Abstract
DNA nanostructures are rationally designed, synthetic, nanoscale assemblies obtained from one or more DNA sequences by their self-assembly. Due to the molecularly programmable as well as modular nature of DNA, such designer DNA architectures have great potential for in cellulo and in vivo applications. However, demonstrations of functionality in living systems necessitates a method to assess the in vivo stability of the relevant nanostructures. Here, we outline a method to quantitatively assay the stability and lifetime of various DNA nanostructures in vivo. This exploits the property of intact DNA nanostructures being uptaken by the coelomocytes of the multicellular model organism Caenorhabditis elegans. These studies reveal that the present fluorescence based assay in coelomocytes of C. elegans is an useful in vivo test bed for measuring DNA nanostructure stability.
Keywords: DNA nanostructure, Stability, Fluorescence, Coelomocytes, Caenorhabditis elegans
1. Introduction
Structural DNA nanotechnology aims to exploit the versatility and molecular programmability of DNA as a building block to synthetically fabricate functional nanoarchitectures [1]. These architectures, which self-assemble into either static structures or dynamic nanomachines, rely on the specificity and predictability of Watson–Crick base pairing, engineerability of the DNA scaffold and the ability of DNA to form a gamut of functional modules. Examples of static architectures include DNA polyhedra such as cubes, tetrahedra, octahedra or dodecahedra [2], while dynamic devices include such assemblies as molecular beacons, hybridization-powered molecular motors, DNA walkers and tweezers and aptamer-based switches [3]. Despite the existence of an array of DNA based nanoarchitectures in vitro, well-defined assays to quantify their in vivo stability need to be developed, in order to guide their potential applications in cellulo as well as in vivo. Here we describe a fluorescence-based method to assess the stability of DNA based nanostructures in a living, multicellular organism and demonstrate that this assay quantitatively captures differences between stabilities of different DNA architectures. As proof of principle, we have chosen two well characterized nanostructures: one, a dynamic nanodevice called the I-switch which undergoes a conformational change in response to changes in proton concentration in its environment [4], while the second is a self-assembled static architecture, the DNA icosahedron [5].
The I-switch (IA488/A647) is a dynamic nanaodevice assembled in a one-pot synthesis and comprises only three DNA strands. It consists of two B-DNA duplexes connected to each other by a hinge, carrying terminal cytosine-rich single-strand overhangs [4]. On acidification of the medium, the cytosine-rich overhangs protonate and form an I-motif [6], thus causing a conformational change in the assembly (Fig. 1a). This conformational change in fluorescently labeled I-switch IA488/A647 is monitored by means of fluorescence resonance energy transfer (FRET) between a donor and an acceptor fluorophore placed at the two ends of the single stranded overhangs. This DNA-based pH sensor has previously been shown to function autonomously as well as quantitatively in cultures of Drosophila hemocytes [4] and coelomocytes of Caenorhabditis elegans [7], where it has been shown to map spatiotemporal changes in pH associated with endosomal maturation.
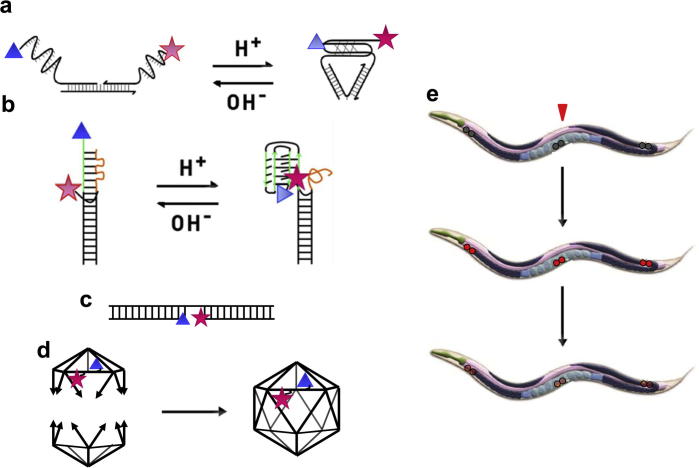
Fig. 1.
