Abstract
Transcription factors (TFs) play central role in normal cellular physiology and their aberrant expression is linked to different diseases. Hepatocyte Nuclear Factors (HNFs) are TFs that have been recognized to play multiple roles in liver physiology. Emerging research has highlighted their function in the sustenance of solid tumors, indicating that HNFs could serve as possible therapeutic targets in cancer. Although, there have been many attempts to develop HNF targeted drugs, the myriad downstream targets associated with these transcription factors, some of which are critical for normal cell homeostasis, led to the realization that HNFs are not easily druggable. Therefore, identifying and optimizing drugs that can selectively inactivate HNFs is a challenge to the pharmaceutical industry. To achieve this, a more in-depth understanding is required of the HNFs binding partners, the protein interaction networks it regulates and the resulting phenotype. This calls for network analysis of the pathways regulated by HNFs and how chemical perturbations can selectively activate or suppress their functions. Network biology is an emerging field of research that is finding applications in cancer drug discovery. Specifically, network pharmacology is cementing its position in cancer research and has various applications such as biomarker identification, in determining synergistic drug pairs and in drug repurposing. Developing a network understanding of HNFs, the target it hits and responses thereof can enhance our ability to design drugs against these TFs. This article reviews how network pharmacology can help in the identification of druggable avenues in TFs and also allow the selection of drugs and their synergistic pairs against HNFs for cancer therapy.
Keywords: Hepatocyte Nuclear Factors, HNFs, HNF1, HNF3, HNF4, HNF4α, Systems Biology, Network Pharmacology, Pancreatic Ductal Adenocarcinoma, p53, HNF4α targeted drug design
1. Introduction
Hepatocyte nuclear factors (HNFs) are a group of phylogenetically unrelated transcription factors (TFs) mainly produced by the liver. HNFs have been recognized to regulate the transcription of a diverse group of genes, and thereby translating to their corresponding proteins [1, 2]. These proteins include blood clotting factors, enzymes and transporters that are directly involved in glucose, cholesterol, and fatty acid transport and metabolism [3, 4]. As the name suggests, HNFs are expressed predominately in the liver [5]. However, HNFs have also been shown to express and play important roles in a number of other tissues so that the name hepatocyte nuclear factor is somewhat a misnomer [6]. Nevertheless, the liver is the only tissue in which a significant number of different HNFs are expressed at the same time. In addition, there are a number of genes which contain multiple promoter and enhancer regions, which are regulated by a different HNFs [7]. Furthermore, efficient expression of these genes requires synergistic activation by multiple HNFs. Hence HNFs main function is to ensure liver specific expression of certain genes. As is the case with many transcription factors, HNFs regulates the expression of a wide variety of target genes, and therefore the tissue-specific functions of HNFs are highly complex. These functions (especially involving the liver) includes the development and metabolic homeostasis of the organism [8]. For example, HNFs influence the expression of insulin gene as well as genes involved in glucose transport and metabolism. In embryonic development, HNF4α is thought to have an important role in the development of the liver [9], kidney [10], and other organs [11]. Their multiple unrelated subtypes, diverse independent as well as overlapping functions, and exponential number of targets makes it difficult to fully understand their role in normal tissue homeostasis as well as their roles in diverse array of disease conditions. The level of the complexity of HNFs mechanisms calls for advanced integrative analyses of their functions using computational tools such as systems biology and network modeling. Recently, using such systems level analysis, we indentified the key role of one HNF (HNF4α) as a biomarker of therapeutic response in pancreatic cancer [12]. In this review, we will first discuss different HNFs, their role in cancer, and then present recent network advancements that have helped to gain deeper understanding of their mechanism(s) of action. We conclude that network tools can help to identify drugs that can indirectly target these pleiotropic TFs. Therefore, it is our expectation that the evidence presented in this review in support of the power of network biology will guide the rational design of future therapeutic strategies by incorporating HNFs targeted drugs in the novel design of therapeutic strategies against cancer.
2. Hepatocyte Nuclear Factors (HNFS)
This section briefly describes the different HNFs and their interacting partners, and some of their known functions.
2.1. HNF1
Members of the HNF1 subfamily contain a pituitary specific pit-1, octamer transcription factor, neural unc-86 (POU)-homeodomain and binds to DNA as homodimers. There are two main subtypes namely, HNF1α also called Transcription Factor 1 (TCF1) or Mature Onset Diabetes of the Young (MODY3) (TCF1) and HNF1β or TCF2 or MODY5 (TCF2). HNF1 homeobox A (hepatocyte nuclear factor 1 homeobox A), also known as HNF1A, is a human gene that encodes a protein highly expressed in the liver, and is involved in the regulation of the expression of several liver-specific genes [13]. The long list of HNF1 interacting partners includes P300/CBP-associated factor (PCAF), rasrelated C3 botulinum toxin substrate 3 (RAC3), CREB binding protein and Src [14]. HNF1 expression is not restricted to hepatocytes because it is also expressed in epithelial cells of several endoderm derived organs and in mesoderm derived kidney tubules as well. Their presence has been linked with liver organogenesis and hepatic differentiation [15].
