Abstract
A common single-nucleotide polymorphism in the human brain-derived neurotrophic factor (BDNF) gene results in a Val66Met substitution in the BDNF prodomain region. This single-nucleotide polymorphism is associated with alterations in memory and with enhanced risk to develop depression and anxiety disorders in humans. Here we show that the isolated BDNF prodomain is detected in the hippocampus and that it can be secreted from neurons in an activity-dependent manner. Using nuclear magnetic resonance spectroscopy and circular dichroism we find that the prodomain is intrinsically disordered, and the Val66Met substitution induces structural changes. Surprisingly, application of Met66 (but not Val66) BDNF prodomain induces acute growth cone retraction and a decrease in Rac activity in hippocampal neurons. Expression of p75NTR and differential engagement of the Met66 prodomain to the SorCS2 receptor are required for this effect. These results identify the Met66 prodomain as a new active ligand which modulates neuronal morphology.
A common single-nucleotide polymorphism (SNP) in the human brain-derived neurotrophic factor (BDNF) gene is highly associated with abnormalities in episodic memory, a reduction in hippocampal volume, and enhanced risk for depression and anxiety disorders in humans1-8. This SNP (rs6265) is observed in more than 25% of the human population (Database of Single Nucleotide Polymorphisms-National Center for Biotechnology Information, National Library of Medicine, 2012), and it leads to a nucleotide change from a guanine to an adenine at position 196 (G196A) that results in a valine (Val) to methionine (Met) substitution at codon 66 (Val66Met).
BDNF is translated as a precursor protein (proBDNF) consisting of an N-terminal prodomain and a C-terminal mature domain (mBDNF). The Val66Met substitution site is within the prodomain (Supplementary Fig. S1). ProBDNF can be cleaved by furin or proconvertases in the trans-Golgi network or secretory vesicles respectively9. Both mBDNF and proBDNF can be secreted from neurons10,11 in an activity-dependent manner after KCl-induced depolarization10 or electrical stimulation11. In addition, proBDNF can be cleaved extracellularly by plasmin generated by tissue plasminogen activator (tPA) or by selective matrix metalloproteinases (MMPs) including MMP3, MMP712 and MMP913 to release mBDNF. The fate of the isolated BDNF prodomain after proBDNF intracellular or extracellular proteolysis is not known. mBDNF binds the tropomyosin-related kinase B (TrkB) receptor to promote neuronal survival, differentiation, neurogenesis and synaptic plasticity. Modest changes in mBDNF levels in rodents result in alterations in hippocampal function and behavioral abnormalities. In contrast, uncleaved proBDNF promotes apoptosis by binding to a receptor complex of the p75 neurotrophin receptor (p75NTR) and sortilin (a Vps10p-domain sorting receptor family member)14. Interestingly, recent reports have shown that proBDNF15 as well as unprocessed pro-nerve growth factor (proNGF)16 induce growth cone retraction in cultured neurons.
The BDNF prodomain is highly conserved with a valine at or near position 66 in more than 70 species examined to date (Supplementary Fig. S1). The prodomain mediates an interaction with sortilin17, which acts as a chaperone to direct intracellular trafficking of proBDNF to the regulated secretory pathway. The Met66 proBDNF variant exhibits decreased binding to sortilin, altered intracellular trafficking, and a reduction in the activity-dependent secretion of mBDNF2,6,17,18. Moreover, a knock-in mouse that expresses Met66 BDNF recapitulates many specific phenotypic properties of the human polymorphism6. Thus, the proposed mechanism by which Met66 BDNF mediates central nervous system effects is indirect, by a reduction in activity-dependent release of mBDNF leading to altered synaptic plasticity. However, the high sequence conservation of the BDNF prodomain (Supplementary Fig. S1), and the recent evolutionarily emergence of the Val66Met polymorphism (only present in humans) prompted us to ask whether the isolated prodomain might function as an independent ligand. We have found that the prodomain is present at high levels in vivo and is secreted from neurons in an activity-dependent manner. We further considered whether the Val66 and Met66 prodomains exhibit different structures to elicit distinct biological activities.
Here we show that structural changes induced by Val66Met substitution confer bioactivity to the prodomain since only the Met66 prodomain acutely alters neuronal morphology. Finally, we demonstrate that the prodomain exerts this effect through differential interaction with SorCS2, the sortilin-related Vps10p-domain sorting receptor 2. Our findings suggest that the Met66 prodomain activity is an additional mechanism that may contribute to the increased incidence of depression and anxiety disorders found in humans with the SNP.
Results
BDNF prodomain is expressed in the hippocampus and secreted
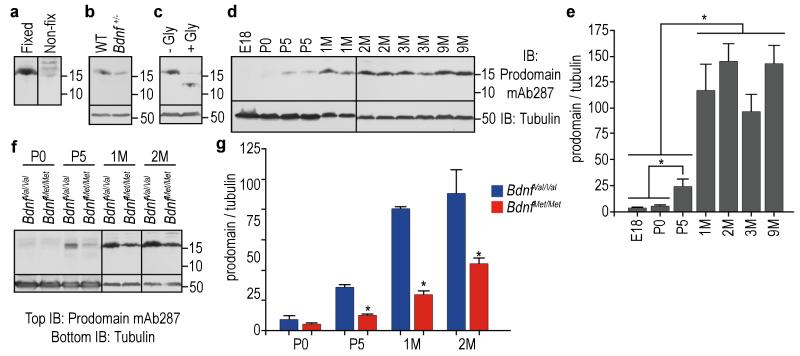
The hippocampus is a key regulator of learning and memory and dysfunction of its neural circuitry underlies the development of memory impairment, depression and anxiety disorders19. To determine if the BDNF prodomain may affect the structure of hippocampal neurons, we first examined whether the prodomain was present in this brain region. Detection of the isolated prodomain in vitro and in vivo has been technically challenging due to low antibody sensitivity. However, glutaraldehyde fixation of proteins to the transfer membranes following sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the use of a BDNF prodomain specific monoclonal antibody previously characterized10, facilitated its detection in the mouse hippocampus as a 15.5 kDa band (Fig. 1a). In order to demonstrate specificity of detection, we observed that the prodomain levels are reduced by half in Bdnf heterozygous (Bdnf+/−) mouse hippocampal lysates compared to wild type (WT) littermates (Fig. 1b). As the prodomain contains an N-linked glycosylation site (Supplementary Fig. S1), we performed enzymatic removal of the N-linked glycosylation, which resulted in migration of the prodomain at 12.3 kDa, consistent with the predicted molecular weight by its amino acid sequence (Fig. 1c).
Figure 1.
The BDNF prodomain is detected at high levels in vivo. (a) Detection of BDNF prodomain (15.5 kDa.) from mice hippocampus after glutaraldehyde fixation of the transfer membrane following SDS-PAGE. (b) The prodomain levels were reduced by half in Bdnf+/− mice hippocampal lysates, compared to wild type animals (WT). (c) Treatment of hippocampal lysates with N-glycanase (+Gly) reduced the molecular weight of the prodomain to 12.3 kDa. (d) BDNF prodomain levels at embryonic day 18 (E18), postnatal days 0 and 5 (P0 and P5), and 1, 2, 3 or 9 months (M) in hippocampal lystes of C57BL/6 mice. (e) Quantification of (d). Bars represent mean ± s.e.m. of prodomain densitometry data normalized to β tubulin. n=3 per group. (f) Comparison of BDNF prodomain levels from hippocampi of BdnfVal/Val and BdnfMet/Met mice at P0, P5, 1 and 2 month of age. (g) Quantification of (f). Bars represent mean ± s.e.m. of prodomain densitometry data normalized to β tubulin. n=4 per group. Statistical comparisons were made by one way analysis of variance test. * p < 0.05.
During late embryonic and early postnatal murine development, the expression of the prodomain in the hippocampus of C57BL/6 mice was negligible (Fig. 1d, e). However, the prodomain was detected at postnatal day 5 (P5), its expression increased significantly at 1 month and plateaued in adult mice (3-9 months) (Fig. 1d, e). The prodomain was detectable in the hippocampi of both wild type BdnfVal/Val and BdnfMet/Met knock-in animals6. However, the levels of expression significantly differ, as BdnfMet/Met mice displayed lower levels of expression as compared to BdnfVal/Val in all the time points studied (Fig. 1f, g). Other commercially available antibodies to the prodomain of BDNF (ANT-006, Alomone, Jerusalem, Israel) failed to detect the endogenous prodomain in isolation and showed very low sensitivity in detecting the recombinant prodomains (Supplementary Fig. S2).