Setting up an assay to assess the stability of DNA based nanostructures in vivo. Schematics illustrating (a) the structure and working of the three-stranded I-switch, IA488/A647. (b) the principle of the re-engineered, two-stranded ITfA488/A647. (c) a DNA duplex, ATMR/A647, formed by three perfectly complementary oligonucleotides. (d) The formation of the DNA icosahedron (ICFITC/A647) from two complementary half icosahedra. In each of these schematics, a blue triangle represents the donor fluorophore, while the acceptor is indicated by a red star. (e) The measurement of DNA architecture stability in vivo using coelomocytes of Caenorhabditis elegans[8], achieved by injecting the nematode with fluorescently labeled DNA nanostructures (red arrow) and then monitoring fluorescence decay in coelomocytes as a function of time.
We have also re-engineered the I-switch to make a two-stranded pH-sensitive DNA nanomachine (ITfA488/A647) (Fig. 1b). ITf possesses a single pH responsive cytosine rich strand which undergoes intramolecular I-motif formation at acidic pH regimes. This cytosine rich strand partially base pairs with a guanine rich overhang in the complementary strand to form a weak duplex. At low pH, this mismatched duplex dissociates due to the formation of an intramolecular I-motif by the C-rich strand. Similar to IA488/A647, ITfA488/A647 relies on FRET between a donor (placed on one end of a DNA oligonucleotide) and an acceptor (incorporated internally into the complementary oligonucleotide) to monitor I-motif formation and dissociation.
To study the effects, if any, of single strand segments on DNA nanostructure stability, we designed a DNA duplex, ATMR/A647, without any overhangs and mismatches (Fig. 1c). This is a stable, perfectly complementary DNA duplex, with a nick in the centre. The nick has a donor, tetra methyl rhodamine (TMR), and an acceptor, Alexa 647 (A647), on the termini abutting the nick. This design enables this structure to be a fluorescently labeled, constitutively FRETting DNA duplex irrespective of the local environment of the structure.
In contrast to a dynamic structure such as the I-switch and a simple DNA duplex like ATMR/A647, the DNA icosahedron (ICFITC/A647) is a complex 3D polyhedron constructed by the stepwise assembly of discrete five-way-junctions (5WJ) (Fig. 1d). Each of these distinct modules is engineered with programmable overhangs for their controlled self assembly into an icosahedron in quantitative yields [5]. This stepwise construction strategy of forming a full icosahedron from two component half icosahedra enables one to physically entrap various nanoscale cargo from bulk solution within the icosahedron during its synthesis. Using this strategy, the DNA icosahedron has been shown to encapsulate molecular cargo such as gold nanoparticles [5] or fluorescent dextrans within its internal void, which have been used as bioimaging agents within living systems like Drosophila hemocytes and coelomocytes of C. elegans [9].
Using these nanostructures as prototypes, we set up an in vivo assay to measure their relative stabilities and half lives (Fig. 1e). The presence of fluorescent moieties on these structures lends them to fluorescence based assays for stability measurements. Reasonably, we chose scavenger cells or coelomocytes of the multicellular model organism C. elegans [10] as our test bed for these in vivo studies. This was primarily due to two reasons: firstly, C. elegans is a transparent organism, which is ideally suited to florescence imaging based measurements. Secondly, DNA nanostructures, once introduced in the pseudocoelom of the worm, are targeted to the coelomocytes due to their negatively charged phosphate backbone and internalized via anionic ligand-binding receptors [7]. Coelomocytes are six oblong macrophage-like cells located in the body cavity of C. elegans. These cells are full of membrane-bound invaginations that are highly active in endocytosis of foreign molecules introduced by microinjection. While the natural molecules that are removed from the body cavity by coelomocytes are unknown, they are known to be completely dispensable to the viability and survival of the worm [10]. Here, we provide a step-by-step protocol for fluorescence-based measurement of stability of any DNA nanostructure using the coelomocytes of C. elegans as a model system.
2. Construction of DNA nanostructures
2.1. Construction of IA488/A647
1. IA488/A647 was constructed using protocols previously described [7].
2.2. Construction of ITfA488/A647
2.2.1. Materials
-
1.
All oligonucleotides (obtained from IBA GmbH, Germany) are high performance liquid chromatography (HPLC) purified and lyophilized. Oligonucleotides are dissolved in Milli Q (MQ) water (Millipore, USA) to prepare 200 μM stocks, aliquoted and all aliquots stored at −20 °C, until further use. Fluorescently labeled oligonucleotides are subjected to ethanol precipitation prior to use to remove any traces of free dye.
-
2.
Ethanol, absolute.
-
3.
3.0 M Potassium acetate solution: 2.94 g CH3COOK dissolved in 10 mL MQ water, and pH adjusted to 5.2.
-
4.