2.2. HNF3
The HNF3 subfamily members also regulate different target genes and contain a winged helix DNA-binding domain that facilitates their binding to DNA as monomers. HNF3α/FOXA1 (forkhead box A1), HNF3β/FOXA2 (forkhead box A2), HNF3γ/FOXA3 [fork [16] and head box A3]. Forkhead box protein A1 (FOXA1 forkhead class of DNA-binding proteins), also known as hepatocyte nuclear factor 3-alpha (HNF-3A) in human, is a protein that is encoded by the FOXA1 gene [17]. These hepatocyte nuclear factors are transcriptional activators for liver-specific transcripts such as albumin and the serum and cerebrospinal fluid carrier of hormone transthyretin [18], and they have also been shown to interact with chromatin. The HNF3 family members in mice have well studied for their roles in the regulation of metabolism and in the differentiation of the pancreas and liver [19]. http://en.wikipedia.org/wiki/FOXA1 - cite_note-entrez-0#cite_note-entrez-0 Apart from investigations on their expression levels in different tissues, HNF3 as been well studied for their roles in different malignancies [20]. In breast cancer, it is highly correlated with estrogen receptor α positive (ERα+), Trans-acting T-cell-specific transcription factor positive (GATA3+), and progesterone receptor positive (PR+) protein expression as well as endocrine signaling [21]. Advanced genomic screening studies have shown that the presence of HNF3 in ERα+ breast cancer patients serves as an indicator of resistant to endocrine therapy [22, 23]. Mutations in HNF3 gene has been reported in prostate cancer [24].
Hepatocyte nuclear factor 3-gamma (HNF-3G), also known as forkhead box protein A3 (FOXA3) or transcription factor 3G (TCF-3G) is a human protein encoded by the FOXA3 gene. Like the HNF3, HNF-3G is a member of the forkhead class of DNA-binding proteins. These hepatocyte nuclear factors are transcriptional activators for liver-specific transcripts such as albumin and transthyretin, and they also interact with chromatin. Similar family members in mice have shown to play important roles in the regulation of metabolism and differentiation of the pancreas and liver [25]. This gene has been linked to sporadic cases of maturity onset diabetes of the young.
2.3. HNF4
HNF4 (Hepatocyte Nuclear Factor 4) is a nuclear receptor protein that is mostly expressed in the liver, gut, kidney, and pancreatic beta cells, and it is critical for liver development. In humans, there are two isoforms of HNF4, alpha and gamma encoded by two separate genes HNF4A and HNF4G, respectively [26]. HNF4 was originally classified as an orphan receptor that exhibits constitutive transactivation activity apparently by being continuously bound to a variety of fatty acids [27]. The existence of a ligand for HNF4 has not been clearly defined and is somewhat controversial, but linoleic acid (LA) has been identified as the reversible endogenous ligand of native HNF4 expressed in mouse liver [28]. The ligand binding domain of HNF4, as with other nuclear receptors, adopts a canonical alpha helical sandwich folding and interacts with multiple co-activator proteins [29-31] whereby HNF4 binds to the consensus sequence AGGTCAAAGGTCA in order to activate transcription. Mutations in the HNF4α gene have been linked to maturity onset diabetes of the young 1 (MODY1). Hepatocyte nuclear factor 4 alpha (HNF4α) also known as NR2A1 (nuclear receptor subfamily 2, group A, member 1) is a nuclear receptor encoded by the HNF4A gene in human, and HNF-4α is a nuclear transcription factor that binds DNA as a homodimer. The encoded protein controls the expression of several genes, including hepatocyte nuclear factor 1 alpha, a transcription factor which regulates the expression of several other hepatic genes. This gene plays a critical role in the development of liver, kidney and intestines. Alternative splicing of this gene results in multiple transcript variants, and thus the regulation and function of this gene is very complex. HNF4A is required for the PXR and CAR-mediated transcriptional activation of CYP3A4. In an interesting study, it was shown that the alkaloid Berberine could upregulate the expression of HNF4α [32]. These findings provided early indications that HNFs can be modulated by chemicals. Whether these modulations can be harnessed for therapeutic benefits is yet to be realized although some pre-clinical evaluations are presented in the next few sections of this review. Mutations in this gene have been shown to be associated with monogenic autosomal dominant non-insulin-dependent diabetes mellitus type II. The protein has been found to be associated with beta-catenin, CREB binding protein [33] MED1 and MED14 [34] and small heterodimer partner [35], and testicular receptor 4 [36]. A more detailed analysis of its role in disease (especially related to pancreatic cancer) and its influence in drug response are presented in subsequent paragraphs.
