Abstract
Dynamic texture quantification, i.e., extracting texture features from the lesion enhancement pattern in all available post-contrast images, has not been evaluated in terms of its ability to classify small lesions. This study investigates the classification performance achieved with texture features extracted from all five post-contrast images of lesions (mean lesion diameter of 1.1 cm) annotated in dynamic breast magnetic resonance imaging exams. Sixty lesions are characterized dynamically using Haralick texture features. The texture features are then used in a classification task with support vector regression and a fuzzy k-nearest neighbor classifier; free parameters of these classifiers are optimized using random sub-sampling cross-validation. Classifier performance is determined through receiver-operator characteristic (ROC) analysis, specifically through computation of the area under the ROC curve (AUC). Mutual information is used to evaluate the contribution of texture features extracted from different post-contrast stages to classifier performance. Significant improvements (p < 0.05) are observed for six of the thirteen texture features when the lesion enhancement pattern is quantified using the proposed approach of dynamic texture quantification. The highest AUC value observed (0.82) is achieved with texture features responsible for capturing aspects of lesion heterogeneity. Mutual information analysis reveals that texture features extracted from the third and fourth post-contrast images contributed most to the observed improvement in classifier performance. These results show that the performance of automated character classification with small lesions can be significantly improved through dynamic texture quantification of the lesion enhancement pattern.
Keywords: Dynamic breast magnetic resonance imaging (MRI), Texture analysis, Gray-level co-occurence matrices, Mutual information, Support vector regression
1. Introduction
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has emerged as an important tool in breast cancer diagnosis. Previous work has established the superiority of DCE-MRI over X-ray mammography and sonography in lesion detection and quantification [1,2]. DCE-MRI has an additional advantage of not using any form of ionizing radiation in the image acquisition process. However, DCE-MRI exams typically require acquisition in both spatial and temporal domains, making subjective evaluation of clinical findings a challenging task for the radiologist. As a result, breast cancer diagnosis using DCE-MRI has been the subject of research in the area of computer-aided diagnosis (CADx) [3–9].
Previous work has investigated the use of dynamic criteria, such as signal intensity (SI) characteristics of the contrast uptake time series, in establishing the malignancy of suspicious breast tissue lesions (rapid contrast enhancement followed by washout) [3–5]. Other investigators have focused on characterizing morphological criteria such as shape, border, or the enhancement pattern and using them to classify unknown breast lesions as benign or malignant [6–9]. The effect of combining morphological and dynamic criteria on the diagnostic accuracy of classifying lesions using DCE-MRI has also been investigated [10–12]. Thus, the ability of CADx to achieve a high diagnostic sensitivity (up to 97%) and reasonable specificity (76.5%) in the task of classifying suspicious lesions from DCE-MRI is well established [13].
However, not many studies have focused on evaluating the value of DCE-MRI for small lesions where clinical findings are unclear. Such lesions can be diagnostically challenging as they may not exhibit typical characteristics of benign and malignant tumors, which are usually easier to discern when the lesions are larger [14,15]. In this regard, Leinsinger et al. reported a diagnostic accuracy of 75% in detecting breast cancer through cluster analysis of signal intensity time curves [14]. More recently, Schlossbauer et al. reported an area under the receiver-operator characteristic (ROC) curve (AUC) of 0.76 when using dynamic characteristics extracted from a dataset of small lesions (mean size of 1.1 cm) in classifying lesion character, and an AUC of 0.61 when using morphological criteria for the character classification task [15]. The primary goal of the present study was to improve the classification performance of such small lesions from DCE-MRI. An example of such a lesion, as localized in a dynamic breast MRI study, is shown in Fig. 1.
Figure 1.