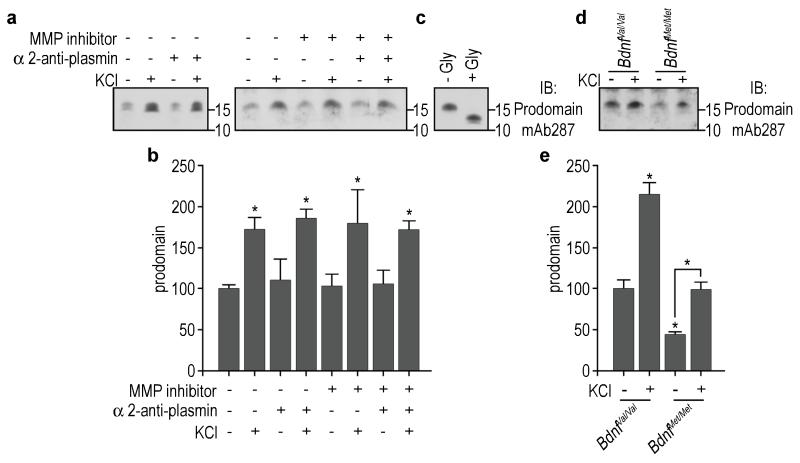
To determine if the prodomain was secreted, we cultured rat hippocampal neurons in conditions to reduce glia contamination and collected the conditioned media of day 6 in vitro (DIV6) cultures. The endogenous prodomain was detected in the media, and prodomain levels were increased significantly after depolarization with 25mM KCl for 6 h (Fig. 2a, b). Lactate dehydrogenase (LDH) assays demonstrated there was no significant release by the cells under these culture conditions after depolarization with KCl (arbitrary LDH release units: control 0.21±0.01; KCl 0.20±0.01). Incubation of hippocampal neuron cultures from DIV3 to DIV6 with α-2 anti-plasmin and/or MMP inhibitor II (which inhibits MMP1, 3, 7 and 9) to prevent extracellular cleavage of secreted proBDNF did not significantly alter the levels of prodomain secreted in the media in basal conditions or after depolarization (Fig. 2a, b). Enzymatic removal of N-linked glycosylation resulted in a reduction of molecular mass of the endogenous secreted prodomain from 15.5 kDa to the predicted 12.3 kDa (Fig. 2c) as described above for the prodomain expressed in the hippocampus. We next compared the activity-dependent secretion of the prodomain from mouse hippocampal-cortical cultures obtained from BdnfVal/Val and BdnfMet/Met knock-in animals6. We found that both Val66 and Met66 prodomains can be secreted after depolarization with KCl; however, the levels of secreted Met66 prodomain were significantly lower as compared to the Val66 prodomain (Fig. 2d, e). These results suggest that the prodomain is detectable in the adolescent and adult murine hippocampus and that neurons secrete it.
Figure 2.
The BDNF prodomain is secreted from cultured neurons. (a) Prodomain was detected in the media of day 6 in vitro rat hippocampal cultures. Cultures were treated with or without α-2 anti-plasmin and/or MMP inhibitor II (MMP1, 3, 7 and 9 inhibitor) in order to prevent extracellular cleavage of secreted proBDNF. Activity-dependent secretion was achieved by depolarization with KCl. (b) Quantification of (a). Bars represent mean ± s.e.m. of prodomain densitometry data. n=3 per group. (c) Treatment with N-glycanase reduced the molecular mass of the secreted prodomain. (d) Prodomain was detected in the media of day 6 in vitro hippocampal-cortical neurons from E18 pups obtained from BdnfMet/+ × BdnfMet/+ mice litters. Level of secreted prodomain differ between BdnfVal/Val and BdnfMet/Met cultures. (e) Quantification of (d). Bars represent mean ± s.e.m. of prodomain densitometry data. n=4 per group. Statistical comparisons were made by one way analysis of variance test. * p < 0.05.
Val66Met substitution alters the BDNF prodomain structure
To evaluate if there are structural differences in the Val66 and Met66 prodomains, we produced both human recombinant prodomains in E. coli (residues 23-113 displayed in red in Supplementary Fig. S1). The purity of the recombinant prodomains was examined by Coomassie blue and silver staining (Supplementary Fig. S3a, b). By SDS-PAGE the prodomains migrate at the predicted molecular weight of 10.2 kDa, with no difference in the detection of the Val66 and Met66 prodomains by the specific prodomain antibody used (Supplementary Fig. S3c). The BDNF prodomain is predicted to be disordered by structure prediction software (Supplementary Fig. S4). This predicted disorder in the BDNF prodomain is consistent with the absence of a defined trace of the related nerve growth factor (NGF) prodomain in the proNGF-p75NTR crystal lattice, indicating a lack of ordered structure20.
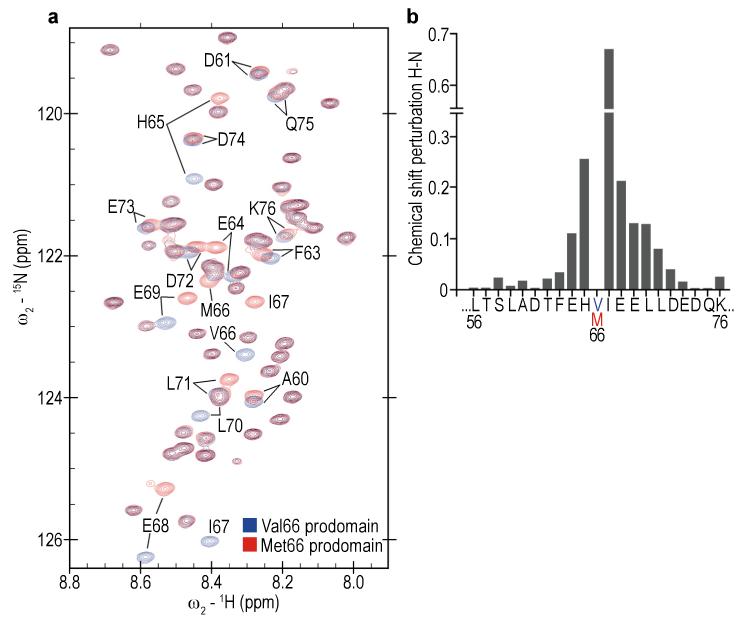
To define the structural effects of the Val66Met polymorphism, we examined the Met66 and Val66 prodomains using nuclear magnetic resonance (NMR). The 1H-15N heteronuclear correlation spectra indicated that the Val66 and Met66 prodomains are intrinsically disordered, lacking stable secondary or tertiary structure (Fig. 3, Supplementary Fig. S5). Secondary chemical shifts analysis for the Val66 prodomain and the Met66 prodomain indicate that both are primarily disordered structures (Supplementary Fig. S6). Moreover, in the NMR conditions employed, both prodomains were monomeric showing no evidence of oligomerization or concentration-dependent change in spectral properties (from 2 to 450μM). To define precise Val66Met structural differences we generated sequence specific 1H/13C/15N heteronuclear NMR assignments for the Val66 and Met66 prodomains using established triple resonance NMR methods21. This technique assigns individual resonance frequencies to specific atoms within each amino acid in the prodomain sequence (chemical shifts displayed as peaks in Fig. 3a and Supplementary Fig. S5a, b). The NMR chemical shifts provide a sensitive indicator of conformational preference within disordered states. Comparison of chemical shift deviations between the Val66 prodomain and the Met66 prodomain indicated that there are significant changes localized to seven residues (E64, H65, I67, E68, E69, L70, L71) neighboring the Val66Met substitution site (Fig. 3a, b). The backbone chemical shifts for the prodomains are deposited in the Biological Magnetic Resonance Bank: Val66 ID number: 19358. Met66 ID number: 19357.
Figure 3.
Impact of the Val66Met substitution on the structure of the BDNF prodomain. (a) Overlay of the heteronuclear single-quantum coherence (HSQC) spectrum on the Val66 (blue) and Met66 (red) prodomains. Each cross-peak (chemical shift) corresponds to one residue within the sequence of the prodomain (one chemical shift for each covalently bonded pair of 1H-15N atoms assigned to specific amides within the prodomain sequence). Full HSQC spectra of Val66 and Met66 prodomains are available in Supplementary Fig. S5. The backbone chemical shifts for the prodomains are deposited in the Biological Magnetic Resonance Bank: Val66 ID number: 19358. Met66 ID number: 19357. (b) Chemical shift deviation (Δδ) between BDNF Val66 and Met66 prodomains showed that changes induced by the substitution are localized to seven residues (E64, H65, I67, E68, E69, L70, L71) neighboring the Val66Met substitution site. The variation in Δδ for residues 23-55, and 77-113 outside the display window was between 0.007 and 0.0037 ppm.