2.4. HNF6
The HNF6 subfamily members contain a cut-homeodomain (ONECUT) and binds to DNA as monomers such as HNF6α/OC-1/ONECUT1 (ONECUT1) and HNF6β/OC-2/ONECUT2 (ONECUT2). Its role in pancreas development was recently evaluated when the transcription factor Pdx1 (Pancreatic and duodenal homeobox 1), also known as insulin promoter factor 1, that is necessary for pancreatic development and β-cell maturation, was shown to co-express HNF6 [37]. However, further in-depth studies are required in order o fully appreciate the role of HNF6 in normal physiology and in disease conditions.
3. HNF4α in Cancer
Shortly after its discovery, the regulatory role of HNF4α was demonstrated in hepatocellular carcinomas. By using a cDNA array representing 14,000 cDNA clusters, Liang Xu and group studied the expression profiles in paired clinical hepatocellular carcinoma (HCC) samples and the distal nontumorous liver tissues from the same patients. Different liver-enriched transcription factors (LETFs), were examined. Among the LETFs, the expression level of CCAAT/enhancer-binding protein (C/EBP) α was downregulated in cancer whereas hepatocyte nuclear factor 1 (HNF-1), HNF-3β, HNF-4α, and HNF-4γ were up-regulated. Thus, the expression profiling data suggested that multiple regulatory pathways are involved in HCC especially that are related to LETFs. In another study, the expression levels of HNF4α in renal cell carcinoma were analyzed [38]. By Western blot analysis and gel retardation assay using HNF4 alpha specific antibodies, the authors showed that in most cases the amount as well as the binding activity of HNF4 was reduced in the tumor samples compared to the corresponding normal tissues. They also found a clear correlation between the HNF4α binding activity and the amount of another transcription factor (HNF1α), which is thought to be transcriptionally activated by HNF4α. Therefore, it has been speculated that disruption of the HNF4α/HNF1α pathway of kidney specific gene expression might be the important molecular mechanism of renal cell carcinogenesis.
In another study, it was shown that differentiated hepatoma cells that stably express an extensive set of adult hepatic functions, express liver-enriched transcription factors including HNF4α, while dedifferentiated cells that have lost the expression of all these hepatic functions no longer express HNF4α and HNF1 [39]. This study concluded that there is a spontaneous dissociation between the expression of these transcription factors and that of the hepatic functions. Cells presenting this phenotype, isolated from differentiated hepatoma cells, cease to accumulate all transcripts coding for hepatic functions but nevertheless maintains the expression of HNF4 and HNF1. Another study examined the expression of HNF4α on ovarian epithelial tumors with immunocytoand immunohistochemistry utilizing monoclonal antibodies that specifically recognize P1 and P2 promoter-driven HNF4α [40]. The authors showed that ovarian mucinous adenoma, mucinous tumors of borderline malignancy, as well as mucinous adenocarcinoma had HNF4α positive nuclear staining. One-third of mucinous tumors showed P1-positive staining while most had P1/P2-positive staining (93%). Nevertheless, the histological subtype of the studied tumors was not correlated with HNF4α expression. Cyto-logical examination showed that cancer cells in the ascites from ovarian mucinous adenocarcinomas were HNF4α positive; however, tumor cells in the ascites from other types of ovarian carcinomas were negative for HNF4α. These findings clearly suggest that HNF4α expression could be a useful marker for histological and cytological diagnosis of ovarian mucinous tumors.
4. Systems and Network Analysis of HNF4α Targets
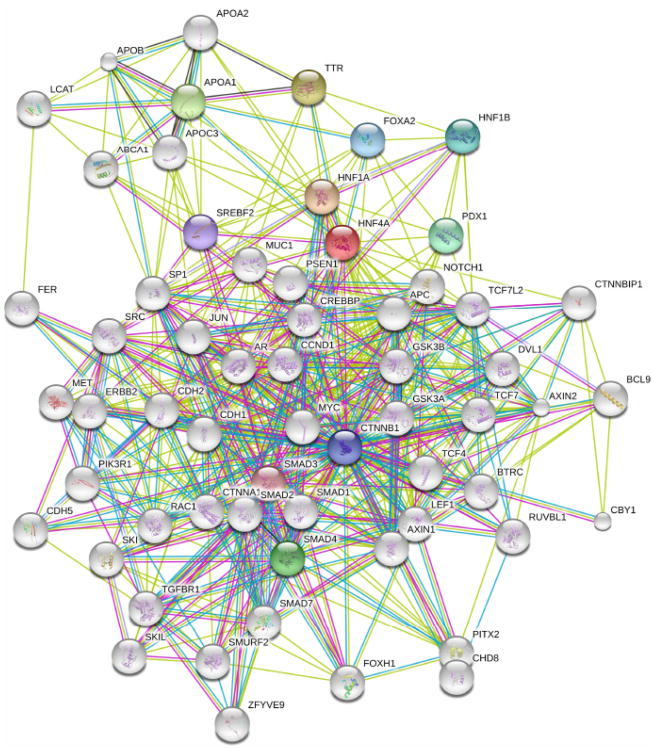
Although HNF4α has been linked to several pathological states and binds to many DNA response elements of its target genes, until recently the complete repertoire of its binding sites and target genes in the human genome was relatively unknown Fig. (1) showing HNF4α network proteins/interacting partners). In a very important study, Bolotin and colleagues utilized protein binding microarrays (PBMs) to examine the DNA-binding characteristics of two HNF4α in two different species [41]. Additionally, they also investigated the binding sites of HNF4α isoforms (HNF4α2 and HNF4α8). Through these high-throughput analysis systems, they identified approximately 1400 new binding sequences. These findings also revealed approximately 240 novel direct HNF4α human target genes, including new functional categories of genes that were not believed to be typically associated with HNF4α (such as gene for cell cycle, immune function, apoptosis, and other cancer-related genes).