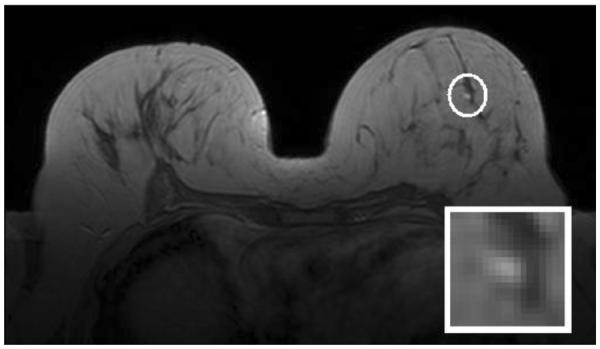
Small contrast-enhanced benign lesion (circled) on the T1-weighted fourth post-contrast image of a dynamic breast MRI study. The annotated lesion is magnified and shown in the lower-right corner of each image; the accompanying histo-pathological report pronounced this lesion benign (fibrosis, possibly induced by degenerative epithelial changes) with a lesion diameter of 0.8 cm.
Texture analysis can be used to quantify image patterns from a specified region of interest (ROI) [16]. The present study uses second-order statistical texture features derived from gray-level co-occurrence matrices (GLCM) as described by Haralick [17,18]. Where the benign or malignant nature of a lesion can be determined through its homogeneous or heterogeneous enhancement appearance [19,20], prior studies have shown that such GLCM-derived texture features can be used to classify lesion character with high diagnostic accuracy [7–9]. However, texture analysis in such studies has focused on extracting texture features from a single post-contrast image (usually the first of five). Providing the classifier with supplementary information derived from texture analysis on later post-contrast images in addition to extracting texture features from the first post-contrast image can significantly improve the performance of lesion character classification, specifically of small, diagnostically challenging lesions, as used in the present study.
In this work, texture analysis using GLCM is performed on all five post-contrast images of a dynamic breast MRI exam and the texture features extracted are combined to form lesion characterizing five-dimensional (5D) texture feature vectors, which are used for lesion classification. The approach is called dynamic texture quantification. For the machine learning task, support vector regression (SVR) [21] is compared to a conventional fuzzy k-nearest neighbor (fkNN) algorithm in terms of its ability to classify diagnostically challenging lesions from DCE-MRI. SVR extends the use of support vector machines to regression analysis and is used in this study as a function approximator that predicts the class label of texture feature vectors extracted from lesions of unknown character. While support vector machines can themselves be used in a classification task [22], they provide binary outputs for class predictions whereas SVR provides fuzzy class labels which can subsequently be used to generate ROC curves, which is the accepted metric of classification performance in previous studies. The contribution of texture features extracted from later post-contrast images of the lesion ROI is analyzed through feature selection.
2. Materials and methods
2.1 Data
Sixty lesions were identified from a representative set of dynamic contrast-enhanced breast MRI exams from 54 female patients by two experienced radiologists (both with > 5 years of experience in reading breast MRI cases), who came to a consensus on the evaluation of clinical findings. The mean patient age was 52 with a standard deviation of 12 and a range of 27 to 78. In all cases, histo-pathologically confirmed diagnosis from needle aspiration/excision biopsy was available prior to this study; 32 of the lesions were diagnosed as benign and the remaining 28 as malignant. The mean lesion diameter was 1.05 cm (standard deviation of 0.73 cm). The histological distribution of the 32 benign lesions is as follows −3 fibroadenoma, 10 fibrocystic change, 5 fibrolipomatous change, 7 adenosis, 1 papilloma, and 6 non-typical benign disease. The histological distribution of the 28 malignant lesions is as follows −18 invasive ductal carcinoma, 5 invasive lobular carcinoma, 3 ductal carcinoma in-situ, and 2 non-typical malignant disease.
Patients were scanned in the prone position using a 1.5-T MR system (Magnetom VisionTM, Siemens, Erlangen, Germany) with a dedicated surface coil to enable simultaneous imaging of both breasts. Images were acquired in the transversal slice orientation using a T1-weighted three-dimensional (3D) spoiled gradient echo sequence with the following imaging parameters: echo repetition time (TR) = 9.1 ms, echo time (TE) = 4.76 ms, and flip angle (FA) = 25°. Acquisition of the pre-contrast series was followed by the administration of 0.1 mmol/kg body weight of paramagnetic contrast agent (gadopentate dimeglumine, MagnevistTM, Schering, Berlin, Germany). Five post-contrast series were then acquired, each with a measurement time of 83 seconds, at intervals of 110 seconds. All breast exams were acquired with informed consent from the patients and were evaluated in this study in a retrospective manner.
