Abstract
Objectives
To determine the fitness effects of various mobile genetic elements (MGEs) in Enterococcus faecium and Enterococcus faecalis when newly acquired. We also tested the hypothesis that the biological cost of vancomycin resistance plasmids could be mitigated during continuous growth in the laboratory.
Methods
Different MGEs, including two conjugative transposons (CTns) of the Tn916 family (18 and 33 kb), a pathogenicity island (PAI) of 200 kb and vancomycin-resistance (vanA) plasmids (80–200 kb) of various origins and classes, were transferred into common ancestral E. faecium and E. faecalis strains by conjugation assays and experimentally evolved (vanA plasmids only). Transconjugants were characterized by PFGE, S1 nuclease assays and Southern blotting hybridization analyses. Single specific primer PCR was performed to determine the target sites for the insertion of the CTns. The fitness costs of various MGEs in E. faecium and E. faecalis were estimated in head-to-head competition experiments, and evolved populations were generated in serial transfer assays.
Results
The biological cost of a newly acquired PAI and two CTns were both host- and insertion-locus-dependent. Newly acquired vanA plasmids may severely reduce host fitness (25%–27%), but these costs were rapidly mitigated after only 400 generations of continuous growth in the absence of antibiotic selection.
Conclusions
Newly acquired MGEs may impose an immediate biological cost in E. faecium. However, as demonstrated for vanA plasmids, the initial costs of MGE carriage may be mitigated during growth and beneficial plasmid–host association can rapidly emerge.
Keywords: antimicrobial resistance, horizontal gene transfer, adaptation, directional selection, plasmids, PAI, Tn916, Tn6000
Introduction
Enterococci are Gram-positive bacteria that normally inhabit the gastrointestinal tract of humans and animals. However, they have become the third most common cause of nosocomial infection due to their ability to develop multidrug resistance and horizontally acquire additional DNA associated with niche adaptation.1,2 Enterococcus faecalis has been the predominant pathogenic enterococcal species in clinical settings, but the relative frequency of Enterococcus faecium is steadily increasing.3,4 Enterococcus spp. are well known for their intrinsic resistance to a number of antimicrobial agents, which, accompanied by their ability to acquire resistance determinants, makes treatment challenging.
Mobile genetic elements (MGEs) are common in enterococci,5 with conjugative transposons (CTns), pathogenicity islands (PAIs) and plasmids all contributing to resistance, virulence and host adaptation. CTns of the Tn916 family are primarily responsible for transferable tetracycline resistance.6 Tn916 has been found in, or introduced into, more than 30 different genera of bacteria,6 whereas Tn6000 and related elements have been found only in Enterococcus spp.7 In addition, different virulence determinants have been shown to be present on multiple PAIs also capable of transfer.8 Plasmids are responsible for, among many adaptive traits, the majority of observed transferable vancomycin resistance.2
Glycopeptide-resistant enterococci (GRE) were first reported in the late 1980s,9 and have since been found worldwide. GRE are troublesome nosocomial pathogens since the glycopeptide vancomycin is considered to be a key therapeutic agent for treating multidrug-resistant, Gram-positive pathogens.5,10 To date, eight types of acquired vancomycin resistance gene clusters have been described in enterococci. The VanA resistance phenotype is the most prevalent,5 and the vanA gene cluster is often plasmid-located and associated with Tn3-like transposons of the Tn1546 prototype.2,11–13
The emergence and spread of antimicrobial resistance determinants by MGEs is not limited to glycopeptide resistance. Enterococci have been termed a ‘hub’ of MGEs,13,14 resulting in concern over the transfer of resistance determinants from the enterococci into more obligate pathogenic bacterial species, including members of the genera Streptococcus, Clostridium and Staphylococcus.15–17 An increasing body of descriptive data on the molecular structure and epidemiology of MGEs in Enterococcus has emerged over recent years. However, a knowledge gap currently exists relating to the specifics of the population dynamics of MGEs in Enterococcus. An increased understanding of how MGEs affect their bacterial hosts is fundamental to understanding the different biological factors controlling the emergence, spread and persistence of antibiotic resistance determinants.2,18
Until the late 1980s, the prevailing dogma was that antibiotic resistance would rapidly disappear from bacterial populations following withdrawal or reduced antibiotic consumption levels of the corresponding antimicrobial drugs. This was due to the assumption that, in the absence of selective pressure from antimicrobial agents, less-fit resistant bacteria would be outcompeted by their more-fit but susceptible counterparts in the population. However, studies from the late 1980s and throughout the 1990s pointed out that bacteria with acquired resistance kept the mechanisms of resistance and alleviated the fitness costs of such traits through compensatory evolution, instead of reverting to susceptibility.2,19–21
It is now clear that, in the absence of selective pressure caused by antibiotics, newly acquired resistances often reduce bacterial fitness when compared with their antibiotic-susceptible counterparts in the population. This has been termed ‘the biological cost of resistance’.22 It is also clear that the time needed for reversal of this is a function of the magnitude of these biological costs. Theory also suggests that the biological cost of resistance plays an important role in the emergence of resistance during antimicrobial therapy in patients.18 Thus, the biological cost of resistance is a key parameter controlling the frequency of resistance in clinical bacterial populations.19,22,23 Consequently, the magnitude of the biological cost of resistance is a key determinant of the persistence of drug resistance when antimicrobial selective pressures are relaxed, i.e. following interventions.
The main body of work in the field of biological costs of resistance has focused on various chromosomal determinants of antimicrobial resistance.21,24–28 The methodological approaches developed in these studies are also highly relevant to investigating the biological costs associated with MGEs harbouring antimicrobial resistance determinants.
The host fitness effects caused by MGE-encoded resistance determinants can be considered at different levels. The considerations of fitness cost can include the MGE backbone,20,29,30 or resistance costs can be measured separately from it.31 Whereas the latter approach specifically addresses the biological costs of the resistance determinant itself, it is important to acknowledge that bacteria with newly acquired MGEs face not only the cost of the resistance element per se, but also the additional cost of other functions encoded by their vectors. Co-evolution is therefore expected to occur between the MGE and the bacterial host in a process not limited to the resistance determinants alone. For instance, such co-evolutionary processes can, if they lead to an increase in relative fitness, directly affect the potential for the persistence and further spread of resistance determinants, even in situations where the resistance determinant alone confers no benefit to the host.
In this study, we experimentally measured the fitness cost of different MGEs in E. faecium and E. faecalis, including two CTns of the Tn916 family, a PAI and vancomycinresistance plasmids of various origins. We showed that, in strains with a newly acquired PAI or one of the two CTns, the biological cost depended on the host and the insertion locus. Moreover, the majority of vanA plasmids severely reduced host fitness, and these costs were rapidly reduced in actively growing bacterial populations.
Materials and methods
Bacterial strains and populations
The bacterial strains and populations used in this study are listed in Table 1. All strains and populations were grown in brain heart infusion (BHI) broth or on BHI agar plates (Sigma-Aldrich, Germany) at 37°C.
Table 1.