Phosphate buffer: 100 mM KH2PO4:1.36 g KH2PO4 dissolved in 10 mL MQ water. 100 mM K2HPO4:1.74 g K2HPO4 dissolved in 10 mL MQ water. Each potassium phosphate solution is diluted to 10 mM. Add 10 mM KH2PO4 and 10 mM K2HPO4 to obtain a buffer of pH 5.5.
-
5.
3.0 M Potassium chloride solution: 2.23 g KCl dissolved in 10 mL MQ water.
-
6.
Heat block and Water bath (Neo Labs, India).
2.2.2. Sample preparation
-
1.
ITfA488/A647 is generally prepared at 5 μM concentration in a volume of 50 μL. 1.25 μL of Sw1-C3-488-cell and Sw2-C3-647-cell (Table 1, each from a 200 μM stock) with 1.65 μL of 3 M KCl are mixed. The volume is made up to 50 μL by adding 10 mM potassium phosphate buffer of pH 5.5. The solution is briefly vortexed.
-
2.
The solution is heated at 90 °C for 5 min in a heat block, and then cooled to room temperature at a rate of 1 °C per 3 min. Samples are then equilibrated at 4 °C overnight. Samples are used within 7 days of annealing.
Table 1.
Oligonucleotide sequences (5′–3′) used for ITfA488/A647.
| Name | Sequence |
|---|---|
| Sw1-C3-488-CELL | CCCTAACCCTAACCCTAACCCGACTCACTGTTTGTCTGTCGTTCTAGGATATATAT (Alexa 488 at 5′ terminus) |
| Sw2-C3-647-CELL | ATATATATCCTAGAACGACAGACAAACAGTGAGTCTTTGTTATGTGTTATGTGTTAT (Alexa 647 on italicized base, bold sequences are perfectly complementary to each other) |
2.3. Construction of ATMR/A647
2.3.1. Materials
-
1.
All oligonucleotides (obtained from IBA GmbH, Germany) are high performance liquid chromatography (HPLC) purified and lyophilized. Oligonucleotides are dissolved in Milli Q (MQ) water (Millipore, USA) to prepare 200 μM stocks, aliquoted and all aliquots stored at −20 °C, until further use. Fluorescently labeled oligonucleotides are subjected to ethanol precipitation prior to use to remove any traces of free dye.
-
2.
Ethanol, absolute.
-
3.
3.0 M Potassium acetate solution: 2.94 g CH3COOK dissolved in 10 mL MQ water, and pH adjusted to 5.2.
-
4.
Phosphate buffer: 100 mM KH2PO4:1.36 g KH2PO4 dissolved in 10 mL MQ water. 100 mM K2HPO4:1.74 g K2HPO4 dissolved in 10 mL MQ water. Each potassium phosphate solution is diluted to 10 mM. Add 10 mM KH2PO4 and 10 mM K2HPO4 to obtain a buffer of pH 5.5.
-
5.
3.0 M Potassium chloride solution: 2.23 g KCl dissolved in 10 mL MQ water.
-
6.
Heat block and Water bath (Neo Labs, India).
2.3.2. Sample preparation
-
1.
ATMR/A647 is generally prepared at 5 μM concentration in a volume of 50 μL. 1.25 μL of A1-TMR, A2-647 and A3 (Table 2, each from a 200 μM stock) with 1.65 μL of 3 M KCl are mixed. The volume is made up to 50 μL by adding 10 mM potassium phosphate buffer of pH 5.5. The solution is briefly vortexed.
-
2.
The solution is heated at 90 °C for 5 min in a heat block, and then cooled to room temperature at a rate of 1 °C per 3 min. Samples are then equilibrated at 4 °C overnight. Samples are used within 7 days of annealing.
Table 2.
Oligonucleotide sequences (5′–3′) used for ATMR/A647.
| Name | Sequence |
|---|---|
| A1-TMR | AGGCTTTAAATACCGGCATG (TMR at 5′ terminus) |
| A2-647 A3 | CTCGAACCACGATATCTTCG (Alexa 647 at 3′ terminus) CATGCCGGTATTTAAAGCCTTTCGAAGATATCGTGGTTCGAG (Bold sequences are perfectly complementary to A1 and A2, respectively) |
2.4. Construction of DNA icosahedron
1. The DNA icosahedron was constructed using the standard protocol described previously [4]. In order to study the stability of this structure in vivo, the U5WJ of the icosahedron was modified to carry a 5′-fluorescein isothiocyanate (FITC) label positioned on U1 oligo and a 5′-Alexa 647 label positioned on U4 oligo. At these positions in the fully formed icosahedron, the two fluorophores are 3.4 ± 2 nm apart, which is well within the R0 of this FRET pair [11].