In the collection of patient data used in this study, images in the dynamic series were acquired with two different settings of spatial parameters: for 19 patients, the images were acquired as 32 slices per series with a 512 × 512 matrix, 0.684 × 0.684 mm2 in-plane resolution, and 4-mm slice thickness, and in the other cases, the images were acquired as 64 slices per series with a 256 × 256 matrix, 1.37 × 1.37 mm2 in-plane resolution, and 2-mm slice thickness. To maintain uniform image data for texture analysis, the images acquired with a 512 × 512 matrix were reduced to a 256 × 256 matrix through bilinear interpolation.
2.2 Lesion annotation and segmentation
With the exception of two patients, for whom two separate lesions were chosen for analysis, one primary lesion was selected from each patient for analysis. Each identified lesion was annotated with a two-dimensional (2D) square ROI with dimensions of 11 × 11 pixels on the central slice of the lesion. The ROI annotations were made on difference images created by subtracting the fourth post-contrast image from the pre-contrast image; these difference images were acquired as part of the clinical imaging protocol and allowed for better localization of the small lesions through enhancement of lesion tissue. This ROI was subsequently translated to the pre-contrast and all five post-contrast images of the T1 dynamic series. The ROI size was chosen to minimize the included amount of surrounding healthy tissue. A single encapsulating ROI was used to capture the lesion in most cases. Four lesions (3 malignant and 1 benign), whose margins exceeded the ROI boundary, were captured with two non-overlapping ROIs to preserve lesion margin information. Three examples of the small lesions used in this study are shown in Fig. 2.
Figure 2.
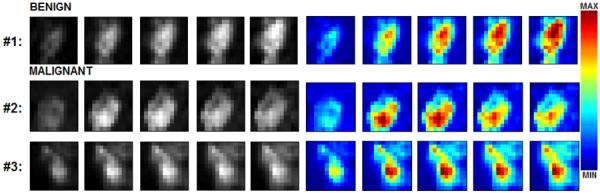
Examples of (1) benign and (2, 3) malignant lesion ROIs at five stages of contrast uptake. The histo-pathological diagnosis for the lesions were as follows: (1) fibrous mastopathy and adenosis, (2) invasive ductal carcinoma and (3) malignant-invasive ductal carcinoma (grade 1). Gray-level images were used in texture analysis after lesion segmentation; the corresponding color images better showcase the dynamic contrast uptake signatures of these lesions ((1) persistent increase for benign; (2) washout and (3) plateau patterns for malignant). The red and blue pixels correspond to high and low intensity values, respectively.
The annotated lesions were then segmented to ensure that the surrounding healthy tissue did not adversely affect subsequent texture analysis. A fuzzy C-means (FCM) approach previously proposed for lesion segmentation in DCE-MRI [23] was used to accomplish this task. The results were verified by the experienced radiologist and corrected if necessary. The FCM algorithm is an unsupervised learning technique that creates fuzzy clustering assignments to separate an input set of data points into a specified number of clusters. In the lesion segmentation problem, the FCM algorithm was used to evaluate fuzzy cluster assignments for each pixel time series (from five post-contrast images of the lesion) in the 2D square ROI as belonging to one of two clusters, lesion or healthy tissue. An empirically chosen threshold was then enforced to assign each pixel to a specific class to obtain an initial segmentation mask. Once created, this initial mask was post-processed with connected component labeling to remove any pixels belonging to blood vessels or noise incorrectly assigned to the lesion class, and to morphological hole filling operations to include any necrotic regions of the lesion that may have been incorrectly assigned to the healthy class owing to their low contrast enhancement profile. A detailed description of the FCM approach to lesion segmentation can be found in [23].
2.3 Pre-processing steps
Lesion enhancement was performed on each post-contrast ROI (Si) as Si = (Si − S0)/S0, where S0 is the corresponding pre-contrast ROI and i = {1, 2, 3, 4, 5}, with the corresponding ROI annotated on the pre-contrast lesion S0, Si = (Si − S0)/S0. This step, while effectively suppressing the healthy tissue that surrounds the lesion in the ROI, can be problematic if patient motion during the acquisition results in improper registration between the various post-contrast and pre-contrast images. Datasets used in this study had only negligible motion artifacts over time and thus, compensatory image registration steps were not required.