Strains and populations used in this study
| Strain/population | Relevant characteristics | MGEa | ST donor | Reference |
|---|---|---|---|---|
| 64/3 | recipient plasmid-free E. faecium strain: RifR, FusR | — | ST21 | 35 |
| 64/3PEV | recipient E. faecium strain 64/3 serially transferred for 400 generations in BHI broth resulting in a mixed evolved ancestral population: RifR, FusR | — | ST21 | this study |
| 64SS | recipient plasmid-free E. faecium strain: StrR, SptR | — | ST21 | 35 |
| OG1RF | recipient plasmid-free E. faecalis strain: RifR, FusR | — | ST1 | 8 |
| 70/90 | clinical E. faecium strain from 1990 (France) | 100 kb rep-inc18 vanA plasmid with axe-txe (PSK) + 45 kb plasmid | ST25 | 35 |
| UW3540 | clinical E. faecium strain from 2002 (Germany) | 80 kb rep-inc18 vanA-erm(B)- vat(D) plasmid | ST145 | 35 |
| H182 | clinical E. faecium strain from 2002 (Portugal) | 92 kb rep-pRUM vanA plasmid | ST18 | 35 |
| UW261 | food E. faecium strain from 1995 (Germany) | 172 kb vanA plasmid of unknown type with ω-ɛ-ζ (PSK) | ND | 35 |
| E0292 | clinical E. faecium strain from 2002 (USA) | 200 kb rep-inc18 vanA plasmid | ST20 | 35 |
| 229710 | clinical E. faecalis strain from 1996 (Portugal) | 85 kb rep-inc18/rep-pAD1 vanA plasmid with par | ST6 | 35 |
| 664.1H1 | E. casseliflavus donor of Tn6000: TetR | 33 kb CTn (Tn6000) | — | 33 |
| BS34A | B. subtilis donor of Tn916: TetR | 18 kb CTn (Tn916) | — | 32 |
| TC1Tn6000 | transconjugant E. faecium 664.1H1 × 64/3: RifR, FusR, TetR | 33 kb Tn6000 | — | this study |
| TC2Tn916 | transconjugant E. faecium BS34A × 64/3: RifR, FusR, TetR | 18 kb Tn916 | — | this study |
| TC3 | transconjugant E. faecium UW3114 × 64/31 with plasmid pLG2, PAI(–): EryR, FusR, RifR | pLG2 | ST21 | 8 |
| TC4PAI | transconjugant E. faecium UW3114 × 64/32 with plasmid pLG2, PAI(+): EryR, FusR, RifR | pLG2, 200 kb PAI | ST21 | 8 |
| TC5 | transconjugant E. faecalis UW3114 × OG1RF1 with plasmid pLG2, PAI(–): EryR, FusR, RifR | pLG2 | ST1 | 8 |
| TC6PAI | transconjugant E. faecalis UW3114 × OG1RF2 with plasmid pLG2, PAI(+): EryR, FusR, RifR | pLG2, 200 kb PAI | ST1 | 8 |
| TCvanA | transconjugant E. faecium 70/90 × 64/3: VanR, FusR, RifR | 100 kb vanA- plasmid + 45 kb plasmid | ST21 | 35 |
| TC8vanA | transconjugant E. faecium UW3540 × 64/3: VanR, FusR, RifR | 80 kb vanA plasmid | ST21 | 35 |
| TC9vanA | transconjugant E. faecium H182 × 64/3: VanR, FusR, RifR | 92 kb vanA plasmid | ST21 | 35 |
| TC10vanA | transconjugant E. faecium UW261 × 64/3: VanR, FusR, RifR | 172 kb vanA plasmid | ST21 | 35 |
| TC11vanA | transconjugant E. faecium E0292 × 64/3: VanR, FusR, RifR | 200 kb vanA plasmid | ST21 | 35 |
| TC12vanA | transconjugant E. faecium 229710 × 64/3: VanR, FusR, RifR | 85 kb vanA plasmid | ST21 | 35 |
| TC7 -1PvanA
EV TC7 -2PvanA EV |
transconjugants E. faecium 70/90 × 64/3 serially transferred for 400 generations in BHI broth without antibiotics resulting in mixed evolved populations: VanR, FusR, RifR | 100 kb vanA plasmid | ST21 | this study |
| TC10-1PvanA
EV TC10-2PvanAEV TC10-3PvanA EV |
transconjugants E. faecium UW261 × 64/3 serially transferred for 400 generations in BHI broth without antibiotics resulting in mixed evolved populations: VanR, FusR, RifR | 172 kb vanA plasmid | ST21 | this study |
| TC12-1PvanAEV TC12-2PvanAEV TC12-3PvanA EV |
transconjugants E. faecium 229710 × 64/3 serially transferred for 400 generations in BHI broth without antibiotics resulting in mixed evolved populations: VanR, FusR, RifR | 85 kb vanA plasmid | ST21 | this study |
| TC7-1vanAEV TC7-2vanA EV |
fitness-representative strains of the evolved populations TC7PEV: VanR, FusR, RifR | 100 kb vanA plasmid | ST21 | this study |
| TC10-1vanA
EV TC10-2vanA EV TC10-3vanA EV |
fitness-representative strains of the evolved populations TC10PEV: VanR, FusR, RifR | 172 kb vanA plasmid | ST21 | this study |
| TC12-1vanA
EV TC12-2vanA EV TC12-3vanA EV |
fitness-representative strains of the evolved populations TC12PEV: VanR, FusR, RifR | 85 kb vanA plasmid | ST21 | this study |
| TC7vanASS | transconjugant E. faecium (70/90 × 64/3) × 64SS: VanR, StrR, SptR | 100 kb vanA plasmid | ST21 | this study |
| TC10vanASS | transconjugant E. faecium (UW261 × 64/3) × 64SS: VanR, StrR, SptR | 172 kb vanA plasmid | ST21 | this study |
| TC12vanASS | transconjugant E. faecium (229710 × 64/3) × 64SS: VanR, StrR, SptR | 85 kb vanA plasmid | ST21 | this study |
Van, vancomycin; Ery, erythromycin; Str, streptomycin (high level of resistance); Spt, spectinomycin (high level of resistance); Fus, fusidic acid; Rif, rifampicin; Tet, tetracycline; ND, not determined; PSK, post-segregation killing system.
aPlasmid content present in both donors and transconjugants is described. Several donors contained more than one plasmid; see Werner et al.35
Two CTns of the Tn916 family that are prevalent in Enterococcus spp.6 were transferred from Bacillus subtilis BS34A (Tn916)32 and Enterococcus casseliflavus 664.1H1 (Tn6000)33,34 into E. faecium 64/3 and E. faecalis OG1RF. Tn6000 encodes tetracycline resistance via tet(S), whereas Tn916 encodes tetracycline resistance with the tet(M) gene. The E. faecium 64/3 transconjugants containing Tn6000 and Tn916 that were selected for further study were designated TC1Tn6000 and TC2Tn916, respectively.
Two PAI-containing transconjugants (designated TC4PAI and TC6PAI) and six vanA plasmid-containing transconjugants (designated TC7vanA to TC12vanA) had previously been generated.8,35 These transconjugants resulted from conjugal transfer of a previously described 200 kb PAI residing in E. faecalis UW3114 as well as plasmids from various donor strains of different geographical and temporal origins into a common genetic background, E. faecium 64/3. The PAI was also transferred into E. faecalis OG1RF.8 In these mating experiments, the PAI was always accompanied by the pheromone-responsive plasmid pLG2. The PAI did not, however, always co-transfer with the plasmid. Two transconjugants harbouring only pLG2 were also used in this study (TC3 and TC5).