Fig. 1. HNF4α Protein Network.
The protein network was built using String 9.0.
The sequencing of the human genome has led to the use of genome-wide approaches that are high throughput for target gene identification such as genome-wide location analysis (ChIP-chip/seq) and expression profiling. Another less well known but highly complementary technique is protein binding microarrays (PBMs) as stated above. PBMs are a high throughput in vitro DNA binding assay that allows for the identification of thousands of distinct DNA binding sequences in a given experiment. Sladek and group applied such high throughput technologies to characterize the DNA binding specificity of native, full length HNF4α in crude nuclear extracts [42]. This led to the identification of > 1400 new binding sequences for HNF4α which was consistent with previous findings discussed above. In our recent analysis, we also searched for the regulatory regions of human genes to identify potential new targets of HNF4α. The PBM data were also used to train a Support Vector Machine (SVM) algorithm to predict additional HNF4α binding sites with high accuracy. Finally, cross referencing with expression profiling data from an HNF4α RNAi knockdown in a human liver cancer cell line (HepG2) and published HNF4α ChIP-chip data identified >240 new direct functional targets of HNF4α. In summary, there are at least 50 CYP, 7 FMO, 21 GST, 13 SULT and 19 UGT (~110 total) human genes that are involved in drug metabolism, are predicted or proven targets of HNF4α. Development of prediction software's such as String 9.0, Ingenuity Pathway Analysis (IPA) and KEGG have facilitated the understanding of different partners of HNFs.
5. Challenges in Developing HNF4α Targeted Drugs: need for Computational Methods
While the function of the endogenous HNF4α ligand remains uncertain, what is clear is that HNF4α contains a ligand binding pocket that is occupied in a reversible fashion dependent upon the feeding state of the host. Hence, it was earlier proposed to be druggable proteins using strategies such as small molecule drugs that could bind to HNF4α and alter its ability to activate transcription of different targets [43]. However, since HNF4α regulates so many different targets, some of which are crucial to normal cell homeostasis, there is a high likelihood of toxic side effects, unless one can develop a drug that is specific to a given HNF4α target. However, this is neither a new problem nor it is unique to HNF4α. As with other nuclear receptors and transcription factors with multiple targets such as NF-κB and p53, minimizing the off target toxicities becomes a daunting task. The major challenge is to develop drugs that are specific to HNF4α regulating one specific target gene or at most only a few genes. This specificity could be found in the DNA sequence of the response elements that we now known although such sequences can vary greatly between different target genes [44].
Researchers have applied both basic and computational methods to identify novel genes regulated by HNF4α. For example, transient transfection of HNF4α into a human hepatoma cell line, a rat insulinoma cell line, and a human kidney cell line [45] has been reported. Additionally, findings with conditional knock-outs of HNF4α were also reported [46]. Notably, in the study of Odom et al. the genome-wide identification of binding sites for HNF4α, HNF1α, and HNF6 has been reported by using the ChIP-chip assay with a 13,000 human promoter sequence containing microarray [47]. In the case of HNF4α, the number of contacted promoters was unexpectedly high; 1,575 potential HNF4α target genes were identified. In addition, 42% of the genes occupied by RNA polymerase II were also occupied by HNF4α, suggesting that nearly 50% of all liver-expressed genes are regulated by HNF4α alone. Similarly, in another recent ChIP-chip experiment of ENCODE (Encyclopedia of DNA Elements) genomic regions (about 1% of the human genome), 663 novel HNF4α binding sites were identified in 100 genes, which suggests that there are a large number of HNF4α targets (over 60,000 sites in the vicinity of about 10,000 genes) if extrapolated to the entire genome [48]. This unprecedented high number of HNF4α binding sites revealed by the ChIP-chip method raises the question on the functional role of all these sites in the regulation of gene transcription, which has yet to be resolved.