Following lesion enhancement, the ROIs were re-binned to 32 gray-level histogram bins between the minimum and maximum intensity limits. The choice of 32 bins for this free parameter was recommended by a previous work [8] in order to balance the need for improving counting statistics (by reducing the number of gray levels) against the corresponding discriminatory power achieved [7,24]. The minimum and maximum intensity limits were defined globally, i.e., from all lesion ROIs, within each post-contrast set. This is yet another free parameter in the pre-processing step. The global choice for the intensity limits was recommended by a previous work in this field [8].
2.4 Texture analysis
GLCMs were extracted from the lesion ROIs as described in [17]. An inter-pixel distance of d = 1 was used in generating the GLCMs; previous work has shown that including additional distances, e.g., d = {1,2,3,4,5}, does not significantly improve classification performance for most features [25]. On each ROI, GLCMs were generated in the four principal directions, i.e., 0°, 45°, 90°, and 135°. These directional GLCMs were then summed up element-wise to obtain one non-directional GLCM.
The ability of such GLCMs to capture the dynamic behavior of lesions is illustrated in Fig. 3. The GLCMs for a benign lesion show high prevalence of low intensity values initially, though more high-intensity-value regions appear in later post-contrast images. This corresponds to the persistent increase in contrast uptake, which is characteristic of such benign lesions. For a malignant lesion with washout characteristics, the GLCM initially exhibits low-intensity-value regions. During the initial rapid increase in contrast uptake (second post-contrast image), more high-intensity-value regions appear in the GLCM, but are then replaced by low-intensity-value regions in subsequent post-contrast images. The third row represents a malignant lesion with plateau characteristics. There is an increase in the high-intensity-value regions in the GLCMs from the first to the second post-contrast images. These high-intensity-value regions are retained in later post-contrast images.
Figure 3.
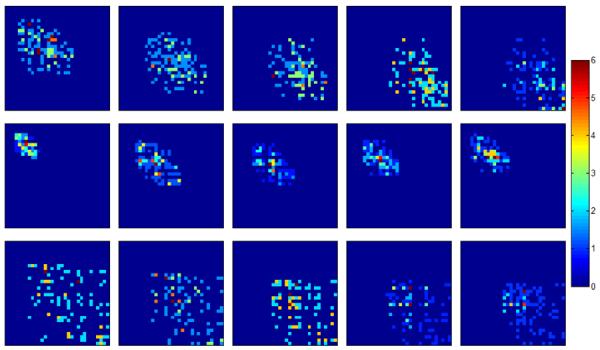
32 × 32 non-directional GLCM matrices shown as color-coded images for the three example lesions shown in Fig. 2. (Top) benign, (middle) malignant-washout, and (bottom) malignant-plateau. Each column corresponds to the respective post-contrast image. The corresponding color bar is shown on the right. Note that the top-left and bottom-right corners represent regions of low and high intensity values, respectively.
The non-directional GLCMs were then used to compute Haralick features f1–f13, as listed in Table 1 and described in [17]. Each texture feature was computed for every post-contrast image and then combined into a texture feature vector; 13 such texture feature vectors with a dimensionality of 5 were computed for each individual lesion ROI. For comparison, these 13 texture features were also extracted from the first post-contrast image alone.
Table 1.