Conjugation experiments
Transconjugants TC1Tn6000 and TC2 Tn916 were generated using a previously described filter mating protocol34 and selected on BHI agar containing 10 mg/L of tetracycline, 32 mg/L of rifampicin and 5 mg/L of fusidic acid.
Strains TC7vanASS, TC10vanASS and TC12vanASS were generated in conjugation experiments in which the evolved plasmids from the fitness-representative strains (TC7-1EV, TC10-1EV and TC12-1EV) were transferred back into the 64/3 E. faecium recipient containing streptomycin and spectinomycin (SS) resistance markers. Conjugation was performed by filter mating as previously described,35,36 with the following modifications. Exponentially growing donor strains and recipients were mixed in 10 : 1 ratio, and a 100 μL sample of mating mixture was spread on a filter and incubated for 18 h at 37°C. Transconjugants were selected on BHI plates supplemented with 10 mg/L of vancomycin, 750 mg/L of streptomycin and 1000 mg/L of spectinomycin (TC7SS, TC10SS and TC12SS). Conjugation frequencies were calculated as the number of transconjugants per donor cfu.
Antimicrobial susceptibility testing and DNA techniques
The MICs of vancomycin, streptomycin, spectinomycin, erythromycin, rifampicin, tetracycline and fusidic acid were determined using Etest strips according to the manufacturer's instructions (bioMérieux, France). Genomic DNA was prepared and macrodigested with SmaI; plasmid DNA was isolated and treated with S1 nuclease as recently described.35 Prepared samples were analysed by PFGE as previously demonstrated.35 SmaI-restricted Staphylococcus aureus 8325 was used as a reference size marker. Southern blot hybridization was performed on HindIII-digested genomic DNA of strain TC2Tn916 using a probe derived from the Tn916 intTn gene (U09422; 16 704–17 725 bp) from within Tn916. Labelling and detection were carried out using the DIG Labeling and Detection Kit (Roche Diagnostics GmbH, Germany) according to the manufacturer's instructions.
Single specific primer (ssp) PCR was performed to determine the target site for the insertion of Tn916. Genomic DNA was digested with HindIII and BamHI and ligated to HindIII- or BamHI-digested and dephosphorylated pUC19. Primers specific for the left end or right end of the transposon (LEO2 5′-GGATAAATCGTCGTATCAAAGCTCATTC-3′ and REO2 5′-CCACTTCTGACAGCTAAGACATGAGG-3′, respectively) and either pUC19F (5′-GGATGTGCTGCAAGGCGATTAAGTTGG-3′) or pUC19R (5′-CTCGTATGTTGTGTGGAATTGTGAGC-3′) were used to amplify the transposon ends and the flanking sequence were ligated to pUC19. LEO2 and REO2 were also used in PCR analysis to amplify the joint of the excised Tn916 in circular form. DNA sequencing was carried out at the UCL Wolfson Institute DNA sequencing facility. Target site determination was carried out on the other transconjugant TC1Tn6000 using primers GWEF5, GWEF7, GWEF8 and GWEF9 as described in Roberts et al.33
Serial transfer experiments
Three original transconjugants, TC7vanA, TC10vanA and TC12vanA, and the recipient strain 64/3 were subjected to serial transfers as previously described,37 with the following modifications. Briefly, 100 μL of stationary-phase culture was transferred into 9.9 mL of fresh BHI every 12 h for 30 days, corresponding to approximately 400 generations. The serial transfers were performed with one parallel for the recipient strain 64/3, two parallels for TC7vanA (one parallel having been lost due to contamination) and three parallels for TC10vanA and TC12vanA, yielding nine evolved populations in total. Growth conditions included incubation at 37°C with shaking (225 rpm). Samples of the mixed populations were stored in aliquots at −80°C for further studies. A few single colonies from each evolved vanA-plasmid-containing population were chosen based on their increased size (an indication of fitness-compensated mutants25,27) and stored. Evolved populations were designated TCPvanA EV. From each evolved population, average-sized single colonies were picked from agar plates, re-streaked and stored at −80°C. The relative fitness of these isolates was measured in head-to-head competition experiments (see below) with the plasmid-free ancestral strain E. faecium 64/3. From each evolved population, one isolate with relative fitness close to the population mean was designated TCvanA EV and used for further analyses of evolved plasmids.
Fitness measurements
The relative fitness (w) of newly acquired and evolved MGEs in E. faecium and E. faecalis (except for TC1Tn6000 and TC2Tn916; see below) was determined in pairwise competition experiments in triplicate and repeated at least twice, as previously described,37,38 with the following modifications. Preconditioned cultures were diluted 1 : 10 in prewarmed BHI broth and OD-adjusted, and a 1 : 1 ratio of each competitor (approximately 3 × 106 cells of each) was then transferred into 2.8 mL of BHI. The mixed culture was incubated for 24 h at 37°C and 225 rpm, and supported the growth of approximately 6.6 generations per competition. Initial (N0) and final (N24) densities of competing strains were measured before the onset of competition and after 24 h by selective and non-selective plating as previously described.38 From the initial and final densities, the population growth of each competitor, known as its Malthusian parameter (m),37 was determined using the equation m = ln(N24/N0). The value of w of each transconjugant was estimated as a ratio of the Malthusian parameter of the transconjugant to that of the recipient counterpart. Since no selective marker was present in the PAI, the cost of the PAI was estimated by subtracting the relative fitness of the PAI-positive TC from the relative fitness of the PAI-negative TC. Clones representative of fitness were determined by a comparison of relative fitness values in competitions between single isolates of each population and E. faecium 64/3, and assumed to be the ones with a relative fitness closest to the mean relative fitness of a given population.
Growth rates were determined during logarithmic growth for multiple transconjugants of Tn6000 and Tn916 using a SpectraMax microplate reader (Molecular Devices, LLC). The relative fitness of TC1Tn6000 and TC2Tn916 versus E. faecium 64/3 was estimated from serial passage experiments as previously described,39 with a few modifications. Briefly, TC1Tn6000 and TC2Tn916 were preconditioned and, at the onset of the experiment, each competitor was mixed in a 1 : 1 ratio (15 μL of each) in 3 mL of cultures containing DNase 1. The cultures were diluted 1 : 100 daily for 7 days, and the ratio of the two competitors was determined each day. The selection coefficient s (referring to the selective difference between the two competitors) was calculated as s = b/ln(1/d)39 where b is the slope of ln(transconjugant density/wild-type density) as a function of transfer, and d is the dilution factor (1 : 100). Wild-type fitness was set to 1.0, and the value of w for the CTn-carrying transconjugants was calculated as w = 1 + SCTn.
Statistical analysis
Statistical tests (mean values, standard deviations and one-sample and independent-sample t-tests) were performed using SPSS (v.19). A straight line was fitted independently to each competition in the serial transfer competition assay using a linear regression model in R (v. 2.15.2), with subsequent test statistics performed as described above on the calculated relative fitness values.
Results
Biological cost of newly acquired Tn916 and Tn6000
The fitness cost of multiple transconjugants of both E. faecalis OG1RF and E. faecium 64/3 containing either Tn916 and Tn6000 were evaluated as described above. All of the E. faecalis transconjugants and the vast majority of E. faecium transconjugants showed no growth difference from the recipient strain (data not shown). However, monoculture growth experiments are suboptimal for detecting smaller differences in relative fitness. To more accurately measure the fitness costs of CTn carriage, two E. faecium transconjugants (TC1Tn6000 and TC2Tn916) were selected for serial transfer pairwise competition experiments with their respective ancestor. Tn6000 was inserted in the same chromosomal locus in all of the transconjugants (see below).33 TC2Tn916 was chosen for further analysis as the most pronounced fitness effect was observed in the monoculture experiments with this transconjugant.