3. C. elegans maintenance and strains
-
1.
C. elegans is grown at 22 °C on nematode growth medium (NGM) containing a lawn of OP50 bacteria.
-
2.
All strains have been obtained from Caenorhabditis Genetics Center (University of Minnesota, USA).
-
3.
Wild type strain: C. elegans isolate from Bristol (strain N2).
4. Microinjections and coelomocyte labeling
4.1. Materials
-
1.
10× medium 1: 4.37 g NaCl, 0.18 g KCl, 0.055 g CaCl2, 0.1 g MgCl2 and 0.95 g HEPES dissolved in 50 mL MQ water and pH adjusted to 7.3. This is filter-sterilized using 0.22 μm membrane filter (Millipore, USA).
-
2.
1× M9 buffer: 0.6 g Na2HPO4, 0.3 g KH2PO4, 0.5 g NaCl and 0.1 g NH4Cl dissolved in 100 mL MQ water and pH is adjusted to 7.3. The buffer is filter-sterilized through a 0.22 μm membrane filter.
-
3.
TE2000-S inverted microscope, equipped with a 40×, 0.75 NA objective (Nikon, Japan) and microinjection setup (Narishige, Japan).
-
4.
Borosilicate glass capillaries (Sutter Instruments, USA).
-
5.
2.0% agarose pads, made on 22 × 60 mm glass coverslips (Blue Star, India).
-
6.
2.0% agarose: 0.2 g agarose (Bangalore Genei, India) dissolved in 10 ml MQ water.
-
7.
Halocarbon oil (Sigma–Aldrich, USA).
4.2. Microinjections in C. elegans
-
1.
For coelomocyte labeling, ICFITC/A647 is diluted to 3 μM, while IA488/A647, ITfA488/A647 and ATMR/A647 are diluted to 500 nM. These dilutions are made using 1x medium 1. These solutions are centrifuged at 12,000 rpm for 10 min in order to precipitate any particulate matter.
-
2.
One day old hermaphrodites grown on NGM plates (+OP50) are mounted on a 2% agarose pad containing a droplet of halocarbon oil.
-
3.
Injections are performed, using borosilicate capillaries, at 50–55 psi in the dorsal side of the pseudocoelom, just opposite the vulva.
-
4.
Injected worms are released using 1× M9 buffer and transferred to NGM plates (+OP50). Plates are incubated at 22 °C. Typically, 10 worms were injected for each time point.
-
5.
After the requisite time, injected worms are mounted on a glass slide containing a 2.0% agarose pad, anesthetized using 30 mM NaN3 in M9 buffer and imaged. Ordinarily, for stability studies, incubation times are varied between 1 h and 36 h.
5. Imaging C. elegans coelomocytes
5.1. Materials
-
1.
Olympus IX81 inverted microcope (Olympus, Japan), equipped with 60×, 1.42 NA objective, XCite Series 2.2Q metal halide illuminator (Olympus, Japan), filters suitable for each fluorophore (Chroma, USA) and an iXon X3 CCD camera (Andor, USA.).
-
2.
MetaMorph image acquisition software (Molecular Devices, USA).
-
3.
ImageJ ver. 1.46 (NIH, freely available from website: http://rsbweb.nih.gov/ij/).
5.2. Ratiometric image acquisition and analysis
5.2.1. IA488/A647 and ITfA488/A647
-
1.
Coelomocytes of worms injected with IA488/A647 or ITfA488/A647 are located and imaged in a single plane so as to focus maximum number of endosomes. Typically, the number of cells imaged was ∼30 to reduce stochastic variation.
-
2.
Care should be taken that exposure times used do not saturate the signal.
-
3.
Fluorescence images of cells are obtained by exciting Alexa 488 and collecting emission using the 530 ± 15 nm emission filter. This yields a donor image (D). The cells are then re-excited at 488 nm and emission of the acceptor, Alexa 647, is acquired using a 710 ± 40 nm filter. This is the FRET image (A). Again, a third image is obtained by directly exciting the acceptor and collecting emission at acceptor emission wavelength. This is the acceptor image (AC).
5.2.2. ATMR/A647
-
1.
Coelomocytes of worms injected with ATMR/A647 are located and imaged in a single plane so as to focus maximum number of endosomes.
