.). The authors gratefully acknowledge Alexandra Besser (SIO) for performing theHCT-116 bioassay and Ariane Jansma (UCSD) for assistance obtaining NMR spectra. T. Narender acknowledges the Department of Science and Technology (DST), New Delhi, India for the award of BOYSCAST fellowship to visit the University of California San Diego.
Footnotes
Accessory Publication
All one and two-dimensional NMR spectroscopic data of compounds 1–6 are available on the Journal’s website.
References
- 1.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M. Int. J. Syst. Evol. Microbiol. 2005;55doi:1759. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 3.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2003;42:355. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 4.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fei Z, McDonald FE. Org. Lett. 2005;7:3617. doi: 10.1021/ol0509742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uyeda M, Yokomizo K, Ito A, Nakayama K, Watanabe H, Kido Y. J. Antibiot. 1997;50:828. doi: 10.7164/antibiotics.50.828. [DOI] [PubMed] [Google Scholar]
- 7.Tietze LF, Gericke KM, Singidi RR. Angew. Chem. Int. Ed. 2006;45:6990. doi: 10.1002/anie.200601992. [DOI] [PubMed] [Google Scholar]
- 8.Tietze LF, Singidi RR, Gericke KM. Org. Lett. 2006;8:5873. doi: 10.1021/ol062492b. [DOI] [PubMed] [Google Scholar]
- 9.Hansen M, Yun S, Hurley L. Chem. Biol. 1995;2:229. doi: 10.1016/1074-5521(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 10.Sun D, Hansen M, Clement JJ, Hurley LH. Biochemistry. 1993;32:8068. doi: 10.1021/bi00083a003. [DOI] [PubMed] [Google Scholar]
- 11.Yoon JH, Lee CS. Mol. Cells. 2000;10:71. doi: 10.1007/s10059-000-0071-z. [DOI] [PubMed] [Google Scholar]
- 12.Hansen MR, Hurley LH. Acc. Chem. Res. 1996;29:249. [Google Scholar]
- 13.Hansen MR, Hurley L. J. Am. Chem. Soc. 1995;117:2421. [Google Scholar]
- 14.Nakatani K, Okamoto A, Matsuono T, Saito I. J. Am. Chem. Soc. 1998;120:11219. [Google Scholar]
- 15.Bennett GN. Nucleic Acids Res. 1982;10:4581. doi: 10.1093/nar/10.15.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara M, Takiguchi T, Ashizawa T, Gomi K, Nakano H. J. Antibiot. 1991;44:33. doi: 10.7164/antibiotics.44.33. [DOI] [PubMed] [Google Scholar]
- 17.Jackson M, Karwowski JP, Theriault RJ, Hardy DJ, Swanson SJ, Barlow GJ, Tillis PM, McAlpine JB. J. Antibiot. 1990;43:223. doi: 10.7164/antibiotics.43.223. [DOI] [PubMed] [Google Scholar]
- 18.Kanai Y, Ishiyama D, Senda H, Iwatani W, Takahashi H, Konno H, Tokumasu S, Kanazawa S. J. Antibiot. 2000;53:863. doi: 10.7164/antibiotics.53.863. [DOI] [PubMed] [Google Scholar]
- 19.Itoh J, Shomura T, Tsuyuki T, Yoshida J, Ito M, Sezaki M, Kojima M. J. Antibiot. 1986;39:773. doi: 10.7164/antibiotics.39.773. [DOI] [PubMed] [Google Scholar]
- 20.Schumacher RW, Davidson BS, Montenegro DA, Bernan VS. J. Nat. Prod. 1995;58:613. doi: 10.1021/np50118a024. [DOI] [PubMed] [Google Scholar]
- 21.Kanda N. J. Antibiot. 1971;24:599. doi: 10.7164/antibiotics.24.599. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz H, Crook KE, Jr, Bush JA. Antimicrob. Agents Chemother. 1966;6:606. doi: 10.1128/AAC.6.5.606. [DOI] [PubMed] [Google Scholar]
- 23.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. Chem. Biol. 2004;11:959. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. ISME J. 2009;3:1193. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gontang EA, Fenical W, Jensen PR. Appl. Environ. Microbiol. 2007;73:3272. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.