The long-term competition experiments revealed that acquisition of Tn916 severely reduced the relative fitness of the new host by 17% (w = 0.83 ± 0.017, P ≤ 0.001). The increased sensitivity compared with the monoculture growth experiments revealed that a newly acquired Tn6000 reduced fitness by 3% (w = 0.97 ± 0.017, P = 0.016) relative to the CTn-free E. faecium 64/3 (Figure 1). One of the six parallel TC1Tn6000 competitions against E. faecium 64/3 was removed from the analyses due to a lack of linearity (i.e. a poor fit of linear regression to the data points; data not shown). Southern blot hybridization analyses revealed the presence of single intTn gene copies in both TC1Tn6000 and TC2Tn916. Sequencing of the ssp-PCR amplicons demonstrated insertions of Tn916 in the RNA-binding S4 protein gene (ID = AFK57627.1) and in the L31 ribosomal gene for Tn6000. Circular Tn6000 and Tn916 intermediates were detected by PCR in TC1 and TC2, demonstrating that the elements remained active (data not shown). TC1Tn6000 and TC2Tn916 displayed MICs of tetracycline of 16 mg/L and 24 mg/L, respectively.
Figure 1.
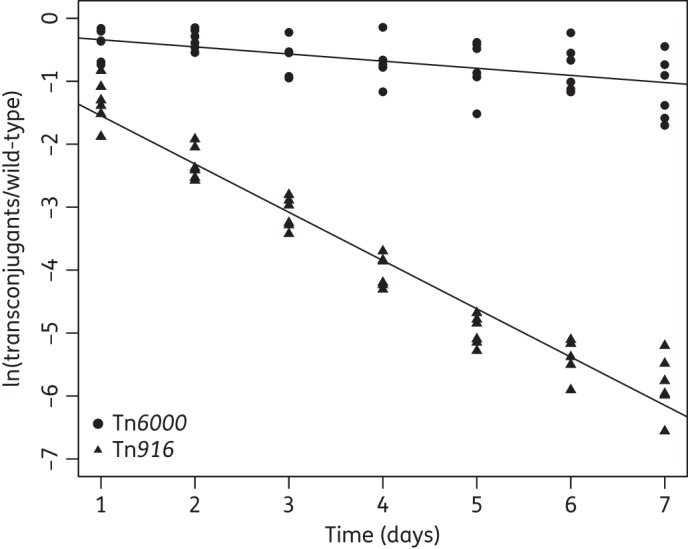
Pairwise serial competitions between the transconjugants TC1Tn6000 and TC2Tn916 and the wild-type 64/3 E. faecium. Changes in ln(transconjugants/wild-type) over time with average regression lines for both competitions displayed over their corresponding data points. The negative slope values reflect an initial fitness cost of harbouring the Tn6000 (TC1Tn6000; circles) or Tn916 (TC2Tn916; triangles) of 3% (one-sample t-test, P = 0.016) and 17% (one-sample t-test, P ≤ 0.001), respectively.
Biological cost of newly acquired PAI in E. faecium and E. faecalis
Horizontal mobility of an E. faecalis PAI encoding several virulence determinants, including enterococcal surface protein gene (esp), has recently been demonstrated.8 The E. faecalis PAI of ∼200 kb was integrated into a tRNAlys gene (E. faecium recipient strain 64/3, resulting in TC4PAI) or into a non-coding region flanked by an open reading frame (ORF) of unknown function (E. faecalis recipient strain OG1RF, resulting in TC6PAI).
Figure 2 shows the results from competition experiments between E. faecium transconjugants TC3 and TC4PAI versus recipient strain 64/3. TC3 contains pLG2 only, whereas TC4PAI contains both pLG2 and the PAI (Table 1). The fitness cost of harbouring PAI alone was estimated to be approximately 9% in E. faecium 64/3 [obtained by subtracting the relative fitness of TC3 (w=0.96) from that of TC4PAI (w=0.87)]. This should be considered to be an underestimate of PAI costs since the 4% (w = 0.96) biological cost of pLG2 carriage alone was not significantly different from 1 (P ≤ 0.3). In E. faecalis transconjugants TC5 and TC6PAI, using the same approach described above yielded no differences in relative fitness, suggesting a low cost, if any, of PAI acquisition as well as of pLG2 carriage (Figure 2).
Figure 2.
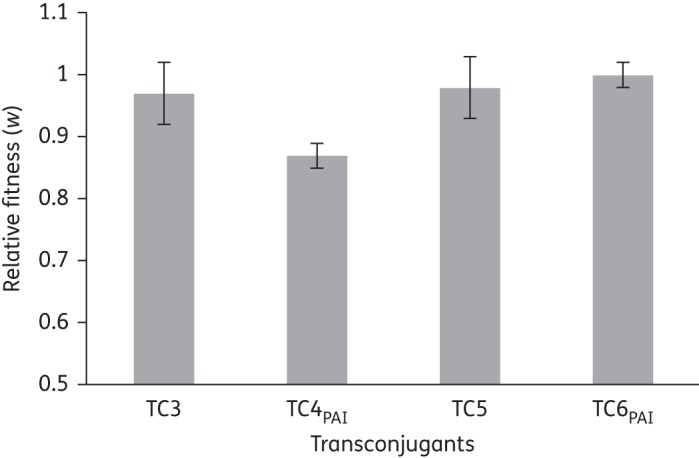
Relative fitness effects of plasmid pLG2 with or without the presence of PAI in E. faecium (TC3 and TC4PAI) and E. faecalis (TC5 and TC6PAI). The difference in fitness between TC3 and TC4PAI is statistically significant (P ≤ 0.001), consistent with an estimated 9% fitness cost of PAI carriage in E. faecium. There is no statistically significant difference in fitness between TC5 and TC6PAI (P ≤ 0.3) suggesting a low fitness cost, if any, of PAI carriage in E. faecalis.
Cost of newly acquired vanA plasmids in E. faecium 64/3
Six vanA plasmid-containing E. faecium 64/3 transconjugants from a previous report were used in this study.35 The vanA plasmid donors were vancomycin-resistant E. faecium of various origins isolated at different timepoints in Europe and the USA. The plasmids differed in size (80 to 200 kb), gene composition and class determined by rep genes.35 One transconjugant (TC7vanA) harboured an additional 45 kb plasmid.