Val66 and Met66 prodomains transient secondary structure
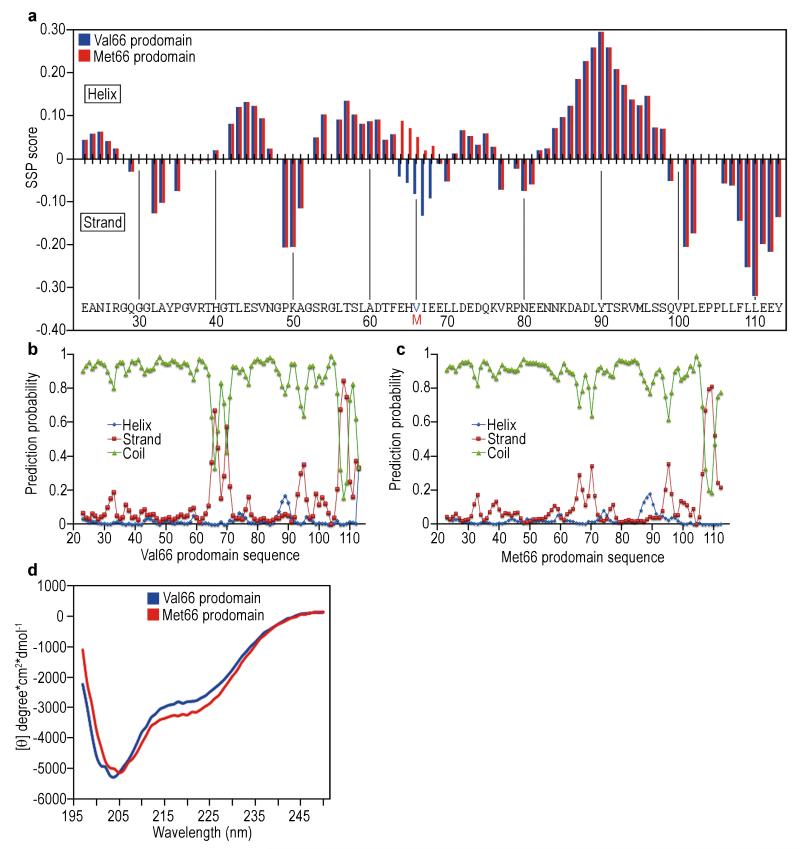
We estimated the prodomain secondary structure tendency based on the heteronuclear chemical shifts using the secondary structure propensity (SSP) score22. The positive SSP scores estimate regions of helical preference, while the negative scores indicate the residues with β-structure propensity. Upon Val66Met substitution, a shift in conformation from β-strand to helical conformation was observed around the position 66 (Fig. 4a). Outside the substitution area, the Val66 and Met66 prodomain SSP scores were coincident with regions of transient helix centered at residues 24, 44, 57, and 90, and β-structure that appears neighboring residues 32, 50, 80, 101 and 110 (Fig. 4a). Chemical shift analysis using Talos+ secondary structure analysis software23 showed a strong preference for β-strand around position 66 in the Val66 prodomain. The Met66 prodomain has reduced tendency to adopt β-strand as compared to the Val66 prodomain (Fig. 4b, c) consistent with the SSP score analysis.
Figure 4.
Val66 and Met66 prodomains differ in transient secondary structure.(a) Secondary structure propensity (SSP) score obtained using the backbone 1H, 15N, 13C chemical shifts from the BDNF prodomain Val66 (blue bars) and Met66 (red bars). The SSP score identified regions of transient structure formation (positive values = α-helix, negative values = β-sheet). (b)(c) Graphs illustrating secondary structure prediction by TALOS+ analysis using the heteronuclear backbone chemical shifts of Val66 (b) and Met66 (c) prodomains. α-helix in blue spheres, β-strand in red squares, and disorder in green triangles. TALOS+ analysis showed decreased β-sheet propensity in the Met66 prodomain compared to the Val66 prodomain, result that is consistent with the SSP score. (d) Ultraviolet circular dichroism (CD) spectra of 30 μM of the Val66 (blue) and Met66 (red) prodomains collected in10 mM NaH2PO4 and 50 mM NaCl pH 7.0 at 23°C. The negative peak around 200 nm revealed the natively unfolded conformation of both prodomains. However, the lower absorption at 222 nm for Met66 prodomain is consistent with increased tendency to helical conformation compared to the Val66 prodomain. Each spectrum is representative of 4 averaged scans and is normalized to the spectrum of buffer alone.
In order to corroborate the SSP analysis and Talos+ results with an NMR-independent technique, we performed circular dichroism (CD) spectroscopy. The negative peak around 200 nm for both prodomains CD spectra revealed the predominant natively unfolded conformation, while the changes in CD intensity at 222 nm are characteristic of helix propensity (Fig. 4d). Met66 prodomain CD spectra showed a more negative absorption at 222 nm consistent with an increased helical tendency as compared to the Val66 prodomain (Fig. 4d), a result that is in agreement with the SSP and Talos+ data. To summarize, both NMR and CD analysis demonstrate that the Val66 prodomain has increased tendency to β-structure in the residues adjacent to the position 66, while the Met66 prodomain displays increased helical propensity.
BDNF Met66 prodomain induces growth cone retraction
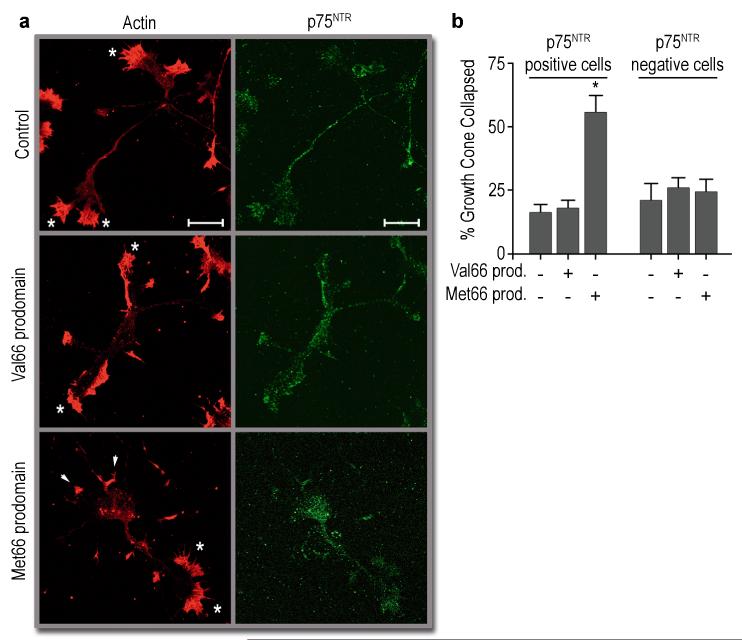
The formation of neuronal networks depends upon the growth and retraction of nascent neuronal processes. Alterations in synaptic network organization influence the occurrence and progression of neuropsychiatric disorders19,24. Therefore, we assessed the bioactivity of Val66 and Met66 prodomains in mediating acute morphological changes in hippocampal neurons using a growth cone retraction assay16. Unprocessed pro-nerve growth factor (proNGF) has been recently identified as a ligand that initiates acute retraction of growth cones, an effect mediated by the expression of p75NTR and SorCS2, a member of the sortilin family of receptors16. A recent study indicates that proBDNF also induces growth cone retraction15, an effect that we have confirmed (Supplementary Fig. S7a). Thus, we examined whether BDNF prodomain in isolation could induce the same effect. We treated primary mouse hippocampal neurons with recombinant Val66 or Met66 prodomains. Surprisingly, the Met66 prodomain induced rapid growth cone retraction over a period of 20 minutes, while the Val66 prodomain was inactive (Fig. 5 a, b). This effect was achieved with 1 nanomolar (10ng/ml) concentration of the Met66 prodomain. At 50 ng/ml, the Val66 prodomain also remained inactive without showing significant growth cone retraction (Supplementary Fig. S8). As with proNGF16 and proBDNF15 (Supplementary Fig. S7a), the growth cone retraction effect induced by the Met66 prodomain was limited to p75NTR positive cells within the culture (Fig. 5b).
Figure 5.