Even though the ChIP-chip assay serves as a highly advanced method for the genome-wide search and identification of transcription factor binding sites (TFBSs), nonetheless, it suffers from unacceptably high false positive rates. In the study of Odom et al described above, 252 (16%) false positive binding sites were predicted. Another major drawback with this method is that only a small fraction of identified ChIP fragments possesses the canonical binding motif for the corresponding TF [49]. These limitations must be overcome, and thus it is highly desirable to identify functional binding sites relevant for the regulation of gene transcription. Furthermore, in existing studies, there is often no rationale for the selection of promoters spotted on the array; for example, no bioinformatics approach has been applied to identify relevant sequences for the design of the ChIP-chip assay. To address this problem computational approach based on a novel machine learning technique, which enabled the identification of genome-wide TFBSs were applied recently [50]. This method was applied to search for HNF4α gene targets. A genetic algorithm and an exhaustive feature selection algorithm were trained on 73 known and well characterized HNF4α target sequences in the promoters and enhancers of different mammalian genes Fig. (2). By genome-wide scanning of all human gene promoters, the authors identified novel genes targeted by HNF4α. Then, a subset of predicted binding sites was confirmed by electrophoretic mobility shift assay (EMSA). They also interrogated promoter sequences for HNF4α binding sites identified by the ChIP-chip assay. Expression of genes targeted by HNF4α was further analyzed and a good correlation between computationally annotated HNF4α binding sites and the expression of targeted genes was observed. Notably, ChIP-chip experiments tend to report a rather high number of TFBSs in promoters of genes whose regulation by HNF4α was not subsequently observed, whereas their computational method for the prediction of HNF4α regulatory sites enabled them to improve the specificity with this method encompassing rules for the regulation of gene expression.
Fig. 2. Computational Analysis of HNF4α binding sequences.
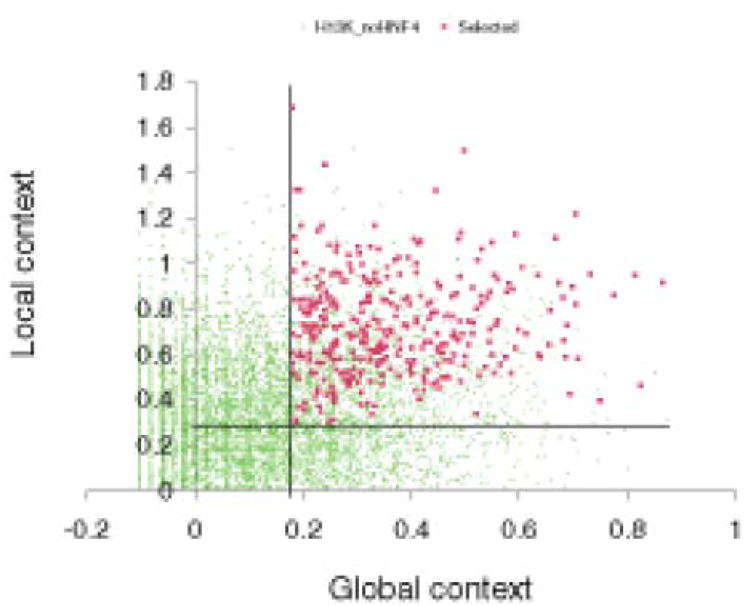
Plot of the distribution of global and local contexts in the 375 sequences (red squares) selected from the ‘positive’ set of ChIP-chip results reported by Odom et al. versus all 10,852 sequences from the ‘negative’ (not binding; set (green dots) reported for the same experiment. The selected sequences are characterized by the highest global and local context scores whereas the majority of the ‘negative’ sequences are characterized by low values for these two scores. The vertical and horizontal lines show two thresholds chosen for the global context score (0.28) and the local context score (0.18). Figure is adopted from article Kel et al. Genome Biology 2008 9: R36 doi:10.1186/gb-2008-9-2-r36. Genome Biology is an open access journal and grants unrestricted permission to re-distribution and re-publication of the work.
Overall, it was demonstrated that computational approaches are necessary in identifying novel genes targeted by HNF4α. Their machine learning technique significantly improved the overall recognition and, therefore, the identification of faithful HNF4α targets. This method enabled refinement of TF site predictions based on the ChIP-chip assay and identification from among them of potentially functional sites, as reported here. Furthermore, such computational method can easily be applied to the genome-wide identification of genes targeted by not only HNFα, but also any mammalian TF and it is not limited to promoter sequences alone, resulting in an overall success of approximately 80% based on experimental confirmation and validation.
6. Targeting the HNF4α Network
As mentioned above that due to the complexity arising from HNF4α interactions with multiple binding partners, regulated inhibition of this important protein in cancer becomes next to impossible. Nevertheless, newer computational tools have emerged that can help in conquering targets as un-druggable as HNF4α. Computational tools have been applied to target, if not the protein itself directly, and its aberrant network in pathological states has also been examined. Concepts such as systems biology and network modeling are routinely being utilized to develop drugs that hit the most relevant up- or downstream network of a difficult target (non-druggable target) for achieving treatment success.