List of 13 Haralick texture features used in this study.
| Feature label | Feature name |
|---|---|
| f1 | Angular Second Moment |
| f2 | Contrast |
| f3 | Correlation |
| f4 | Sum of Squares: Variance |
| f5 | Inverse Difference Moment |
| f6 | Sum Average |
| f7 | Sum Variance |
| f8 | Sum Entropy |
| f9 | Entropy |
| f10 | Difference Variance |
| f11 | Difference Entropy |
| f12 | Information Measures of Correlation I |
| f13 | Information Measures of Correlation II |
2.5 Feature selection
Feature selection involves identifying a subset of features from the input feature space that makes the most relevant contribution to separating the two classes of data points in the machine learning step. In this study, mutual information (MI) analysis was used for this step; the details of this algorithm can be found in [26]. MI is a nonlinear approach to feature selection that measures the information content of each feature with regards to the decision task to be performed. In this study, MI was used to identify a subset of dimensions for each of the 13 5D texture feature vectors that best contributed to the lesion character classification. This was akin to selecting a smaller subset of the post-contrast images from the available five to extract texture features from, in order to get the best classification performance from the machine learning task. Specifically, the performance obtained using all five dimensions (or post-contrast images) of the texture feature vectors was compared to that obtained using the best two and the best three dimensions of the feature vectors, as determined by MI analysis.
MI is a measure of the general independence between random variables [27]. For two random variables X and Y, the MI is defined as:
| (1) |
where entropy H(•) measures the uncertainty associated with a random variable.
The MI I(X,Y) estimates how the uncertainty of X is reduced if Y has been observed. If X and Y are independent, their MI is zero. For the ROI dataset in this study, the MI between the single texture features fi(sp) and the corresponding class labels yi was calculated by approximating the probability density function of each variable using histograms P(•):
| (2) |
Here, the number of classes nc = 2 was used; the number of histogram bins for the texture features nf was determined adaptively according to:
| (3) |
where κ is the estimated kurtosis and N is the number of ROIs in the dataset [26].
2.6 Classification
The extraction of texture features and subsequent feature selection was followed by a supervised learning step where the lesion patterns were classified as benign or malignant. In this work, the suitability of three classifiers, namely (1) SVR with a radial basis function kernel (SVRrbf), (2) SVR with a linear kernel (SVRlin), and (3) a fkNN classifier, was evaluated. In the machine learning task, SVR treats the texture features as dependent variables and their labels as the independent variable and acts as a function approximator; this function is then used in conjunction with the texture features of the test data points to predict their labels. The fkNN classifier proposed by Keller et al. [28], which models learning through density estimation, was used as a baseline for comparison with SVR.
In this study, 70% of the data was used for the training phase and the remaining 30% served as an independent test set. The training data was sub-sampled from the complete dataset in such a manner that at least 40% of each class (benign and malignant) was represented. Special care was taken to ensure that lesion ROIs extracted from a given patient were used either as training or test data to prevent any potential for biased training. To ensure the integrity of the independent test set, global intensity limits for pre-processing were determined using lesion ROIs from the training data alone. The best dimensions of the texture feature vectors were selected by evaluating the mutual information criteria of the training data alone; this ensured that label information for the test data was not used prior to the classification task. Pre-processing the two smallest lesions in this dataset with 32 bins and global intensity limits resulted in a constant image. Extracting texture features related to correlation, i.e., f3 and f12, as defined in [17], yielded undefined values. For these two features alone, the problematic lesion ROIs were excluded from the classification task and results reported in such instances are marked accordingly.
In the training phase, models were created from labeled data by employing a random sub-sampling cross-validation strategy, where the training set is further split into 70% training samples and 30% validation samples. The purpose of the training was to determine the optimal classifier parameters, i.e., those that best capture the boundaries between the two classes of lesion patterns. The free parameters for the classifiers used in this study were the number of nearest neighbors k for fkNN, the cost parameter for SVRlin and SVRrbf, and the shape parameter of the radial basis function kernel of SVRrbf. During the testing phase, the optimized classifier predicted the label (benign or malignant) of lesion ROIs in the independent test dataset. An ROC curve was generated and used to compute the AUC, which served as a measure of classifier performance. This process was repeated 100 times, resulting in an AUC distribution for each feature set.
2.7 Statistical analysis
A Wilcoxon signed-rank test was used to compare two AUC distributions. Significance thresholds were adjusted for multiple comparisons using the Holm-Bonferroni correction to achieve an overall type I error rate (significance level) of less than α (where α = 0.05) [29,30]. Texture, classifier, and statistical analysis were implemented using Matlab 2008b (The MathWorks, Natick, MA).