The Met66 prodomain induces growth cone retraction in cultured hippocampal neurons. (a) Neurons were treated with Val66 or Met66 prodomains (10 ng/ml) for 20 min, fixed, and stained for actin and p75NTR. Arrows indicate retracted growth cones; asterisks indicate intact growth cones. Scale bar: 20 μm. (b) Quantification of growth cone retraction in p75NTR positive cells shown in (a), compared to p75NTR negative cells. Prodomain abbreviated as prod. (b) n= 4 independent experiments. Bars represent mean ± s.e.m. Statistical comparisons were made by one way analysis of variance test. * p <0.05.
Met66 prodomain binds differentially to SorCS2
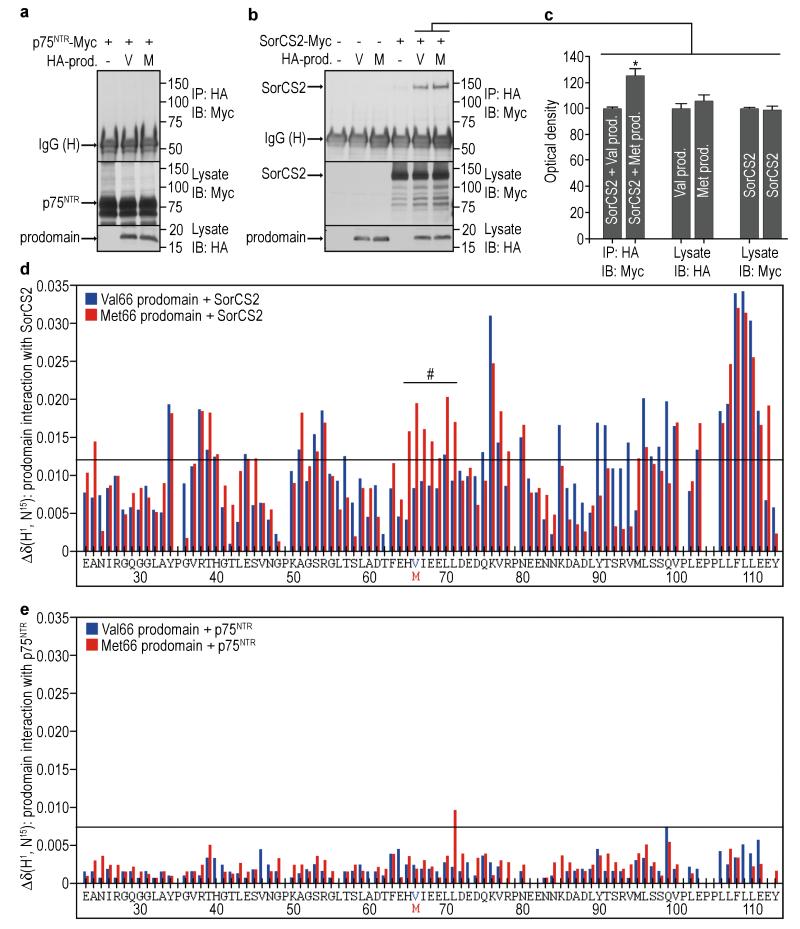
Prior experiments have established that p75NTR and sortilin interact to form a receptor complex on the cell surface25, and that SorCS2 facilitates the interaction of p75NTR with downstream signaling proteins to promote growth cone retraction16. We were unable to detect sortilin in these culture conditions, but SorCS2 was expressed in p75NTR positive cells16. Thus, we tested whether the prodomain interacts with SorCS2 and p75NTR using co-immunoprecipitation analysis of cells expressing the prodomain with either of the receptors. We were unable to detect interaction of the prodomains with p75NTR (Fig. 6a). However, both Met66 and Val66 prodomains interact with SorCS2 (Fig. 6b). By quantitative analysis, the Met66 prodomain interacted more effectively with SorCS2 than the Val66 prodomain (Fig. 6b, c). Moreover, we performed NMR spectroscopy of the Val66 prodomain or the Met66 prodomains with human recombinant SorCS2 to determine if the prodomains interacted differently with this receptor. Consistent with the co-immunoprecipitation result, both Val66 and Met66 prodomains interacted with SorCS2 as detected by chemical shift perturbation in the prodomains upon addition of the receptor (Fig. 6d, Supplementary Fig. S9a, b). Amino acids 34, 38-40, 51, 53-54, 76-77, 80, 96 and 100-111 of both Val66 and Met66 prodomains interact with SorCS2 as shown by chemical shift deviation upon addition of the receptor (Fig. 6d). Surprisingly, the Met66 prodomain displayed greater chemical shift changes upon interaction to SorCS2 between residues 65 and 71, while the same residues in the Val66 prodomain do not engage the receptor (Fig. 6d, under *). This represents a novel region of interaction in the prodomain that is conferred by the Met66 substitution. These larger chemical shift changes in the Met66 prodomain upon SorCS2 binding may reflect a tighter interaction or a different binding conformation. Thus, although both prodomains interact with SorCS2, the interactions are distinct, and this difference can result in the differential activity of the prodomains.
Figure 6.
The Val66 and Met66 prodomains binds differently to SorCS2 but do not bind to p75NTR. (a, b) HEK293T cells were transfected with the indicated constructs, the lysates were immunoprecipitated with anti-HA antibody, followed by detection with indicated antibodies. V=Val66 prodomain and M=Met66 prodomain constructs. (a) We were unable to detect interaction of either prodomains with p75NTR by co-immunoprecipitation. Representative blot of 4 independent experiments. (b) Both Val66 and Met66 prodomains co-immunoprecipitated with SorCS2, however Met66 prodomain interacted ~23% more than the Val66 as quantified in (c). (c) n=9 independent experiments. Bars represent mean ± s.e.m. Statistical comparisons were made by one way analysis of variance test. * p <0.05. Interaction of Val66 and Met66 prodomains with SorCS2 (d) or p75NTR (e) was also assessed by NMR spectroscopy. Δδ = chemical shift deviation between Val66 and Met66 prodomains together with the receptors. Interaction of the prodomain with SorCS2 (d) was assessed at 4.5μM for both proteins and data were collected at 600 MHz. The Met66 prodomain displayed greater chemical shift changes upon interaction to SorCS2 between residues 65 and 71 (shown under #) compared to the Val66 prodomain. Interaction of the prodomain with p75NTR (e) was assessed at 6.6μM for both proteins and data were collected at 800 MHz. Interaction of the prodomain with p75NTR was undetectable by NMR. Standard deviation (s.d.) was calculated for all the analysis and a threshold line equivalent to 2 s.d. was drawn to show the limit of what is considered interaction.
To further confirm association of the Val66 or Met66 prodomains with SorCS2, we performed diffusion NMR measurements. Measurements of translational diffusion are highly sensitive to changes in hydrodynamic shape which can be influence by changes in intramolecular conformation and intermolecular associations. Association of the prodomains (10.2 kDa) with the ectodomain of SorCS2 (113 kDa) will influence the prodomain solution mobility and the apparent radius of hydration. The measured diffusion rates of Val66 and Met66 prodomains were 1.02 × 10−10 m2sec−1 and 1.04 × 10−10 m2sec−1, respectively. Upon addition of SorCS2, Val66 display a slight decrease in diffusion DTVal66=0.99× 10−10 m2 sec−1, whereas the Met66 displayed significant reduction in diffusion rates to DTMet66=0.74 × 10−10 m2 sec−1, corresponding to more than a doubling of the apparent hydrodynamic volume (Supplementary Table S1). This indicates that the Met66 prodomain interacts with SorCS2 in a greater temporal extent as compared to the Val66 prodomain, and confirms that the Val66 and Met66 prodomains interact differentially with SorCS2.
We could not detect significant structural changes in SorCS2 after either prodomain binding using intrinsic tryptophan fluorescence spectroscopy. Using tryptophan excitation at 285nm, emission spectra with maxima at 329nm was collected with SorCS2 alone as a control. The addition of Val66 or Met66 prodomain to SorCS2 showed emission maxima at 329nm of 98.1% and 97.1% of the control, respectively. This is consistent with the lack of structural changes in Sortilin after neurotensin binding26. Significant chemical shift changes of either prodomain were not observed after the addition of p75NTR by NMR (Fig. 6e, Supplementary Fig. S9c, d) confirming the lack of binding found by co-immunoprecipitation analysis. In contrast to the isolated prodomains, proBDNF was able to interact with SorCS2 and p75NTR assessed by co-immunoprecipitation analysis (Supplementary Fig. S7b).