-
2.
Care should be taken that exposure times used are compatible with unsaturation of signal.
-
3.
Fluorescence images of cells are obtained by exciting TMR and collecting emission using the 570 ± 15 nm emission filter. This yields a donor image (D). The cells are then re-excited at 540 nm and emission of the acceptor, Alexa 647, is acquired using a 710 ± 40 nm filter. This is the FRET image (A). A third image is obtained by directly exciting the acceptor and collecting emission at acceptor emission wavelength. This is the acceptor image (AC).
5.2.3. ICFITC/A647
-
1.
Similar to IA488/A647 and ITfA488/A647, coelomocytes of worms injected with ICFITC/A647, are located and imaged in a single plane so as to focus maximum number of endosomes.
-
2.
Care should be taken that exposure times used are compatible with unsaturation of signal.
-
3.
Fluorescence images of cells are obtained by exciting fluorescein isothiocyanate (FITC) and collecting emission using the 530 ± 15 nm emission filter. This yields a donor image (D). The cells are then re-excited at 488 nm and emission of the acceptor, Alexa 647, is acquired using a 710 ± 40 nm filter. This is the FRET image (A). A third image is obtained by directly exciting the acceptor and collecting emission at acceptor emission wavelength. This is the acceptor image (AC).
5.3. Image analysis
-
1.
Autofluorescence of each image (D, A and AC) is calculated by measuring mean pixel intensity over an adjacent cell-free area in that image. This autofluorescence is subtracted from the corresponding image, prior to all image processing.
-
2.
Each endosome in the donor channel (D) is selected by the ROI plugin in ImageJ, and total and mean intensities in each endosome are measured and recorded.
-
3.
Each saved ROI is recalled and total and mean intensities of the corresponding vesicles in the FRET image (A) and the acceptor image (AC) are measured.
-
4.
Dividing the mean intensity of each endosome in the donor image by the corresponding intensity in the FRET image provides a donor/acceptor (D/A) ratio for that endosome. Taking all the endosomes of a single coelomocyte into consideration, a mean D/A value for each cell is calculated. This is repeated for all the cells imaged (each reading is obtained from coelomocytes of 10 worms). These values are then used to calculate a mean D/A ratio for the corresponding time point.
-
5.
Concomitantly, the integrated acceptor intensity for each endosome is also calculated from the acceptor image (AC). Each of these endosomal intensity values are added up for a single cell, and the total cellular intensity is thus calculated. These values are then used to calculate mean acceptor intensity for that time point.
6. Results and discussion
Here, using C. elegans as an in vivo test bed, we have addressed the stability of DNA nanostructures and exploited the negative charge on the DNA backbone that is responsible for them being targeted to anionic ligand-binding receptors (ALBRs) on the coelomocyte surface [7]. The DNA nanostructures chosen to set up and check the feasibility of this assay were two well characterized dynamic pH-sensitive DNA nanomachines [4] and the static architecture, the DNA icosahedron [5]. We used a simple fluorescently labeled DNA duplex as a control. Once inside the ceoelomocyte, these fluorescently labeled architectures form bright puncta of ∼0.8 μm size. These puncta, which have previously been characterized as endosomes [7], mature to reach the lysosomal network, which is known to be the seat of cellular degradation [12]. Thus, the fluorescent signal of the architectures was used to quantitatively assess the stability and persistence of these structures within the endo-lysosomal pathway.