3. Results
Figure 4 shows the classification performance obtained using the fkNN, SVRlin, and SVRrbf classifiers for texture features f4 and f6, which exhibited the best overall AUC values (0.82). The SVRrbf classifier significantly outperformed the other classifiers when these texture features were extracted from all five post-contrast images. When texture features were extracted from the first post-contrast image alone, the performance of fkNN was comparable to that of SVRrbf. Since the highest AUC values were observed with the SVRrbf classifier, all other results reported in this study used SVRrbf for the classification task.
Figure 4.
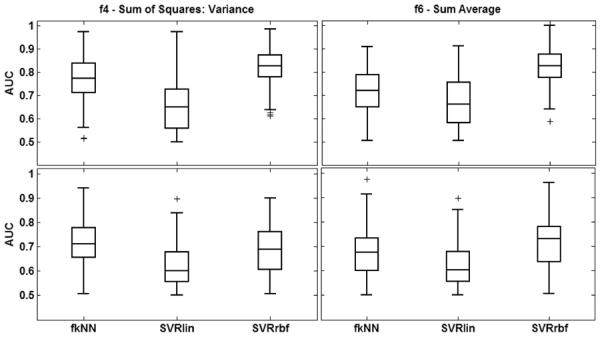
Classification performance of texture features f4 (Sum of Squares: Variance) (left) and f6 (Sum Average) (right) for different classifiers when all five post-contrast images are used for the classification task. The top and bottom rows correspond to texture feature vectors extracted using all five post-contrast image (ALL) and texture features extracted from the first post-contrast image alone (P1), respectively. For each distribution, the central mark corresponds with the median and the edges are the 25th and 75th percentiles. SVRrbf significantly outperforms the other classifiers (p < 0.05).
Table 2 compares the classification performance obtained with texture features extracted from all five post-contrast images to that obtained when texture analysis involves the first post-contrast image alone. Six of thirteen texture features showed statistically significant improvements in classification performance (p < 0.05) when the dynamic texture quantification approach was used to characterize lesion enhancement. In particular, texture features f4 and f6 had AUC values of 0.82, the highest observed in this study. Only texture feature f2 significantly deteriorated in performance when the dynamic texture quantification approach was used.
Table 2.
Classification performance (mean AUC ± standard deviation) of texture features extracted from the 1st post-contrast image (P1) compared to that of texture feature vectors extracted from all five post-contrast images (ALL).
| Feature | P1 | ALL | p-value | threshold |
|---|---|---|---|---|
| f1 | 0.66 ± 0.09 | 0.66 ± 0.09 | 0.2868 | -- |
| f2 | 0.68 ± 0.12 | 0.63 ± 0.09 | 0.0002 | 0.0071 |
| f3 | 0.60 ± 0.08 | 0.60± 0.08x | 0.4172 | -- |
| f4 | 0.69 ± 0.10 | 0.82 ± 0.08 | < 0.0001 | 0.0042 |
| f5 | 0.69 ± 0.10 | 0.69 ± 0.10 | 0.4006 | -- |
| f6 | 0.72 ± 0.10 | 0.82 ± 0.08 | < 0.0001 | 0.0038 |
| f7 | 0.67 ± 0.10 | 0.79 ± 0.09 | < 0.0001 | 0.0045 |
| f8 | 0.61 ± 0.08 | 0.72 ± 0.11 | < 0.0001 | 0.0050 |
| f9 | 0.68 ± 0.09 | 0.72 ± 0.11 | < 0.0001 | 0.0063 |
| f10 | 0.67 ± 0.11 | 0.64 ± 0.10 | 0.0345 | -- |
| f11 | 0.73 ± 0.10 | 0.74 ± 0.08 | 0.6182 | -- |
| f12 | 0.62 ± 0.08 | 0.64 ± 0.09x | 0.3120 | -- |
| f13 | 0.60 ± 0.07 | 0.67 ± 0.10 | < 0.0001 | 0.0056 |
Significantly higher AUC values in each row are marked in bold.
Results marked with an 'x' indicate numbers that were obtained after excluding two lesions for reasons mentioned in the text. Significantly better classification performance is observed for 6 of 13 features when texture features extracted from all five post-contrast images are combined as feature vectors.