SorCS2 is required for Met66-induced growth cone retraction
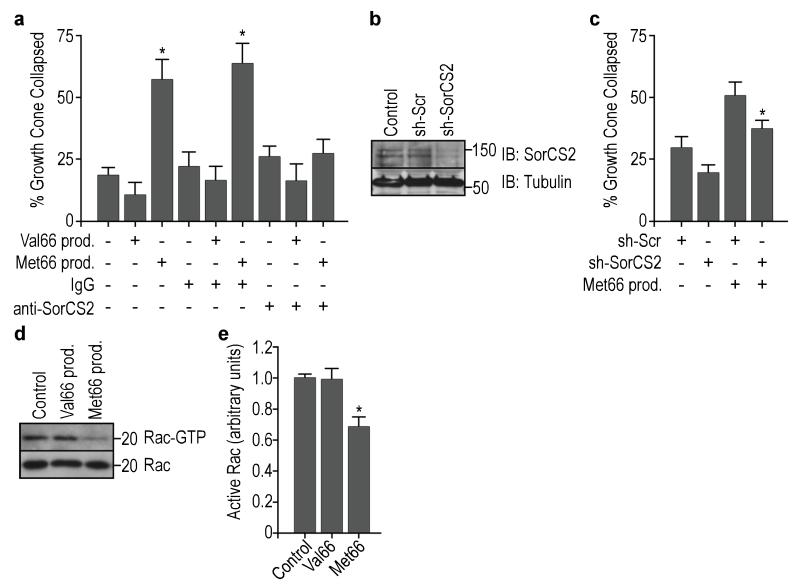
As the prodomains interact with SorCS2, we evaluated whether SorCS2 is involved in Met66 prodomain-induced retraction of growth cones. To address this, we pre-incubated hippocampal cultures with function blocking antibodies directed against the SorCS2 ectodomain before the addition of the ligands. Addition of antibodies did not affect neuronal morphology, but blocked the ability the Met66 prodomain to induce growth cone retraction (Fig. 7a). Control IgGs did not impair the effect (Fig. 7a). Moreover, anti-SorCS2 antibodies also blocked the ability of proBDNF to induce growth cone retraction (Supplementary Fig. S7a). To further confirm the involvement of SorCS2 in growth cone retraction triggered by the Met66 prodomain, we infected the hippocampal cultures with lentivirus expressing small hairpin RNA (shRNA) targeting SorCS2. SorCS2 shRNA (sh-SorCS2) expression resulted in a partial but significant decrease in SorCS2 protein levels as compared with the scrambled shRNA (sh-Scr) infection (Fig. 7b). The infection with scrambled shRNA resulted in higher growth cone retraction in basal conditions without ligand addition (Fig. 7c). However, the shRNA-induced SorCS2 decrease resulted in reduced growth cone retraction after Met66 prodomain administration, a result that is in agreement with the effects of function-blocking SorCS2 antibody experiments.
Figure 7.
Met66-SorCS2 interaction is required to induce growth cone retraction. (a) SorCS2 antibodies were able to block the Met66 prodomain-induced growth cone retraction. Neurons were pre-incubated with anti-SorCS2 (or control IgGs), treated with Val66 or Met66 prodomains (10 ng/ml) for 20 min, and followed by growth cone retraction analysis. Quantification assessed in p75NTR positive cells from 3 independent experiments. (b) Representative blot showing SorCS2 expression in cultured hippocampal neurons after knock-down using SorCS2 shRNA (sh-SorCS2) lentivirus infection, as compared to uninfected controls or scramble shRNA (sh-Scr) infected cells. (c) SorCS2 partial down regulation achieved with SorCS2 shRNA was able to partially prevent the Met66 prodomain-induced growth cone retraction. Quantification of 4 independent experiments. (d) Only Met66 prodomain administration, but not the Val66, induced a decrease in Rac activity in cultured hippocampal neurons. Hippocampal neurons were incubated with Val66 or Met66 prodomain for 20 min, and cell lysates were incubated with GST-PAK-CRIB beads to isolate activated Rac (Rac-GTP). (e) Quantification of (d). Activated Rac was measured by densitometry and normalized to total Rac in the input. Quantification assessed in 4 independent experiments. Bars represent mean ± s.e.m. Statistical comparisons were made by one way analysis of variance test. * p <0.05.
Growth cones are rich in actin filament-containing structures such as lamellipodia and filopodia. The dynamic extension and retraction of these actin structures is regulated by Rho GTPases family members including Rac27. We previously showed that proNGF induces displacement of the guanine nucleotide exchange factor (GEF) Trio from p75NTR and SorCS2, thereby decreasing local activity of Rac to induce growth cone retraction16. To determine whether the Val66 prodomain, the Met66 prodomain or proBDNF lead to differential signaling downstream of SorCS2, we isolated and quantified activated Rac using the Cdc42/Rac interactive binding (CRIB) domain of the Rac effector p21-activated kinase (PAK-CRIB). Surprisingly, the Met66 prodomain or proBDNF exposure led to a significant decrease in Rac activity in cultured hippocampal neurons, as compared to the absence of effect in Val66 prodomain-treated or in untreated neurons (Fig. 7d, e; Supplementary Fig. S7c).
The results strongly suggest that the helical preference in the Met66 prodomain is advantageous in interacting with SorCS2 and inducing active binding that is reflected in down-regulation of Rac activity and growth cone retraction. In contrast the Val66 prodomain with β structure tendency remains inactive.
Discussion
We found that the variant Met66 prodomain acutely alters neuronal morphology as it induces growth cone retraction. Mechanistically, we showed that the Val66Met substitution induces a shift in transient secondary structure from β sheet to a helical conformation in the residues surrounding the Val66Met substitution. This shift confers bioactivity to the Met66 prodomain through differential interaction with SorCS2, the sortilin-related Vps10p-domain sorting receptor 2. Moreover, we have demonstrated that the BDNF prodomain is detectable at high levels in vivo and that it is secreted in an activity-dependent manner from cultured hippocampal neurons.
In previous studies the isolated BDNF prodomain was undetectable10. It is known that low molecular weight proteins can be detected more efficiently by cross-linking the peptides to the transfer membrane using glutaraldehyde28,29. Glutaraldehyde fixation of the proteins to the transfer membrane following SDS-PAGE and the use of a prodomain specific monoclonal antibody previously characterized10, allow reliable detection of the prodomain in the murine central nervous system. However, we cannot rule out the possibility that the fixative promotes an antigen retrieval effect unmasking the prodomain epitopes. We corroborate that both the intracellular and the secreted prodomains are N-linked glycosylated as predicted by the amino acid sequence. Furthermore, the developmental production of the prodomain parallels the expression of mBDNF10,30,31.
The hippocampal expression of the prodomain in the BdnfMet/Met mice is less than the observed in control BdnfVal/Val mice, which is in agreement with a prior study that reported diminished hippocampal BDNF expression for the BdnfMet/Met mice by enzyme-linked immunosorbent assay32. The Val66Met polymorphism leads to a decrease in the trafficking of BDNF to secretory vesicles and the subsequent impairment of activity-dependent release of BDNF2,17,18. The decreased levels of prodomain could in part be due to decreased transport and/or secretion from afferent inputs to the hippocampus such as the cortex or sub-cortical areas. Alternatively, it was reported previously that the Met66 BDNF transcripts show decreased dendritic trafficking of BDNF mRNA33, and this may result in decreased local translation and secretion in the hippocampus. We cannot exclude the possibility that the Met66 proBDNF precursor exhibits decreased cleavage to its mature counterpart and prodomain, or that there is preferential degradation of the Met66 prodomain. Nonetheless, we were able to detect the Met66 prodomain in the BdnfMet/Met mice hippocampus indicating that it may exert local bioactivity.
Both mBDNF34 and proBDNF10,11 are secreted in an activity-dependent manner. Similarly, here we demonstrate that the release of the prodomain from neurons is also regulated by activity. Inhibition of extracellular cleavage of secreted proBDNF using α-2 anti-plasmin and an inhibitor of numerous MMPs did not significantly change levels of secreted prodomain in our culture conditions, suggesting that most of the cleavage of proBDNF to the prodomain and mBDNF may occur in intracellular compartments. It has been reported that there is impaired regulated secretion of mBDNF from BdnfMet/Met neuronal cultures6. In agreement with these studies, we found that there is a significant decrease in the activity dependent release of the endogenous Met66 prodomain as compared with the Val66 prodomain. However, regardless of the secreted levels, both are detected in the media suggesting that they may act as ligands for paracrine or autocrine signaling.