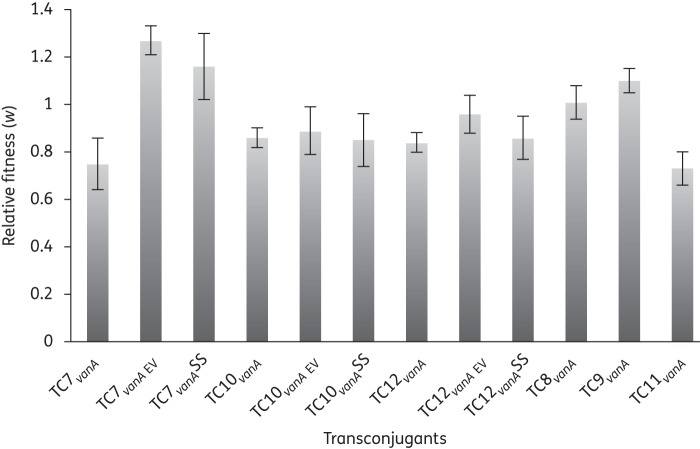
The biological cost of newly acquired vanA plasmids was estimated in mixed culture competition experiments (Table 2). The relative fitness changes in transconjugant isolates TC7vanA to TC12vanA relative to the plasmid-free otherwise isogenic E. faecium 64/3 varied from highly detrimental (w = 0.73 ± 0.07, TC11vanA) to close to selectively neutral (w = 1.01 ± 0.07, TC8vanA) and even beneficial (w = 1.10 ± 0.05, TC9vanA). In TC7vanA, the 100 kb plasmid isolated in France in 1990 initially severely reduced the fitness of the new host by 25%. The 172 kb plasmid isolated from food in Germany in 1996 initially reduced the relative fitness of its new host (TC10vanA) by 14%. Finally, in TC12vanA, the 85 kb plasmid originally isolated from Portugal in 1996 reduced host fitness by 16%.
Table 2.
Relative fitness effects of various MGEs in E. faecium or E. faecalis
| Competing strains/populations | Relative fitness mean | SD |
|---|---|---|
| TC1Tn6000 × 64/3a | 0.97 | 0.02 |
| TC2Tn916 × 64/3a | 0.83 | 0.02 |
| TC3 × 64/3 | 0.96 | 0.06 |
| TC4PAI × 64/3a | 0.87 | 0.09 |
| TC5 × OG1RF | 0.98 | 0.06 |
| TC6PAI × OG1RF | 1.01 | 0.01 |
| TC7vanA × 64/3a | 0.75 | 0.11 |
| TC8vanA × 64/3 | 1.01 | 0.07 |
| TC9vanA × 64/3 | 1.10 | 0.05 |
| TC10vanA × 64/3a | 0.86 | 0.04 |
| TC11vanA × 64/3a | 0.73 | 0.07 |
| TC12vanA × 64/3a | 0.84 | 0.05 |
| TC7-1PvanA EV × 64/3 | 2.54 | 0.60 |
| TC7-2PvanA EV × 64/3a | 1.87 | 0.32 |
| TC10-1PvanA EV × 64/3 | 2,10 | 1.10 |
| TC10-2PvanA EV × 64/3 | 2.00 | 0.89 |
| TC10-3PvanA EV × 64/3a | 1.90 | 0.69 |
| TC12-1PvanA EV × 64/3 | 1.37 | 0.32 |
| TC12-2PvanA EV × 64/3 | 1.46 | 0.42 |
| TC12-3PvanA EV × 64/3a | 1.41 | 0.32 |
| TC7-1PvanA EV × 64/3PEV | 1.20 | 0.36 |
| TC7-2PvanA EV × 64/3PEV | 1.25 | 0.22 |
| TC10-1PvanA EV × 64/3PEV | 0.91 | 0.20 |
| TC10-2PvanA EV × 64/3PEVa | 0.74 | 0.17 |
| TC10-3PvanA EV × 64/3PEV | 0.92 | 0.25 |
| TC12-1PvanA EV × 64/3PEV | 0.99 | 0.25 |
| TC12-2PvanA EV × 64/3PEV | 0.96 | 0.12 |
| TC12-3PvanA EV × 64/3PEV | 0.94 | 0.15 |
| TC7-1vanA EV × 64/3a | 2.33 | 0.13 |
| TC7-2vanA EV × 64/3a | 1.88 | 0.17 |
| TC10-1vanA EV × 64/3a | 2.08 | 0.30 |
| TC10-2vanA EV × 64/3a | 1.99 | 0.17 |
| TC10-3vanA EV × 64/3a | 1.98 | 0.18 |
| TC12-1vanA EV × 64/3a | 1.36 | 0.12 |
| TC12-2vanA EV × 64/3a | 1.46 | 0.28 |
| TC12-3vanA EV × 64/3a | 1.41 | 0.15 |
| TC7-1vanA EV × 64/3PEVa | 1.25 | 0.14 |
| TC7-2vanA EV × 64/3PEV | 1.28 | 0.25 |
| TC10-1vanA EV × 64/3PEVa | 0.92 | 0.06 |
| TC10-2vanA EV × 64/3PEVa | 0.81 | 0.09 |
| TC10-3vanA EV × 64/3PEV | 0.93 | 0.10 |
| TC12-1vanA EV × 64/3PEV | 0.99 | 0.10 |
| TC12-2vanA EV × 64/3PEV | 0.95 | 0.10 |
| TC12-3vanA EV × 64/3PEV | 0.92 | 0.12 |
| TC7vanASS × 64SSa,b | 1.16 | 0.14 |
| TC10van SS × 64SSa | 0.85 | 0.11 |
| TC12van SS × 64SSa | 0.86 | 0.09 |
aRelative fitness of the strain/population is significantly different from that of the ancestral strain (which is by default equal to 1).
bRelative fitness is significantly different from that of the original transconjugant (P ≤ 0.01).
Two transconjugant isolates (TC8vanA and TC9vanA) revealed no obvious cost of plasmid carriage (Figure 3). No apparent correlation was seen between fitness cost and size of the newly acquired plasmids, which ranged from 80 to 200 kb.
Figure 3.
Fitness effects of newly acquired and evolved vanA plasmids in E. faecium 64/3. TC7vanA, TC10vanA and TC12vanA with newly acquired vanA plasmids were competed against the plasmid-free but otherwise isogenic E. faecium 64/3. The evolved TC7vanA EV, TC10vanA EV and TC12vanA EV were competed against the evolved plasmid-free E. faecium 64/3. The initial fitness costs of plasmid carriage were reduced in TC7vanA EV (P ≤ 0.01) and TC12vanA EV (P ≤ 0.04). Bars for TC7vanASS, TC10vanASS and TC12vanASS show the fitness effects of evolved plasmids reintroduced into the ancestral E. faecium 64/3SS. A relative fitness below 1 indicates a fitness burden associated with plasmid carriage. Competitions were performed in triplicate and repeated at least twice.
Cost of evolved vanA plasmids in E. faecium 64/3 and 64SS
To test the hypothesis that the fitness cost of plasmid carriage would be mitigated during experimental evolution, three transconjugant isolates (TC7vanA, TC10vanA and TC12vanA) with plasmids conferring an initial high fitness cost were subjected to 400 generations of serial transfer in antibiotic-free BHI broth. Competition experiments between the evolved vancomycin-resistant populations of transconjugants (TCPvanA EV) and the ancestor revealed strong adaptation to the growth medium, with means ranging from w = 1.37 ± 0.32 (TC12-1PvanA EV) to w = 2.54 ± 0.63 (TC7-1PvanA EV). The evolved plasmid-containing populations were also competed with the evolved but plasmid-free E. faecium 64/3PEV population. The mean relative fitness between these populations ranged from w = 0.74 ± 0.17 (TC10-2PvanA EV) to w = 1.25 ± 0.22 (TC7-2PvanA EV). For a summary of results, see Table 2. The large variation around the mean in these experiments suggested a heterogeneous population structure and a variable occurrence of adaptive mutations.