Table 3 compares the classification performance obtained with texture features extracted from all five post-contrast images to that obtained with texture features from the best 2 and best 3 post-contrast images, as determined by MI criteria. Of particular interest are texture features f4 and f6, whose classification performance did not significantly deteriorate when certain post-contrast images were dropped from the texture analysis. In fact, none of the features exhibited significant changes when extracted from a subset of post-contrast images compared to being extracted from all available post-contrast images.
Table 3.
Classification performance (mean AUC ± standard deviation) of texture features extracted from all five post-contrast images (ALL) compared to that of texture feature vectors extracted from the best two (Best 2) and best three (Best 3) post-contrast images identified using mutual information.
| Feature | ALL | Best 2 | Best 3 |
|---|---|---|---|
| f1 | 0.66 ± 0.09 | 0.67 ± 0.09 | 0.67 ± 0.10 |
| f2 | 0.63 ± 0.09 | 0.64 ± 0.10 | 0.64 ± 0.10 |
| f3 | 0.60 ± 0.08x | 0.59 ± 0.08x | 0.60 ± 0.07x |
| f4 | 0.82 ± 0.08 | 0.82 ± 0.09 | 0.83 ± 0.09 |
| f5 | 0.69 ± 0.10 | 0.69 ± 0.10 | 0.69 ± 0.10 |
| f6 | 0.82 ± 0.08 | 0.82 ± 0.09 | 0.81 ± 0.09 |
| f7 | 0.79 ± 0.09 | 0.80 ± 0.09 | 0.80 ± 0.08 |
| f8 | 0.72 ± 0.11 | 0.72 ± 0.10 | 0.71 ± 0.10 |
| f9 | 0.72 ± 0.11 | 0.72 ± 0.11 | 0.73 ± 0.10 |
| f10 | 0.64 ± 0.10 | 0.63 ± 0.10 | 0.63 ± 0.09 |
| f11 | 0.74 ± 0.08 | 0.74 ± 0.08 | 0.74 ± 0.09 |
| f12 | 0.64 ± 0.09x | 0.64 ± 0.10x | 0.63 ± 0.09x |
| f13 | 0.67 ± 0.10 | 0.66 ± 0.09 | 0.66 ± 0.09 |
Significantly higher AUC values in each row are marked in bold.
Results marked with an 'x' indicate numbers that were obtained after excluding two lesions for reasons mentioned in the text. Classification performance does not deteriorate when texture features are extracted from the best two or three post-contrast images alone.
Since MI criteria were used to rank the contribution of each post-contrast image to the classification task, the number of times each post-contrast image was ranked in the top 2 or top 3 was recorded for 100 different sub-sampled sets of training data during the classification task. These results are presented as histograms in Fig. 5 for texture features f4 and f6 to better understand the contribution of different post-contrast images to the classification task. As shown in the figure, when selecting the three best features, the third and fourth post-contrast images seem to be selected most frequently and often in combination with either the first or the second post-contrast image. Similar trends were observed while selecting the two best features, where again the third and fourth post-contrast images were selected most often.
Figure 5.

Histogram analysis for texture features f4 (Sum of Squares: Variance) (top, black) and f6 (Sum Average) (bottom, gray), showing the number of times each post-contrast image is ranked 1st, 2nd, or 3rd by MI criteria in 100 sets of sub-sampled training data (used in the machine learning task).
4. Discussion
The primary goal of this study was to improve the performance of classifying diagnostically challenging lesions, specifically those considered small (mean lesion diameter of 1.05 cm), from DCE-MRI. Previous approaches to CADx have involved quantifying the lesion enhancement pattern of breast lesions on the first post-contrast image alone using texture features [7–9]. However, this approach does not provide the best classification performance for small lesions (as confirmed in this study), where improved performance can have a significant clinical value. To address this problem, a dynamic texture quantification method where the lesion enhancement pattern is quantified dynamically by extracting texture features from all five post-contrast images of the lesion was proposed here. The results show that such an approach can significantly improve the performance of the lesion character classification task. Improved classification performance can contribute to reducing (1) the likelihood of performing false-positive biopsies of benign lesions, thereby eliminating the surgical risks associated with the biopsy, and (2) missed breast cancers developing from misdiagnosed malignant lesions, while also enabling earlier diagnosis of suspicious lesions.