The Val66 and Met66 prodomains were determined to be intrinsically disordered with transient structural features using both NMR and CD spectroscopy. We detected specific conformational changes in the residues neighboring the Val66Met substitution area, from β-structure propensity in the Val66 prodomain to helical conformation tendency in the Met66. It is known that small changes in local structure can substantially alter protein activity. This is particularly true in the case of intrinsically disordered proteins which play prominent roles in signal transduction, antigen-antibody recognition, intracellular trafficking and as hormones, as well as in a number of neurodegenerative diseases such as Alzheimer’s, Huntington’s, Parkinson’s, prion and frontotemporal dementia diseases35,36. For instance, structural changes of similar magnitude to the ones observed for Val66Met substitution are observed in the A53T substitution in α-synuclein which has been correlated to impact the kinetics of aggregation of this protein and toxicity in Parkinson’s disease37. The structural changes between Val66 and Met66 prodomain impact on their interaction with SorCS2 receptor, as the Met66 with more helical propensity, shows greater local chemical shifts perturbation during binding. We document that both Val66 and Met66 prodomains interact with SorCS2, however they engage differentially as observed by NMR (Fig. 6d). Residues 65-71 of the Met66 prodomain interact with SorCS2 while the same residues in the Val66 prodomain do not engage the receptor, and this represents a novel region of interaction in the prodomain that is conferred by the Met66 substitution. This differential binding can explain the different bioactivity between the prodomains, however, other differences such as differential aggregation, bio-availability of the prodomains, differential trafficking or interaction of the prodomains with unknown factors may also play a role. This differential binding was confirmed by NMR diffusion measurements and co-immunoprecipitation experiments, and resulted in Rac inactivation only after Met66 addition to the cultured neurons.
The development, maturation and plasticity of neuronal networks rely on the emergence, path finding and retraction of neuronal processes. Alterations in synaptic network organization influence the occurrence and progression of neuropsychiatric disorders19,24,38. ProNGF has been recently identified to activate p75NTR and SorCS2 to promote fascin phosphorylation and its dissociation from actin filaments resulting in neurite retraction16. ProBDNF is also able to induce growth cone retraction as shown in supplementary Fig. S7a confirming previously published studies15. Here we describe that the Met66 prodomain is sufficient to initiate growth cone retraction. As shown by NMR, the Met66 prodomain engages differently with SorCS2, but not with p75NTR. However, Met66 prodomain is only active in p75NTR expressing cells suggesting that p75NTR acts in a complex with SorCS2 or is downstream of SorCS2.
ProBDNF interacts with both SorCS2 and p75NTR and induces growth cone retraction. This may contribute to the bioactivity of proBDNF in this assay, while the Val66 prodomain, which binds in a disadvantageous manner to SorCS2, is inactive. In early postnatal ages both proBDNF and processed products (mBDNF and prodomain) are present in the hippocampus. During adulthood, mBDNF and the prodomain are the most abundant forms, while proBDNF levels are decreased10. From this we can infer that the actions of proBDNF may predominate in early postnatal time points, but the actions of the prodomain may prevail during adulthood.
Most growth factors are synthesized as precursors, and prodomains are thought to function primarily to promote protein folding, intracellular sorting, or to shield the mature domain from receptors to limit bioavailability39. Here we have demonstrated that the endogenous BDNF prodomain is detected in the hippocampus, and is secreted in an activity-dependent manner from neurons. Most importantly, the evolutionarily recent SNP that results in the Val66Met substitution in the prodomain promotes growth cone retraction in hippocampal neurons with involvement of p75NTR and binding to SorCS2, in contrast to inactive Val66 prodomain. A function of the Val66 prodomain remains to be elucidated and future studies with mature neuronal networks are required to understand other prodomain actions.
The endophenotypes related to neuropsychiatric disorders associated with the Val66Met polymorphism have been linked with decreased sorting and secretion of BDNF, thus impairing neuronal differentiation and plasticity via reduced TrkB activation32,33. Here we show that the Met66 prodomain is a newly identified ligand that selectively activates SorCS2 to acutely alter neuronal morphology, and we propose that this effect is an additional mechanism that contributes to altered neural plasticity in humans with the SNP.
Methods
Animals
C57BL/6 male mice from different ages (as specified in the figures and captions) were maintained with a 12 h light/dark cycle and with free access to water and food. Val66Met knock-in mice (BdnfMet/Met) and wild-type (BdnfVal/Val) were used as reported previously6. Briefly, the BDNF coding region was replaced by a targeting vector with the BDNF sequence with or without a point mutation (G196A) which was introduced to result in the Val66Met substitution. These BDNF knock-in alleles are regulated by the endogenous BDNF promoters. Heterozygous BdnfVal/Met mice were intercrossed to yield BdnfVal/Val, BdnfVal/Met and BdnfMet/Met offspring at Mendelian rates. Heterozygous Bdnf+/− mice (Bdnftm1Jae) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Sprague Dawley pregnant rats and C57BL/6 pregnant mice were purchased from Taconic Farms (Hudson, NY, USA) and Charles River (Wilmington, MA, USA), respectively. Animal care was in accordance with Weill Medical College of Cornell University IACUC.
SDS-PAGE and Western Blot
Dissected hippocampi or HEK293T (ATCC, Manassas, VA, USA) cells were lysed in 1% triton X-100 (Sigma, Saint Louis, MO, USA), 1% nonidet P-40 (Roche, Indianapolis, IN, USA), 10% glycerol, in buffer Tris-buffered saline pH 7.4 supplemented with protease inhibitor cocktail (Sigma). Protein concentration was determined by Bradford (Bio-Rad, Hercules, CA, USA). For deglycosylation, 100μg of lysates or 100μl of conditioned media were incubated with 1μl of N-glycanase (ProZyme, San Leandro, CA, USA) for 1h at 37C. The lysates or conditioned media were run in SDS–PAGE, the proteins were transferred to a 0.45μm polyvinylidene fluoride membrane (PVDF) (Millipore, Billerica, MA, USA), and the membranes were fixed with 2.5% glutaraldehyde (Sigma) in PBS pH 7.4 unless otherwise written. After blocking with 5% bovine serum albumin (BSA) in Tris-buffered saline with 0.1% Tween 20, membranes were incubated with the prodomain antibody10 (mAb287, 1:2000, 12-16hs at 4C, GeneCopoeia, Rockville, MD, USA), anti-β tubulin (Sigma, 1:15000, 1h at 20-25C) or anti-SorCS2 (R&D Systems, Minneapolis, MN, USA, 1:1000, 12-16hs at 4C) followed by anti-mouse horse radish peroxidase (HRP) secondary antibody (1:5000 for prodomain, or 1:15000 for β tubulin, 1h at 20-25C, Calbiochem-Millipore) or anti-sheep HRP (1:5000 for SorCS2, 1h at 20-25C, Calbiochem-Millipore). Bands were analyzed by enhanced chemiluminescence (Amersham-GE, Pittsburgh, PA, USA), and the densitometry was analyzed using Image J (1.45, NIH, Bethesda, MD, USA, http://rsbweb.nih.gov/ij/). Quantization was normalized to β tubulin for each individual sample. Full gel scans are provided in Supplementary Fig. S10.
Hippocampal cultures for prodomain secretion analysis
Primary hippocampal neurons were isolated from E18 Sprague Dawley rat embryos. Neurons were dissociated with 0.05% trypsin at 37°C for 20 min followed by trituration with fire-polished glass pipettes. Cells plated on poly-D-lysine (Sigma) coated dishes were grown in Neurobasal medium (Gibco, Grand Island, NY, USA) containing B27, 1mM pyruvate (Gibco), 2mM glutamine (Gibco), and 10 mM 5-fluorodeoxyuridine (Sigma). From day 3 in vitro to day 6, α-2-anti-plasmin (100μM, Calbiochem-Millipore) and/or MMP Inhibitor II (10μM, Calbiochem-Millipore; MMP1 IC50 = 24 nM, MMP3 IC50 = 18.4 nM, MMP7 IC50 = 30 nM, and MMP9 IC50 = 2.7 nM.) were added to the cultures. Day 6 in vitro media was collected and protease inhibitor cocktail (Sigma) was added. For depolarization experiments, 25mM KCl was added to the cultures for 6 h. To study the secretion of the endogenous Val66 and Met66 prodomains, we cultured hippocampal-cortical neurons from E18 pups obtained from BdnfMet/+ x BdnfMet/+ litters. Each pup dissected area was plated in a different well and genotyped as described6. Subsequently, depolarization and harvesting of the culture media were performed as described for rat hippocampal cultures above.