As previously reported,40 fitness-representative isolates were obtained from each of the evolved populations (i) to more accurately enable a measurement of the fitness cost of evolved plasmid carriage through competitions with the evolved ancestral population, and (ii) to transfer specific evolved plasmids back into the ancestral genetic background in order to determine whether changes in relative fitness were due to genetic changes in the plasmids or alternatively in the chromosome. The relative fitness of the single isolates (TCvanA EV) was determined in mixed culture competitions between the plasmid-free ancestral E. faecium 64/3 and the evolved susceptible population 64/3PEV. The results presented in Table 2 show that when these fitness-representative isolates were competed against the naive E. faecium 64/3, the differences in mean relative fitness were in the same ranges as previously described with the evolved vanA plasmid-containing populations. However, the variation around the mean [expressed as the standard deviations (SD)] was strongly reduced (Table 2).
To further investigate whether the initial fitness cost of plasmid carriage was reduced, the evolved vanA-plasmid-containing isolates were competed against the evolved but plasmid-free E. faecium 64/3 (64/3PEV). For the evolved TC7vanA, the mixed culture competitions between the two evolved fitness-representative isolates and the evolved ancestral plasmid-free E. faecium 64/3 population showed that the vanA plasmid-containing strains had on average a 27% fitness advantage (P < 0.01). These data clearly suggest that a beneficial host–plasmid association had evolved during 400 generations of growth in the laboratory. To test whether these changes were plasmid specific or whether the adaptive changes resided in the evolved genome, we transferred the evolved plasmids from the fitness-representative strain (TC7-1vanA EV) back into the ancestral E. faecium 64SS strain (TC7vanASS). Against this genetic background, the plasmids (TC7vanA containing a 45 kb plasmid in addition to the 100 kb vanA plasmid) still increased the relative fitness of the host by on average 16% (w = 1.16 ± 0.14) (Figure 3).
For the three evolved TC10vanA strains, head-to-head competitions with the evolved ancestral population revealed no average changes in relative fitness after 400 generations of serial dilutions (0.89 ± 0.1, P ≤ 0.4) (Figure 3). When the evolved plasmid from the fitness-representative TC10-1vanA EV was transferred back into the ancestral E. faecium 64SS, the plasmid still reduced fitness by 15% (w = 0.85 ± 0.11).
For the three evolved TC12vanA strains, the initial fitness costs were on average almost completely mitigated following 400 generations of experimental evolution (w = 0.96 ± 0.1; P ≤ 0.04) (Figure 3). Transfer back into the ancestral background (TC12-1vanA EV to 64/3SS) demonstrated that the relative fitness of the evolved plasmid was unchanged compared with that of the unevolved plasmid (w = 0.86 ± 0.09), suggesting that chromosome-specific adaptive mutations were responsible for the reduced cost of carrying the plasmid.
All evolved and unevolved plasmids were isolated, treated with S1 nuclease and analysed on a PFGE gel. The plasmid profiles remained intact throughout the serial transfer experiments, as demonstrated in Figure 4, suggesting that major reductions in size did not account for the improvements in fitness that were observed. Moreover, the determination of MIC revealed an identical resistance profile of all descendent isolates during the experimental evolution period (data not shown). To test whether the observed differences in the fitness costs of plasmid carriage were due to altered transfer capabilities, conjugation experiments were performed before and after experimental evolution for TC7vanA and TC12vanA, in both cases with E. faecium 64SS as a common recipient. For TC7vanA and TC12vanA, the conjugation frequencies before and after serial transfer remained unchanged, at 1 × 10−4 and 3 × 10−4 transconjugants per donor, respectively. This strongly suggests that alterations in the initial fitness costs of plasmid carriage described above were not linked to modifications of the conjugation machinery.
Figure 4.
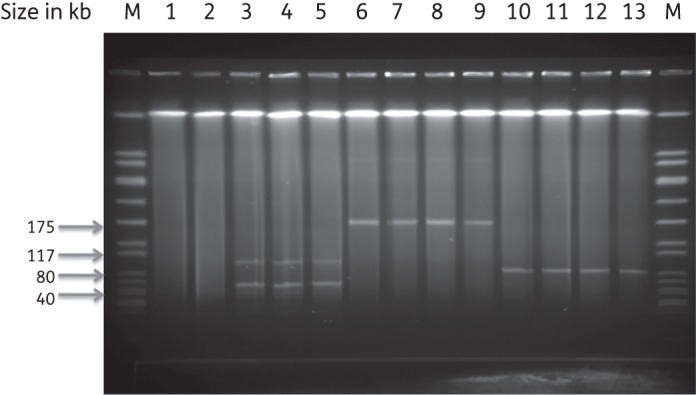
S1 nuclease-digested plasmid DNA extracted from E. faecium strains. Lane M, size marker (SmaI-digested S. aureus NCTC 8325); lane 1, 64/3; lane 2, 64SS; lane 3, TC7-1vanA EV; lane 4, TC7-2vanA EV; lane 5, TC7vanASS; lane 6, TC10-1vanA EV; lane 7, TC10-2vanA EV; lane 8, TC10-3vanA EV; lane 9, TC10vanASS; lane 10, TC12-1vanA EV; lane 11, TC12-2vanA EV; lane 12, TC12-3vanA EV; lane 13, TC12vanASS.
Discussion
In this study, we have estimated the fitness effects of newly acquired MGEs harbouring clinically relevant determinants of antimicrobial resistance and virulence in E. faecium and E. faecalis. We measured the relative fitness costs imposed by two CTns, one PAI and six different Tn1546-containing plasmids when newly transferred into common, plasmid-free strains of both E. faecium and E. faecalis. The transconjugants differed from their isogenic counterparts only by the presence of the transferred MGEs.
When transferred into the E. faecium recipient, Tn6000 reduced fitness by approximately 3%. As demonstrated here, the CTn integrated into the same target site as previously described,33 further demonstrating the site-specific integration of this element. Tn916, however, does not display the target site specificity described above. When transferred into E. faecium 64/3, it severely reduced fitness by 17% in one of the transconjugants. We determined the target site and copy number of the element in this strain and it was shown to be a single copy inserted into a ribosome-binding S4 gene. We also showed that this element was still capable of excision in the transconjugant, and therefore the mutation generated was likely to be alleviated periodically in each cell over time. This has, however, been determined to be at very low levels,41 strongly suggesting the observed fitness cost is due to disruption of this gene. It is thus feasible that the choice of target site in this transconjugant accounts for the difference in fitness between the other transconjugants, none of which contains Tn916 at this target site.
The previous conjugation experiments between E. faecalis strains in which the movement of this element was originally described yielded transconjugants with the PAI inserted into a non-coding region of the E. faecalis OG1RF genome flanked by an ORF of unknown function.8 The locus of insertion was identical to the integration site of PAI in the donor E. faecalis strain, demonstrating a high preference for site-specific integration in this species.5,8 When inserted into this particular locus, the PAI was not found in our studies to significantly reduce host fitness. The detection limit in the 24 h competition experiments is an ∼1% difference in relative fitness.37 When transferred into E. faecium, the previous analyses of the flanking sequences of the PAI demonstrated insertion into the essential tRNAlys gene.8 Our studies revealed that, when present at this locus, the PAI severely reduced fitness in E. faecium (a 9% fitness cost). Similar results were obtained with the two CTns included in this study.