Lately, important concepts such as network pharmacology, have emerged that are helping in the development of network targeted drugs. According to network pharmacology theory, modulating multiple nodes simultaneously, is often required for modifying phenotypes [51]. The concept includes network topological properties, such as network centrality, clustering coefficient; network functional properties, such as lethality/essentiality; and dynamical properties, such as entropy, fractal, robustness and complexity. Some of the properties which differentiate a protein as a candidate drug target are the ability to bind to small molecules, overlap with disease [52], connectivity (the number of other proteins with which it interacts) and between-ness (shortest path between two networks). These properties make it important to study them from the network biology perspective. As discussed below, we investigated this concept using Ingenuity Pathways Analysis, and assessed the changes in HNF4a protein network in response to a highly potent small molecule inhibitor, and together with combination strategy using conventional therapeutics.
7. Network Biology Reveals HNF4α as A Biomarker of Therapeutic Response in Pancreatic Cancer
Our laboratory previously investigated a potent combination involving MDM2-p53 interaction inhibitor (MDM2i-MI-219) and platinum based chemotherapy regimen in drug-resistant pancreatic tumor models [53]. This combination showed high anti-tumor efficacy in pancreatic tumor xenografts. We undertook a systems and network approach in order to thoroughly examine the underlying principles behind such unusual synergistic efficacy. Our investigations revealed that the MDM2-p53 inhibitor and chemotherapy combination efficacy was resulting from the biological synergy between overlapping networks (both MDM2 and p53) that drove enhanced p53 driven apoptosis. Most striking was the observation that this synergy was also replicated in other solid tumors as well.
In addition, we observed that the biological synergy was concomitant with consistent down-regulation of HNF circuitry. Pathway network modeling on MDM2i MI-219-oxaliplatin treated Capan-2 pancreatic cancer cells not only demonstrated biological synergy between the two drugs involved an interplay of many secondary neighboring networks augmenting p53 re-activation mediated events, and also strengthened the role of HNF4α in our experimental system. We found a dramatic down-regulation of HNF4α expression along with its target genes. These target sets were distinct but directly linked to p53 and MDM2 [please see Fig. (5) of our published article [54] for details if one is interested]. The identification of HNF4α as a key player was certainly very interesting because the role of HNFα in pancreas cancer was not previously well defined [Capan-2 (wt-p53)].
HNF4α is kown to interact with the p53 positive regulator CREBBP [55] and also confirmed its role in augmenting apoptotic effects in this synergic combination. Therefore, our findings not only provided the connection between HNF4α and p53-MDM2 loop, these findings also informed us on the possible druggable avenue that can be exploited in future drug discovery strategies. These observations are consistent with previous investigations where it was shown that p53 down-regulates HNF4α expression [56]. In the earlier study, the authors proposed that in view of the role of p53 in inhibiting different nuclear receptors, and also that over re-expressed p53 correlates with poor differentiation, then wild-type p53 should also affect the function of HNF4α. They elegantly showed that HNF4α-mediated transactivation was repressed by p53 but the mechanism of repression was not due to the inhibition of HNF4α DNA binding, which calls for further in-depth investigations. The authors also found that p53 in human embryonic kidney whole-cell extracts preferentially bound to the ligand-binding domain of HNF4α, and that the activation function 2 region was required for this binding. These results suggest that p53, like other transcriptional repressors, inhibits transcription by multiple mechanisms, one of which involves the interaction between the ligand-binding domain of HNF4α and the recruitment of histone deacetylase activity. Therefore, we anticipate that HNF4α can serve as a therapeutic response marker for assessing the p53 re-activating clinical strategy in the future.
8. Conclusions and Future Directions
HNFs are a set of transcription factors that were discovered decades ago and were recognized for their diverse role in tissue development. It was only recently, scientists made the observations linking HNFs with the maintenance of prostate cancer, hepatoma and also its role in the development of early stage pancreatic cancer. These putative causal links have led to the proposal that HNFs could serve as possible therapeutic targets in cancer. There has been a new drive to develop HNF target drugs and numerous pharmaceutical companies have investigated candidate small molecules that indirectly target HNFs. Nevertheless, due in part, to the myriad targets of these transcription factors, some of which are critical for normal cell function, a switch-on and switch-off strategy may not be successful. The use of a highly specific small molecule drug that completely blocks HNFs function may not be a feasible option, and thus strategies that will allow cancer selective inhibition of HNFs is expected to be successful. Small molecule library screening for potent compounds against HNFs cannot address this problem as discussed in this review, and thus we anticipate that network pharmacological approaches are needed for selective targeting of HNFs. Using such combinatorial network pharmacology strategies, our group had already shown that a combination of MDM2-p53 inhibitor and platinum compound could efficiently suppress HNF4α and it target genes, resulting in enhanced cell killing in highly resistant pancreatic cancer cell line models. These studies prove that successful strategies against HNFs can be achieved through in-direct targeting of these important TFs by hitting associated nodes in their target network. In future, we expect that using systems and network principles, other pre-clinical and/or approved drugs can be re-purposed and developed against HNFs. Further investigations that focus on developing newer combinatorial strategies targeting of HNFs will help in the treatment of different malignancies especially pancreatic cancer for which newer treatments are urgently needed.
