Fig. 1.
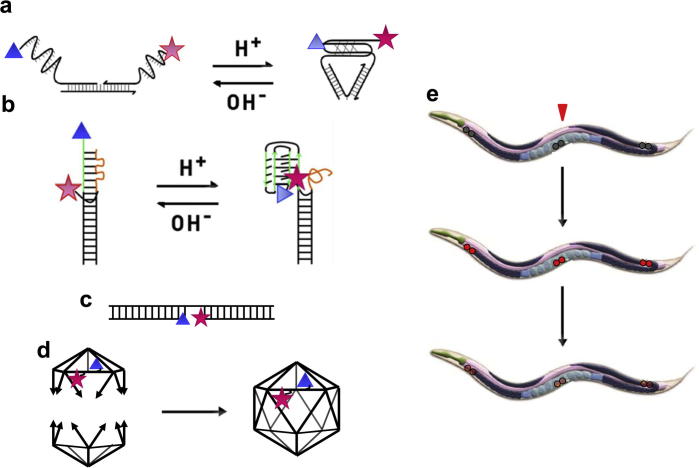
Setting up an assay to assess the stability of DNA based nanostructures in vivo. Schematics illustrating (a) the structure and working of the three-stranded I-switch, IA488/A647. (b) the principle of the re-engineered, two-stranded ITfA488/A647. (c) a DNA duplex, ATMR/A647, formed by three perfectly complementary oligonucleotides. (d) The formation of the DNA icosahedron (ICFITC/A647) from two complementary half icosahedra. In each of these schematics, a blue triangle represents the donor fluorophore, while the acceptor is indicated by a red star. (e) The measurement of DNA architecture stability in vivo using coelomocytes of Caenorhabditis elegans[8], achieved by injecting the nematode with fluorescently labeled DNA nanostructures (red arrow) and then monitoring fluorescence decay in coelomocytes as a function of time.