Arguably, site-specificity has evolved in some MGEs with chromosomal targets to reduce the effects on host fitness. The data presented here on the PAI when it was acquired by E. faecalis, as well as Tn6000 (E. faecium), support the hypothesis of a trade-off between a low impact on host fitness (due to target site specificity) and the promiscuity of these MGEs. Moreover, the data on the effects of Tn916 integration on host fitness are consistent with a recent report on the vanB-containing CTn Tn1549.31 Using the same E. faecium 64/3 recipient as was used here, these authors demonstrated that, of 19 different integration sites, two significantly reduced host fitness. Taking this together with our data as well as observations made by Elena and Lenski,42 we conclude that the fitness effects of MGEs with reduced target site specificity depend on the integration site.
Whereas a good correlation exists between in vitro and in vivo measurements of relative fitness,25 the quantitative results presented here suggest that measurements of the biological cost of newly acquired vanA plasmids are poor predictors of prevalence in clinical settings. The TC11vanA plasmid, isolated from a urine sample in the USA in 2002, was originally present in an outbreak strain35 and caused the highest reduction in relative fitness (27%) of the host. It is, however, possible that the plasmid content in TC11vanA would impose a different fitness cost on the 2002 outbreak strain. Transconjugants TC9vanA and TC8vanA harbour large plasmids of 92 kb and 80 kb, respectively, but neither of these reduced the relative fitness of their host. The vanA plasmid in TC8vanA was originally isolated from a clinical ST18 strain in 2002 in Portugal, where it is widespread.35 It is plausible that the low biological cost, in combination with determinants of antibiotic resistance, accounts at least partly for the success of this plasmid. Large plasmids may, however, contain additional determinants that could alter the plasmid–host fitness association in various environments. Alternatively, the experimental conditions may have favoured the plasmid–strain association, and the results might have been different under more stressful conditions, as demonstrated for Escherichia coli.43 It also remains to be elucidated whether there is a correlation between (replicase-determined) plasmid classes and fitness effects since our experiments, performed with only a limited number of strains and plasmids, do not allow us to draw general conclusions.
It is also important to consider that the initially high fitness costs of MGEs can rapidly drop due to the emergence of compensatory mutations. This has been shown for numerous chromosomally encoded resistance determinants19 and for a few plasmids.20,29
We tested the hypothesis that the initial high fitness costs of vanA plasmid carriage could be ameliorated during serial transfer experiments in the laboratory. Three original transconjugants containing vanA plasmids with a size ranging from 85 kb to 175 kb were subjected to 400 generations of experimental evolution. Our data show that the initial cost of the vanA plasmid was altered in two out of three evolved transconjugants (TC7vanA EV and TC12vanA EV). Transconjugant TC7vanA contained two plasmids: a 100 kb vanA-containing plasmid as well as an additional plasmid of 45 kb. This plasmid load initially reduced the fitness of the transconjugant by 25%. After 400 generations in continuous culture, we isolated fitness-representative strains from the evolved vancomycin-resistant populations and competed them against the evolved plasmid-free, otherwise isogenic E. faecium genetic background. Surprisingly, the fitness of the plasmid-carrying strains relative to the plasmid-free evolved ancestor population had changed from detrimental to highly beneficial. The average relative fitness benefit was approximately 27%.
We then set up filter mating experiments between the evolved plasmid-containing strains and the ancestral E. faecium genetic background, differing from the original recipient strain (E. faecium 64/3) only by two additional selective markers (SS resistance). This new transconjugant received two intact plasmids, as evident from S1 nuclease treatments and PFGE analyses. In the ancestral, unevolved genetic background, the plasmids increased fitness by on average 16%. Thus, in TC7vanA, adaptive mutations are expected to have occurred both in the plasmids and in the host chromosome. The data presented clearly demonstrate that a beneficial plasmid–host association had evolved in vitro after only 400 generations.
In transconjugant TC12vanA, the uptake of a single 85 kb plasmid initially reduced host fitness by 16%. This initial reduction of relative fitness was almost completely restored to wild-type fitness levels following experimental evolution. However, re-transfer of the evolved plasmid into the ancestral background revealed that the evolved plasmid still reduced fitness by approximately 15%, suggesting the presence of plasmid-specific compensatory mutations in the host chromosome.
Our findings of rapid reductions in the initial biological costs of vanA plasmid carriage, as well as the evolved advantageous effect of plasmid carriage on host fitness, are consistent with the seminal report of Bouma and Lenski,20 in which a beneficial association evolved after 500 generations of experimental evolution between E. coli B and the plasmid-cloning vector pACYC184. Dionisio et al.44 later reported similar results, with the multiresistance plasmid R1 increasing fitness after 420 generations in both its original E. coli host and in Salmonella enterica. The data presented in this report differ from the data in those two studies20,44 in that at no point did we use antibiotic selection to maintain the plasmids during the experimental evolution. Other reports have demonstrated that the initial fitness costs of plasmids can rapidly be mitigated and/or that the biological costs of plasmids are both plasmid and host dependent.29,45–48 This report is the first to demonstrate that a beneficial plasmid–host association can evolve rapidly under simple experimental conditions in the absence of antibiotic selection, even when it was initially very costly.
In this report, we have demonstrated that newly acquired MGEs may impose an immediate biological cost in E. faecium. However, as demonstrated for vanA plasmids, the initial costs may be rapidly mitigated during serial transfer experiments. Our observations are worrying from the perspective of the reversibility of vancomycin resistance following reduced consumption levels of glycopeptide antibiotics since vancomycin is still considered a drug of last resort for the treatment of serious Gram-positive infections. If similar plasmid–host associations evolve in clinical settings, interventions to reduce consumption levels of glycopeptide antibiotics may have a limited impact on the short-term frequency of resistance. Our results also provide a possible explanation for how plasmid-mediated vanA resistance determinants have persisted at low but detectable frequencies 12 years after the ban on the animal growth promoter avoparcin in Norway and Denmark.2,49
Funding
This work was supported by the Tromsø Research Fund (grant number A5870 to P. J. J.), the University of Tromsø (awarded to P. J. J. and K. M. N.) and the Research Council of Norway (grant number 204263/F20 to P. J. J.). Initial mating experiments with vanA plasmids received funding from EU project ACE (grant no. LSHE-CT-2007-37410). This research was funded by the Commission of the European Communities, specifically the Infectious Diseases research domain of the Health theme of the 7th Framework Programme, contract 241446, ‘The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites’ (awarded to A. P. R.) and M. A.-H. is the recipient of a Marie Curie Intra European Research Fellowship; grant number 272914, BioCHARGE; Investigation into the Biological Cost and Adaptation of the Host to Antibiotic Resistance on Mobile Genetic Elements in Enterococcus Species.
Transparency declarations
None to declare.
Acknowledgements
We thank Julia Kloos, Anne Hilde Conradi, Carola Fleige and Vera Diachkova for technical assistance. G. W. would like to thank Ana Tedim and Teresa M. Coque for sharing unpublished experimental results on plasmids and fitness in Enterococcus spp. and is very grateful for the stimulating and fruitful discussions with them and Fernando Baquero on this topic in the context of EU ACE project (grant no. LSHE-CT-2007-037410).