Acknowledgments
NIH Grant to Dr. RM Mohammad 5R01CA10939 05 is acknowledged.
Footnotes
The authors confirm that this article content has no conflicts of interest.
References
- 1.Costa RH, Kalinichenko VV, Holterman AX, et al. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–47. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 2.Rehman J, Traktuev D, Li J, et al. Secretion of angiogenic and antiapoptotic factors by human adipose stromal cells. Circulation. 2004;109:1292–8. doi: 10.1161/01.CIR.0000121425.42966.F1. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell SM, Gloyn AL, Owen KR, et al. The role of the HNF4alpha enhancer in type 2 diabetes. Mol Genet Metab. 2002;76:148–51. doi: 10.1016/S1096-7192(02)00027-6. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell SM, Frayling TM. The role of transcription factors in maturity-onset diabetes of the young. Mol Genet Metab. 2002;77:35–43. doi: 10.1016/s1096-7192(02)00150-6. [DOI] [PubMed] [Google Scholar]
- 5.Kuo CJ, Conley PB, Hsieh CL, et al. Molecular cloning, functional expression, and chromosomal localization of mouse hepatocyte nuclear factor 1. Proc Natl Acad Sci USA. 1990;87:9838–42. doi: 10.1073/pnas.87.24.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brimo F, Herawi M, Sharma R, et al. Hepatocyte nuclear factor-1beta expression in clear cell adenocarcinomas of the bladder and urethra: diagnostic utility and implications for histogenesis. Hum Pathol. 2011;42:1613–9. doi: 10.1016/j.humpath.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RK, Vatamaniuk MZ, Lee CS, et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–15. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RK, Gao N, Gorski RK, et al. Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4alpha. Genes Dev. 2007;21:756–69. doi: 10.1101/gad.1535507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu X, Lam E, Doughman YQ, et al. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 2007;26:4445–56. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanazawa T, Konno A, Hashimoto Y, et al. Hepatocyte nuclear factor 4 alpha is related to survival of the condensed mesenchyme in the developing mouse kidney. Dev Dyn. 2010;239:1145–54. doi: 10.1002/dvdy.22276. [DOI] [PubMed] [Google Scholar]
- 11.Kanazawa T, Konno A, Hashimoto Y, et al. Expression of hepato-cyte nuclear factor 4alpha in developing mice. Anat Histol Embryol. 2009;38:34–41. doi: 10.1111/j.1439-0264.2008.00889.x. [DOI] [PubMed] [Google Scholar]
- 12.Azmi AS, Wang Z, Philip PA, et al. Proof of concept: network and systems biology approaches aid in the discovery of potent antican-cer drug combinations. Mol Cancer Ther. 2010;9:3137–44. doi: 10.1158/1535-7163.MCT-10-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtois G, Morgan JG, Campbell LA, et al. Interaction of a liver-specific nuclear factor with the fibrinogen and alpha 1-antitrypsin promoters. Science. 1987;238:688–92. doi: 10.1126/science.3499668. [DOI] [PubMed] [Google Scholar]
- 14.Soutoglou E, Papafotiou G, Katrakili N, et al. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J Biol Chem. 2000;275:12515–20. doi: 10.1074/jbc.275.17.12515. [DOI] [PubMed] [Google Scholar]
- 15.Tronche F, Yaniv M. HNF1, a homeoprotein member of the hepatic transcription regulatory network. Bioessays. 1992;14:579–87. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- 16.Wu KL, Gannon M, Peshavaria M, et al. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–13. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bingle CD, Gowan S. Molecular cloning of the forkhead transcription factor HNF-3 alpha from a human pulmonary adenocarcinoma cell line. Biochim Biophys Acta. 1996;1307:17–20. doi: 10.1016/0167-4781(96)00058-9. [DOI] [PubMed] [Google Scholar]
- 18.Costa RH, Lai E, Darnell JE., Jr Transcriptional control of the mouse prealbumin (transthyretin) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol. 1986;6:4697–708. doi: 10.1128/mcb.6.12.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu KL, Gannon M, Peshavaria M, et al. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–13. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin L, Miller CT, Contreras JI, et al. The hepatocyte nuclear factor 3alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 isamplified and overexpressed in esophageal and lung adenocarci-nomas. Cancer Res. 2002;62:5273–9. [PubMed] [Google Scholar]
- 21.Mehta RJ, Jain RK, Leung S, et al. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res Treat. 2012;131:881–90. doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- 22.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha G, Roth A, Lai D, et al. Integrative analysis of genome-wide loss of heterozygosity and mono-allelic expression at nucleotide resolution reveals disrupted pathways in triple negative breast cancer. Genome Res. 2012 doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HJ, Hwang M, Chattopadhyay S, et al. Hepatocyte nuclear factor-3 alpha (HNF-3alpha) negatively regulates androgen receptor transactivation in prostate cancer cells. Biochem Biophys Res Commun. 2008;367:481–6. doi: 10.1016/j.bbrc.2007.12.162. [DOI] [PubMed] [Google Scholar]