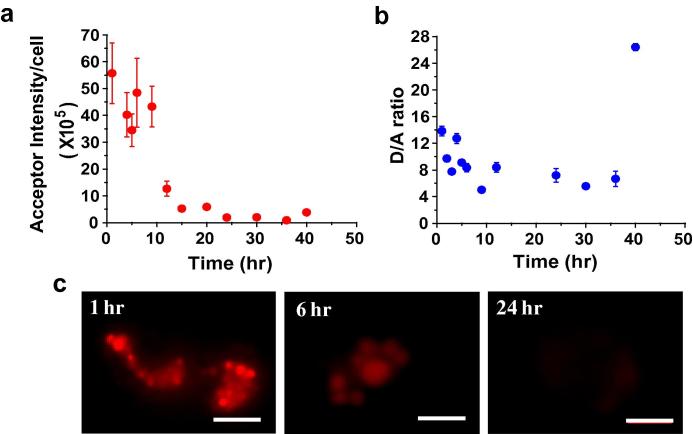
The I-switch is a rationally designed, dynamic, DNA based pH sensor, where a change in donor and FRET intensities is a read-out of the environmental acidity. This precludes the use of D/A ratio to study the stability of this nanodevice, since this ratio would report on the acidification of endosomes [7], and not intactness of structure. As a result, the decay of acceptor intensity (AC) was chosen to assess the stability of this architecture in the endo-lysosomal system. On injecting dually labeled I-switch, IA488/A647 and following average acceptor intensity per cell as a function of time, it was found that IA488/A647 shows a well-behaved degradation profile over time (Fig. 2a). The data clearly revealed that IA488/A647 has a half-life of ∼8 h within coelomocytes. It was observed that the D/A ratios (and hence pH values), on the contrary, did not change appreciably over time (Fig 2b). This reveals that degraded IA488/A647 does not contribute to the signal, because the D/A ratio of the I-switch in these endosomes remains identical to the value at 45 min post-injection. Thus it is likely that when the I-switch is degraded inside the lysosomes, it is digested into very low molecular weight fragments with or without attached fluorophores that leak out of the lysosomal membranes. This degradation was clearly reflected at later time points as endosomal fluorescence within coelomocytes was palpably fainter (Fig. 2c).
Fig. 2.
Assessing the in vivo stability of the three-stranded I-switch, IA488/A647. Plots showing (a) mean acceptor intensity per cell and (b) mean donor/FRET (D/A) ratio of the I-switch as a function of time. Error bars indicate s.e.m. (n = 10 worms, ⩾10 cells) (c) Representative images of coelomocytes labeled with IA488/A647 at various time points. Scale bar: 5 μm.
An alternative form of I-switch consisting of a two-stranded pH-sensitive DNA assembly, ITfA488/A647, was used next. This was based on the prediction that a structure with only one single-stranded overhang, as opposed to two single-stranded overhangs in IA488/A647, would be more resistant to lysosomal degradation. This prediction was correctly borne out by the observation that while D/A ratios for ITfA488/A647 remain similarly constant over time, acceptor intensities per cell show a reduced rate of decrease, with an increased half-life of ∼11 h (Fig. 3). Thus, a rational re-designing of a nanomachine led to an increased stability of the device, resulting in an increase in temporal regimes over which it can be used to give quantitative read-outs of compartment function.
Fig. 3.
Assessing the in vivo stability of the pH sensitive two-stranded I-switch, ITfA488/A647. Plots showing (a) mean acceptor intensity per cell and (b) mean donor/FRET (D/A) ratio as a function of time. Error bars indicate s.e.m. (n = 10 worms, ⩾20 cells) (c) Representative images of coelomocytes labeled with ITfA488/A647 at various time points. Scale bar: 5 μm.
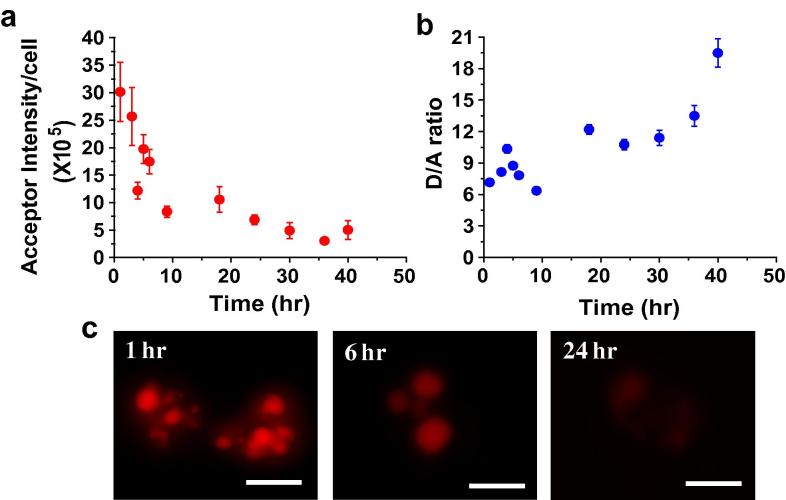
We also designed a DNA duplex, ATMR/A647, without any overhangs and mismatches. Such a duplex was designed keeping in mind the effects of single strands, such as in IA488/A647 and ITfA488/A647, on degradation. The fluorophores on this duplex were placed in a position to enable FRET irrespective of the local environment of the structure. When injected into the pseudocoelom of C. elegans, it is observed that it shows very high donor (D) as well as acceptor (A) intensities within 2 h of injection, which show a decrease over time (Fig. 4a and c). Its half life within lysosomes of coelomocytes is noticeably higher (∼14 h) than IA488/A647 and ITfA488/A647. This increase in half life is a clear indication of the overriding contribution of single stranded segments to DNA architecture stability. D/A ratios, on the other hand, remain steady till 24 h, again pointing to the absence of signal contribution from the degraded structure (Fig. 4b).