Lactate dehydrogenase release assay
Conditioned media (50ul) from DIV6 neurons in culture (with or without depolarization with KCl) was harvested and analyzed for LDH levels using a colorimetric assay following the manufacturer’s protocol (CytoTox 96; Promega, Madison, WI, USA).
Expression and purification of recombinant BDNF prodomains
The plasmid pET28 (Novagen, Madison, WI, USA) containing the gene of human BDNF prodomain Val66 or Met66 (amino acids 23 to 113 preceded by an N-terminal His-tag and SUMO)40, was transformed in BL21(DE3)pLysS competent cells (Invitrogen, Grand Island, NY, USA). Proteins were produced using the Marley media swap method for isotopic labeling (15N and 13C/15N) of recombinant proteins41. Prodomain samples were purified using a nickel resin column (Ni-NTA, Invitrogen). SUMO was cleaved by Ulp-1 (His6 tagged) proteolysis. Subsequently, the sample was negatively selected in a second nickel resin column where the now untagged BDNF prodomain was obtained in the flow though fraction. The resulting protein was precipitated using acetone, resuspended and dialyzed in 50mM NaH2PO4 pH 7.0. The sample purity was assessed by SDS-PAGE using Coomassie blue and silver staining methods (Supplementary Fig. S3). The prodomains concentrations were calculated using random-coil UV absorbance in water (ε(280) = 4470 M−1cm−1)42, and confirmed using the Bradford method.
NMR spectroscopy
NMR samples were prepared in 93% H2O/7% D2O, 50 mM NaH2PO4 100 mM NaCl pH 7.0 at concentrations varying from 2 μM to 450 μM. NMR spectra were acquired on a Bruker Avance 500 MHz, a Varian INOVA 600 MHz spectrometer both at Weill Cornell Medical College, and a Bruker Avance 800 MHz spectrometer at the New York Structural Biology Center. The 600 MHz and 800 MHz spectrometers were equipped with triple-resonance cryogenic probes. The 500 MHz spectrometer was equipped with a BBO room temperature probe and NMR chemical shift assignments were performed at 7C. NMR data were processed using the software nmrPipe43 and analyzed using Sparky44. Secondary structure predictions based on the observed chemical shifts were performed using the SSP22 and TALOS+23 programs. Chemical shift deviation: Δδ=[(Δδ1H)2+(0.153*Δδ15N)2]1/2, where Δδ1H and Δδ15N are the 1H and 15N chemical shifts change, and 0.153 is a weighting factor for 15N shifts. Interaction of the prodomain with recombinant human SorCS2 (R&D Systems) was assessed at 4.5μM for both proteins and data were collected at 600 MHz. Interaction of the prodomain with human recombinant p75NTR (R&D Systems) was assessed at 6.6μM for both proteins and data were collected at 800 MHz. Data were processed and analyzed using Topspin software (Bruker Instruments version 2.1). Sequential backbone resonance assignments were achieved using standard three dimensional HNCO, HNCA, HN(CO)CA, HNCACB and CBCA(CO)NH resonance heteronuclear experiments45.
NMR diffusion measurements
Gradient diffusion measurements were acquired using the BPP-LED pulse sequence46 at 500 MHz in a Bruker Avance spectrometer, at 25C. Samples were all run in a volume constricted Shigemi tube, using a 20 mm sample height between plunger and tube bottom, to minimize convection artifacts. Data were collected at 298K using 32 gradient experiments with linearly increasing z-gradient strengths from 2% to 95% of the maximum 55.7 G/cm, while maintaining a constant 6 ms gradient delay over the 100 ms diffusion period. The hydrodynamic radius was calibrated using 10mM dioxane as an internal standard47. The data were processed and analyzed using Bruker topspin software version 2.1. The hydrodynamic radius is calculated using the Stokes-Einstein equation DT = kBT(6πηRh)−1 where DT is the translational diffusion coefficient, kb is Boltzmann constant, T is temperature in Kelvin, η is the solvent viscosity and Rh is the hydrodynamic radius, by using dioxane as an internal reference the radius can be calculated Rh = (DTdioxane/DT)Rhdioxane, assuming Rhdiovane = 2.12 Å.
Intrinsic tryptophan fluorescence spectroscopy
Fluorescence data were collected on a PTI spectrophotometer (Photon Technology International) at 20°C with measurements from 305 nm to 500 nm in 1 nm increments. SorCS2, Val66 prodomain and Met66 prodomain were used at 0.17μM in 50 mM Tris, 100mM NaCl pH 7.0.
Circular dichroism spectroscopy
CD spectra were acquired on an Aviv 62DS (Aviv Associates, Lakewood, NJ, USA) circular dichroism spectropolarimeter, using a 0.1cm path length cuvette with 30 μM prodomains in 10 mM NaH2PO4 50 mM NaCl pH 7.0. The wavelength dependence of molar ellipticity, [θ], was monitored at 23 °C as the average of 4 scans, using a 5 second integration time with 1.0 nm wavelength increments. Spectra were baseline-corrected against buffer alone.
Growth cone retraction assay
Primary hippocampal neurons were isolated from E15 C57BL/6 mouse embryos. Neurons were dissociated with 0.05% trypsin (Gibco) at 37°C for 8 min followed by trituration with fire-polished glass pipettes. Cells plated on poly-D-lysine (Sigma) coated dishes were grown in Neurobasal medium (Gibco) containing B27 (Gibco) and 0.5 mM glutamine (Gibco). Val66 proBDNF was collected from supernatants of HEK293FT cells (ATCC) transfected with a construct encoding human proBDNF (furin cleavage site mutated: RR128AA). proBDNF concentration was estimated by western blot (using BDNF antibody from Santa Cruz) comparing to known concentrations of recombinant BDNF (PeproTech, Rocky Hill, NJ, USA). For the assay, we added 10 or 50 ng/ml of the Val66 prodomain, Met66 prodomain, or proBDNF to hippocampal neurons for 20 min before fixation. In order to block SorCS2, we pre-incubated cells with anti-SorCS2 (20 μg/ml, R&D Systems) or control sheep IgG for 20 min on ice before addition of the prodomains or proBDNF at 37°C. DIV3 neurons were fixed with ice-cold methanol for 10 min. Coverslips were blocked with 10% normal donkey serum, 2% bovine serum albumin, and 0.25% fish skin gelatin in Tris-buffered saline for 30 min; incubated with anti-Actin (1:10000, Sigma, clone AC-74; anti-p75NTR 1:1000, R&D Systems; both for 30min at 20-25°C); followed by secondary antibodies (anti-goat Alexa 488, and anti-mouse Alexa 555, 1:1000, Invitrogen) mixed with Hoechst (1:10000) for 30min at 20-25°C; and mounted with Mowiol488 (Calbiochem-Millipore). Cells were imaged with an LSM 510 laser-scanning confocal microscope (with a 40× Plan Neofluor, numerical aperture 1.3 DIC oil immersion objective, Carl Zeiss, Oberkochen, Germany). Images were processed with LSM 510 software (Carl Zeiss) and Image J.
Co-immunoprecipitation experiments in HEK293T cells
HEK293T cells (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (Gibco) and penicillin-streptomycin. Cells were transfected using Lipofectamine 2000 (Invitrogen) with Myc-tagged p75NTR (a kind gift of Dr. Phil Barker), Myc-tagged SorCS2, and HA/Flag-tagged Val66 prodomain, Met66 prodomain (both produced by PCR using human pcDNA3.1 proBDNF as a template), and proBDNF constructs. 48 h after transfection, cells were lysed, protein concentration was determined by Bradford (Bio-Rad). Lysates were pre-cleared with protein A-sepharose (Sigma), immunoprecipitated using an HA antibody (Sigma), followed by protein A-sepharose resin (Sigma). Following SDS-PAGE, the membranes were incubated with Myc antibody (1:15000, 1hr at 20-25°C, Bethyl) followed by anti-rabbit HRP secondary antibody (1:15000, 1hr at 20-25°C, Calbiochem-Millipore), or with HA.11 antibody (1:10000, 1hr at 20-25°C, Covance) followed by anti-mouse HRP secondary antibody (1:10000, 1hr at 20-25°C, Calbiochem-Millipore).