References
- 1.Billstrom H, Lund B, Sullivan A, et al. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int J Antimicrob Agents. 2008;32:374–7. doi: 10.1016/j.ijantimicag.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 2.Johnsen PJ, Townsend JP, Bohn T, et al. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect Dis. 2009;9:357–64. doi: 10.1016/S1473-3099(09)70105-7. [DOI] [PubMed] [Google Scholar]
- 3.Willems RJ, Bonten MJ. Glycopeptide-resistant enterococci: deciphering virulence, resistance and epidemicity. Curr Opin Infect Dis. 2007;20:384–90. doi: 10.1097/QCO.0b013e32818be63d. [DOI] [PubMed] [Google Scholar]
- 4.Willems RJ, Top J, van Santen M, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–8. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegstad K, Mikalsen T, Coque TM, et al. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–54. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 6.Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev. 2011;35:856–71. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- 7.Novais C, Freitas AR, Silveira E, et al. Different genetic supports for the tet(S) gene in enterococci. Antimicrob Agents Chemother. 2012;56:6014–8. doi: 10.1128/AAC.00758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laverde Gomez JA, Hendrickx AP, Willems RJ, et al. Intra- and interspecies genomic transfer of the Enterococcus faecalis pathogenicity island. PLoS One. 2011;6:e16720. doi: 10.1371/journal.pone.0016720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uttley AH, Collins CH, Naidoo J, et al. Vancomycin-resistant enterococci. Lancet. 1988;1:57–8. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 10.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42(Suppl 1):S25–34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 11.Arthur M, Molinas C, Depardieu F, et al. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–27. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonsen GS, Myhre MR, Dahl KH, et al. Typeability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb Drug Resist. 2000;6:49–57. doi: 10.1089/mdr.2000.6.49. [DOI] [PubMed] [Google Scholar]
- 13.Willems RJ, Top J, van Schaik W, et al. Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio. 2012;3:e00151–12. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner G, Coque TM, Franz CMAP, et al. Enterococci-tales of a drug resistance gene trafficker. Int J Med Microbiol. 2013 doi: 10.1016/j.ijmm.2013.03.001. doi:10.1016/j.ijmm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhu W, Clark NC, McDougal LK, et al. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob Agents Chemother. 2008;52:452–7. doi: 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems RJ, Hanage WP, Bessen DE, et al. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasni AS, Mullany P, Hussain H, et al. Demonstration of conjugative transposon (Tn5397)-mediated horizontal gene transfer between Clostridium difficile and Enterococcus faecalis. Antimicrob Agents Chemother. 2010;54:4924–6. doi: 10.1128/AAC.00496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin BR, Lipsitch M, Perrot V, et al. The population genetics of antibiotic resistance. Clin Infect Dis. 1997;24(Suppl 1):S9–16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]
- 19.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 20.Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–2. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 21.Schrag SJ, Perrot V. Reducing antibiotic resistance. Nature. 1996;381:120–1. doi: 10.1038/381120b0. [DOI] [PubMed] [Google Scholar]
- 22.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–93. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 23.Martinez JL, Baquero F, Andersson DI. Predicting antibiotic resistance. Nat Rev Microbiol. 2007;5:958–65. doi: 10.1038/nrmicro1796. [DOI] [PubMed] [Google Scholar]
- 24.Bjorkholm B, Sjolund M, Falk PG, et al. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci USA. 2001;98:14607–12. doi: 10.1073/pnas.241517298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorkman J, Nagaev I, Berg OG, et al. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287:1479–82. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 26.Gagneux S, Long CD, Small PM, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–6. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 27.Maisnier-Patin S, Berg OG, Liljas L, et al. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol Microbiol. 2002;46:355–66. doi: 10.1046/j.1365-2958.2002.03173.x. [DOI] [PubMed] [Google Scholar]
- 28.Rozen DE, McGee L, Levin BR, et al. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007;51:412–6. doi: 10.1128/AAC.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlberg C, Chao L. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics. 2003;165:1641–9. doi: 10.1093/genetics/165.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnsen PJ, Simonsen GS, Olsvik O, et al. Stability, persistence, and evolution of plasmid-encoded VanA glycopeptide resistance in enterococci in the absence of antibiotic selection in vitro and in gnotobiotic mice. Microb Drug Resist. 2002;8:161–70. doi: 10.1089/107662902760326869. [DOI] [PubMed] [Google Scholar]
- 31.Foucault ML, Depardieu F, Courvalin P, et al. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci USA. 2010;107:16964–9. doi: 10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts AP, Hennequin C, Elmore M, et al. Development of an integrative vector for the expression of antisense RNA in Clostridium difficile. J Microbiol Methods. 2003;55:617–24. doi: 10.1016/s0167-7012(03)00200-8. [DOI] [PubMed] [Google Scholar]
- 33.Roberts AP, Davis IJ, Seville L, et al. Characterization of the ends and target site of a novel tetracycline resistance-encoding conjugative transposon from Enterococcus faecium 664.1H1. J Bacteriol. 2006;188:4356–61. doi: 10.1128/JB.00129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouwer MS, Mullany P, Roberts AP. Characterization of the conjugative transposon Tn6000 from Enterococcus casseliflavus 664.1H1 (formerly Enterococcus faecium 664.1H1) FEMS Microbiol Lett. 2010;309:71–6. doi: 10.1111/j.1574-6968.2010.02018.x. [DOI] [PubMed] [Google Scholar]
- 35.Werner G, Freitas AR, Coque TM, et al. Host range of enterococcal vanA plasmids among Gram-positive intestinal bacteria. J Antimicrob Chemother. 2011;66:273–82. doi: 10.1093/jac/dkq455. [DOI] [PubMed] [Google Scholar]
- 36.Dahl KH, Mater DD, Flores MJ, et al. Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptide selection. J Antimicrob Chemother. 2007;59:478–86. doi: 10.1093/jac/dkl530. [DOI] [PubMed] [Google Scholar]
- 37.Lenski RE, Rose MR, Simpson SC, et al. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–41. [Google Scholar]
- 38.Ray JL, Harms K, Wikmark OG, et al. Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-fold over the genome. Genetics. 2009;182:1165–81. doi: 10.1534/genetics.109.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–97. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starikova I, Harms K, Haugen P, et al. A trade-off between the fitness cost of functional integrases and long-term stability of integrons. PLoS Path. 2012;8:e1003043. doi: 10.1371/journal.ppat.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manganelli R, Romano L, Ricci S, et al. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 42.Elena SF, Lenski RE. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–8. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- 43.Petersen A, Aarestrup FM, Olsen JE. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol Lett. 2009;299:53–9. doi: 10.1111/j.1574-6968.2009.01734.x. [DOI] [PubMed] [Google Scholar]
- 44.Dionisio F, Conceicao IC, Marques AC, et al. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol Lett. 2005;1:250–2. doi: 10.1098/rsbl.2004.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enne VI, Bennett PM, Livermore DM, et al. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother. 2004;53:958–63. doi: 10.1093/jac/dkh217. [DOI] [PubMed] [Google Scholar]
- 46.Humphrey B, Thomson NR, Thomas CM, et al. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol. 2012;12:53. doi: 10.1186/1471-2180-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heuer H, Fox RE, Top EM. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol Ecol. 2007;59:738–48. doi: 10.1111/j.1574-6941.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 48.De Gelder L, Ponciano JM, Joyce P, et al. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology. 2007;153:452–63. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 49.Johnsen PJ, Townsend JP, Bohn T, et al. Retrospective evidence for a biological cost of vancomycin resistance determinants in the absence of glycopeptide selective pressures. J Antimicrob Chemother. 2011;66:608–10. doi: 10.1093/jac/dkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]