Fig. 4.
Measuring the stability of a fluorescently labeled DNA duplex, ATMR/A647 in coelomocytes of C. elegans. Graphs showing (a) mean acceptor intensity per cell and (b) mean donor/FRET (D/A) ratio as a function of time. Error bars indicate s.e.m. (n = 10 worms, ⩾12 cells) (c) Representative images of coelomocytes labeled with ATMR/A647 at various time points. Scale bar: 5 μm.
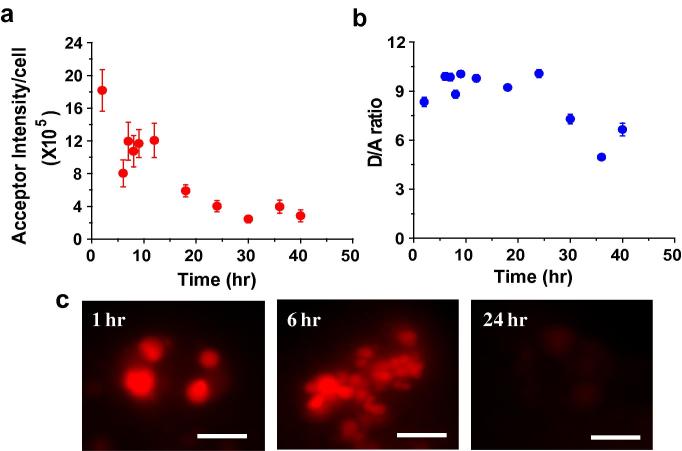
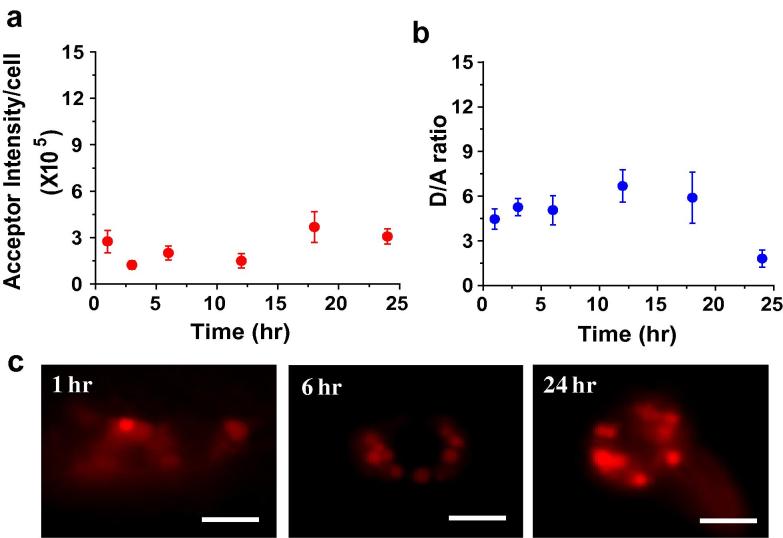
The DNA icosahedron is a self-assembled, fully catenated, platonic solid with no single stranded overhangs or free termini. When modified with FITC and Alexa 647 at appropriate positions for FRET and injected into the body cavity of C. elegans, it shows very high donor (D) as well as FRET (A) intensities. Since degradation in the lysosomes would lead to disassembly of its component strands and subsequent decrease in FRET, the decay of donor/FRET ratios (D/A) was chosen as a measure of stability of the icosahedron. When these D/A ratios are monitored as a function of time, from 1 h to 24 h, it is revealed that there is no change within error over 24 h post-injection. Average acceptor intensity per coelomocyte was also measured over time and this too, showed no change till 24 h (Fig. 5a and b). Beyond 24 h, acceptor intensity actually increases slightly, consistent with greater accumulation of DNA cargo in endosomes due to prolonged, efficient endocytosis of the pseudocoelomic fluid. In fact, the intensities of labeling, FRET intensities, D/A ratios and labeling efficiencies are almost indistinguishable from 1 h to 20 h post-injection (Fig. 5c). The DNA icosahedron shows recalcitrance to such lysosomal degradation consistent with the fact that all the juxtaposed termini are ligated into circularized oligonucleotides. This indicates that the population of DNA icosahedra present inside the endosomes consists predominantly of intact structures up to 24 h, and can well be used to make reliable functional measurements in vivo up to this time.