Lentiviral vectors and transduction
Lentiviral plasmids (pLKO.1) containing shRNA against mouse SorCS2 (targeting sequence: CGCTGAACTCTCATAGAATCA) and a scramble control were obtained from Open Biosystems Thermo Scientific (Waltham, MA, USA). Lentivirus was packaged by co-transfection of shRNA constructs with packaging plasmids pMD2.G and pCMV-dR8.9 using TransIT-LT transfection reagent (Mirus, Madison, WI, USA) into HEK293-FT cells. Media was changed 24 h later, and the media supernatant was collected 48 h after transfection. Supernatants were filtered through a 0.45μm filter and pelleted by centrifugation with PEG-it (System Biosciences, Mountain View, CA, USA). Pellets were re-suspended in Neurobasal medium (Gibco). Mouse primary hippocampal neurons were infected on DIV0 and medium was changed the following day.
Rac activity assay
Rac activity assays were performed as described previously48. Briefly, DIV2 hippocampal neurons were stimulated with Val66 prodomain, Met66 prodomains or proBDNF (10 ng/ml) for 20 min, and cells were lysed in lysis buffer supplemented with 10 mM MgCl2. Cleared lysates were incubated with glutathione S-transferase (GST) or GST-PAK-CRIB beads (20μg per reaction) for 30 min at 4C. Isolated active Rac was analyzed by Western blot using a Rac1 antibody (Millipore, clone 23A8), and normalized to the input. GST and GST-PAK-CRIB proteins were expressed in BL21(DE3)pLysS cells (Invitrogen) followed by incubation with glutathione sepharose. The purity of the recombinant proteins was analyzed by Coomassie blue staining.
Supplementary Material
Acknowledgments
This work was supported by the NIH (NS21072 and HD23315 to M.V.C., NS052819 to F.S.L., NS030687 and NS064114 to B.L.H. and S10-RR023694-01EWOF to C.B.) and the Human Frontier Science Program to K.D. We acknowledge the kind gift of the Myc tagged p75NTR construct from Dr. Phil Barker (McGill University, Montreal, Canada). We thank Jianmin Yang for helpful discussion and revision of the manuscript. Parts of this work were carried out at the New York Structural Biology Center (NYSBC) supported by NIH, USA, the Keck Foundation and New York State Office of Science, Technology and Academic Research. We thank the NYSBC staff for excellent assistance.
Footnotes
Author contributions:
A.A. performed the biochemical analysis of brain lysates, primary cultured cells and transfected heterologous cells. A.A and N.E.W. generated the constructs. A.A. and C.B. synthesized and purified the recombinant prodomains, performed circular dichroism, fluorescence and NMR spectroscopy. K.D. performed the growth cone retraction assay. KI generated the SorCS2 shRNA lentivirus. A.A, K.D., M.V.C., F.S.L., B.L.H. and C.B. designed the experiments and wrote the manuscript.
Competing Financial Interests statement:
The authors declare that they have no conflict of interest.
References
- 1.Hajek T, Kopecek M, Hoschl C. Reduced hippocampal volumes in healthy carriers of brain-derived neurotrophic factor Val66Met polymorphism: meta-analysis. World J Biol.Psychiatry. 2012;13:178–187. doi: 10.3109/15622975.2011.580005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 3.Frielingsdorf H, et al. Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Ann N.Y.Acad Sci. 2010;1208:150–157. doi: 10.1111/j.1749-6632.2010.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soliman F, et al. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhagen M, et al. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry. 2010;15:260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZY, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dincheva I, Glatt CE, Lee FS. Impact of the BDNF Val66Met Polymorphism on Cognition: Implications for Behavioral Genetics. Neuroscientist. 2012 doi: 10.1177/1073858411431646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat.Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- 9.Teng KK, Felice S, Kim T, Hempstead BL. Understanding proneurotrophin actions: Recent advances and challenges. Dev.Neurobiol. 2010;70:350–359. doi: 10.1002/dneu.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. Neuronal release of proBDNF. Nat.Neurosci. 2009;12:113–115. doi: 10.1038/nn.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagappan G, et al. Control of extracellular cleavage of ProBDNF by high frequency neuronal activity. Proc.Natl.Acad Sci U.S.A. 2009;106:1267–1272. doi: 10.1073/pnas.0807322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 13.Mizoguchi H, et al. Matrix metalloproteinase-9 contributes to kindled seizure development in pentylenetetrazole-treated mice by converting pro-BDNF to mature BDNF in the hippocampus. J Neurosci. 2011;31:12963–12971. doi: 10.1523/JNEUROSCI.3118-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng HK, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J.Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, et al. ProBDNF Collapses Neurite Outgrowth of Primary Neurons by Activating RhoA. PLoS.One. 2012;7:e35883. doi: 10.1371/journal.pone.0035883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deinhardt K, et al. Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci Signal. 2011;4:ra82. doi: 10.1126/scisignal.2002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ZY, et al. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J Neurosci. 2005;25:6156–6166. doi: 10.1523/JNEUROSCI.1017-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ZY, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price JL, Drevets WC. Neurocircuitry of mood disorders. Neuropsychopharmacology. 2010;35:192–216. doi: 10.1038/npp.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng D, et al. Molecular and structural insight into proNGF engagement of p75NTR and sortilin. J.Mol.Biol. 2010;396:967–984. doi: 10.1016/j.jmb.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattlera M, Schleucherb J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy. 1999;34:93–158. [Google Scholar]
- 22.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol.NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr.Opin.Neurol. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nykjaer A, et al. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 26.Quistgaard EM, et al. Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain. Nat.Struct.Mol Biol. 2009;16:96–98. doi: 10.1038/nsmb.1543. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Billault C, et al. The role of small GTPases in neuronal morphogenesis and polarity. Cytoskeleton (Hoboken.) 2012;69:464–485. doi: 10.1002/cm.21034. [DOI] [PubMed] [Google Scholar]
- 28.Kurien BT, Scofield RH. A brief review of other notable protein detection methods on blots. Methods Mol Biol. 2009;536:557–571. doi: 10.1007/978-1-59745-542-8_56. [DOI] [PubMed] [Google Scholar]
- 29.Karey KP, Sirbasku DA. Glutaraldehyde fixation increases retention of low molecular weight proteins (growth factors) transferred to nylon membranes for western blot analysis. Anal.Biochem. 1989;178:255–259. doi: 10.1016/0003-2697(89)90634-9. [DOI] [PubMed] [Google Scholar]
- 30.Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolbeck R, Bartke I, Eberle W, Barde YA. Brain-derived neurotrophic factor levels in the nervous system of wild-type and neurotrophin gene mutant mice. J Neurochem. 1999;72:1930–1938. doi: 10.1046/j.1471-4159.1999.0721930.x. [DOI] [PubMed] [Google Scholar]
- 32.Bath KG, et al. BDNF Val66Met Impairs Fluoxetine-Induced Enhancement of Adult Hippocampus Plasticity. Neuropsychopharmacology. 2012 doi: 10.1038/npp.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiaruttini C, et al. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc.Natl.Acad Sci U.S.A. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman LJ, et al. Regulated release and polarized localization of brain-derived neurotrophic factor in hippocampal neurons. Mol Cell Neurosci. 1996;7:222–238. doi: 10.1006/mcne.1996.0017. [DOI] [PubMed] [Google Scholar]
- 35.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu.Rev.Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 36.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem.Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Bussell R, Jr., Eliezer D. Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol.Chem. 2001;276:45996–46003. doi: 10.1074/jbc.M106777200. [DOI] [PubMed] [Google Scholar]
- 38.Dwivedi Y. Brain-Derived Neurotrophic Factor in Suicide Pathophysiology. 2012 [PubMed] [Google Scholar]
- 39.Shi M, et al. Latent TGF-beta structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 41.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol.NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 42.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal.Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol.NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. SPARKY 3. University of Californina; San Francisco: 2002. [Google Scholar]
- 45.Cao W, Bracken C, Kallenbach NR, Lu M. Helix formation and the unfolded state of a 52-residue helical protein. Protein Sci. 2004;13:177–189. doi: 10.1110/ps.03383004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu DHC, A.D., Johnson CS. An Improved Diffusion-Ordered Spectroscopy Experiment Incorporating Bipolar-Gradient Pulses. Journal of Magnetic Resonance, Series A. 1995;115 [Google Scholar]
- 47.Jones JAW, D.K., Smith LJ, Dobson CM. Characterization of protein unfolding by NMR diffusion measurements. J. Biomol. NMR. 1997;119 [Google Scholar]
- 48.Neubrand VE, Thomas C, Schmidt S, Debant A, Schiavo G. Kidins220/ARMS regulates Rac1-dependent neurite outgrowth by direct interaction with the RhoGEF Trio. J Cell Sci. 2010;123:2111–2123. doi: 10.1242/jcs.064055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.