- 25.Wu KL, Gannon M, Peshavaria M, et al. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–13. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chartier FL, Bossu JP, Laudet V, et al. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147:269–72. doi: 10.1016/0378-1119(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 27.Jump DB. Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care. 2011;14:115–20. doi: 10.1097/MCO.0b013e328342991c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan X, Ta TC, Lin M, et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisely GB, Miller AB, Davis RG, et al. Hepatocyte nuclear factor 4is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–34. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 30.Dhe-Paganon S, Duda K, Iwamoto M, et al. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–6. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 31.Duda K, Chi YI, Shoelson SE. Structural basis for HNF-4alpha activation by ligand and coactivator binding. J Biol Chem. 2004;279:23311–6. doi: 10.1074/jbc.M400864200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Xiao X, Feng K, et al. Berberine Moderates Glucose and Lipid Metabolism through Multipathway Mechanism. Evid Based Complement Alternat Med. 2011;2011 doi: 10.1155/2011/924851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–80. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 34.Malik S, Wallberg AE, Kang YK, et al. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 2002;22:5626–37. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimamoto Y, Ishida J, Yamagata K, et al. Inhibitory effect of the small heterodimer partner on hepatocyte nuclear factor-4 mediates bile acid-induced repression of the human angiotensinogen gene. J Biol Chem. 2004;279:7770–6. doi: 10.1074/jbc.M310577200. [DOI] [PubMed] [Google Scholar]
- 36.Lin WJ, Li J, Lee YF, et al. Suppression of hepatitis B virus core promoter by the nuclear orphan receptor TR4. J Biol Chem. 2003;278:9353–60. doi: 10.1074/jbc.M205944200. [DOI] [PubMed] [Google Scholar]
- 37.Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 2003;258:105–16. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 38.Sel S, Ebert T, Ryffel GU, et al. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett. 1996;101:205–10. doi: 10.1016/0304-3835(96)04136-5. [DOI] [PubMed] [Google Scholar]
- 39.Chaya D, Fougere-Deschatrette C, Weiss MC. Liver-enriched transcription factors uncoupled from expression of hepatic functions in hepatoma cell lines. Mol Cell Biol. 1997;17:6311–20. doi: 10.1128/mcb.17.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugai M, Umezu H, Yamamoto T, et al. Expression of hepatocyte nuclear factor 4 alpha in primary ovarian mucinous tumors. Pathol Int. 2008;58:681–6. doi: 10.1111/j.1440-1827.2008.02293.x. [DOI] [PubMed] [Google Scholar]
- 41.Bolotin E, Liao H, Ta TC, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–53. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang B, Mane-Padros D, Bolotin E, et al. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res. 2012;40:5343–56. doi: 10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang-Verslues WW, Sladek FM. HNF4alpha--role in drug metabolism and potential drug target? Curr Opin Pharmacol. 2010;10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulyk ML. Analysis of sequence specificities of DNA-binding proteins with protein binding microarrays. Methods Enzymol. 2006;410:279–99. doi: 10.1016/S0076-6879(06)10013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas H, Senkel S, Erdmann S, et al. Pattern of genes influenced by conditional expression of the transcription factors HNF6, HNF4alpha and HNF1beta in a pancreatic beta-cell line. Nucleic Acids Res. 2004;32:e150. doi: 10.1093/nar/gnh144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battle MA, Konopka G, Parviz F, et al. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA. 2006;103:8419–24. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rada-Iglesias A, Wallerman O, Koch C, et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic mi-croarrays. Hum Mol Genet. 2005;14:3435–47. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 49.Jin VX, Rabinovich A, Squazzo SL, et al. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data--a case study using E2F1. Genome Res. 2006;16:1585–95. doi: 10.1101/gr.5520206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kel AE, Niehof M, Matys V, et al. Genome wide prediction of HNF4alpha functional binding sites by the use of local and global sequence context. Genome Biol. 2008;9:R36. doi: 10.1186/gb-2008-9-2-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–1. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 52.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 53.Azmi AS, Aboukameel A, Banerjee S, et al. MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function. Eur J Cancer. 2010;46:1122–31. doi: 10.1016/j.ejca.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azmi AS, Wang Z, Philip PA, et al. Proof of concept: network and systems biology approaches aid in the discovery of potent antican-cer drug combinations. Mol Cancer Ther. 2010;9:3137–44. doi: 10.1158/1535-7163.MCT-10-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida E, Aratani S, Itou H, et al. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Com-mun. 1997;241:664–9. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 56.Maeda Y, Seidel SD, Wei G, et al. Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol. 2002;16:402–10. doi: 10.1210/mend.16.2.0769. [DOI] [PubMed] [Google Scholar]