Fig. 5.
Measuring the stability of the static architecture, ICFITC/A647 in coelomocytes of C. elegans. Graphs showing (a) mean acceptor intensity per cell and (b) mean donor/FRET (D/A) ratio as a function of time. Error bars indicate s.e.m. (n = 10 worms, ⩾10 cells) (c) Representative images of coelomocytes labeled with ICFITC/A647 at various time points. Scale bar: 5 μm.
Thus, here we have outlined a method to quantitatively assay the stability and lifetime of DNA based architectures in the coelomocytes of the multicellular organism C. elegans. Our studies show that this assay is capable of robustly capturing differences in the stabilities of different DNA nanostructures to lysosomal degradation in vivo and relies on the relative ease with which any DNA based assembly can be introduced in the worm pseudocoelom and subsequently, into coelomocytes. A key feature of this approach is the use of fluorophore tags on the DNA assembly, which can be programmed precisely according to the architecture under investigation; measurements of fluorescence decay as a function of time can then be chosen and tailored accordingly. This study also yields insight into factors that may potentially govern DNA nanostructure stability in vivo. Our investigations reveal that the stability of a nanostructure depends on the number of single strand segments it has; due to the conformational flexibility of these segments and ease of accessibility by various nucleases, these regions are more prone to degradation, leading to overall structure clearance. This is supported by our experiments with ITfA488/A647, which has only one single strand, and has a higher half life (∼11 h) compared to IA488/A647 (∼8 h), which comprises two single stranded overhangs. By removing these overhangs and creating a simple linear assembly, as in ATMR/A647, we have increased the stability of this structure in vivo (∼14 h). On the other hand, fabricating a designer nanostructure like the DNA icosahedron which has been ligated to have no free termini, results in an extremely stable structure which does not undergo degradation up to 24 h. This is analogous to having chemically resistant synthetic DNA counterparts, which can then be incorporated into designer nanostructures to have greater stabilities than ATMR/A647. This assay then becomes a good test bed to investigate the relative stabilities of larger numbers of diverse DNA nanostructures. Such measurements are an essential precursor to the application of rationally designed DNA nanostructures and are exceedingly important for such structures to be used as investigative tools in biological systems.
Acknowledgements
We thank Sandhya P. Koushika for inputs on experiments, Central Imaging Facility at NCBS, the Caenorhabditis Genetics Center (funded by NIH-NCRR) for nematode strains, DBT and the Nanoscience and Technology Initiative of DST for funding. S.S. and D.B. acknowledge the CSIR and Y.K. acknowledges the Innovative Young Biotechnologist Award and Wellcome Trust-DBT India Alliance for fellowships.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Krishnan Y., Simmel F.C. Angew. Chem., Int. Ed. 2011;50:3124–3156. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia D., Sharma S., Krishnan Y. Curr. Opin. Biotechnol. 2011;22:475–484. doi: 10.1016/j.copbio.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Bath J., Turberfield A.J. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 4.Modi S., Shwetha M.G., Goswami D., Gupta G.D., Mayor S., Krishnan Y. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia D., Mehtab S., Krishnan R., Indi S.S., Basu A., Krishnan Y. Angew. Chem., Int. Ed. 2009;48:4134–4137. doi: 10.1002/anie.200806000. [DOI] [PubMed] [Google Scholar]
- 6.Guéron M., Leroy J.L. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 7.Surana S., Bhat J.M., Koushika S.P., Krishnan Y. Nat. Commun. 2011;2:340. doi: 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- 8.Z.F. Altun and D. Hall, Handbook of C. elegans anatomy, 2008, in WormAtlas. http://www.wormatlas.org/hermaphrodite/hermaphroditehomepage.html.
- 9.Bhatia D., Surana S., Chakraborty S., Koushika S.P., Krishnan Y. Nat. Commun. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 10.Fares H., Greenwald I. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawski A. Photochem. Photobiol. 1983;38:487–508. [Google Scholar]
- 12.de Voer G., Peters D., Taschner P.E.M. Biochim. Biophys. Acta. 2008;1782:433–446. doi: 10.1016/j.bbadis.2008.04.003. [DOI] [PubMed] [Google Scholar]