In this work, 13 Haralick texture features were extracted from all five post-contrast images of the lesion and combined to form 13 5D lesion characterizing vectors. As shown in Table 2 and Fig. 2, such texture feature vectors significantly outperform texture features extracted from the first post-contrast image alone. This can be attributed to the ability of the dynamic quantification approach to capture variation in textural information as the contrast uptake dynamics change. As shown in Fig. 6 (first column), features f4 and f6 do not provide any discrimination between the benign and malignant lesions shown in Fig. 1 when extracted from the first post-contrast image alone. However, when these features are computed as a function of time (or contrast uptake), more distinct differences between the two classes of lesions are observed. This is believed to be the primary reason behind the improvements observed in lesion character classification.
Figure 6.
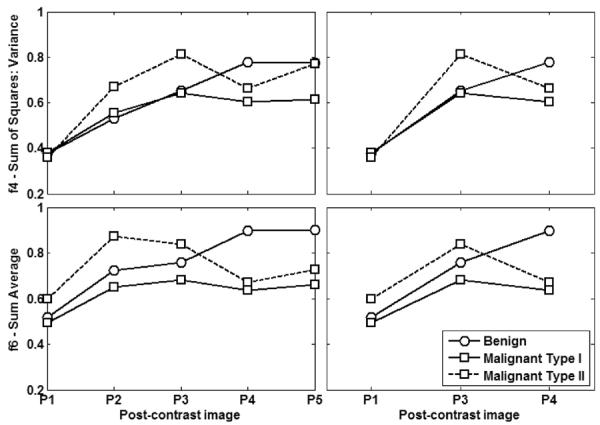
Left column shows features f4 (Sum of Squares: Variance) (top) and f6 (Sum Average) (bottom) as computed from all five post-contrast images (P1–5) for the lesions shown in Fig. 2. The lesions are harder to distinguish when texture features are extracted from the first post-contrast image alone. However, the behavior of these features over time is distinctly different in benign and malignant lesions. When such feature curves include P1, P3, and P4 alone (right column), the differences in the behavior of both lesion classes is much more obvious. Note that malignant type I corresponds to washout (#2 on Fig. 2) and malignant type II corresponds to plateau (#3 on Fig. 2).
Other researchers have characterized lesion enhancement through dynamic evaluation of spatial variation. Gilhuijs et al. described lesion enhancement through descriptors such as margin enhancement and radial gradient analysis as a funtion of space and time [31]. Zheng et al. performed a discrete fourier transform (DFT) of the pixel time series and created enhancement maps using the first three DFT coefficients; texture features were subsequently extracted from these enhancement maps [32]. Buelow et al. computed a serial enhancement ratio (SER) for each lesion pixel and then used features describing the variation in SER values to predict lesion character [33]. The results reported in the present study are expected to be poorer owing to the collection of small lesions used in the character classification task. However, they still present an improvement over previous work with a similar dataset of small lesions [14,15]. Of the six features that showed an improvement with the proposed dynamic quantification approach, f6 (Sum Average) and f9 (Entropy) had been previously identified as texture features that are associated with homogeneity (f6) and heterogeneity (f9) of the enhancement pattern [9]. Features f4 (Sum of Squares: Variance), f7 (Sum Variance), and f8 (Sum Entropy) are all variance or entropy measures, suggesting that homogeneity/ heterogeneity of the enhancement pattern is a key distinguishing factor between benign and malignant lesions. While it is expected that some of these texture features are correlated, previous work has shown that f4 and f9 are uncorrelated [9]. Given the small size of lesions in this dataset, where characteristics of the enhancement pattern may not be easily perceived, these results strongly motivate the use of such texture features in a CADx approach to assist radiologists in diagnosing small lesions.
Different classifiers were evaluated in terms of their classification performance when used in conjunction with these high-dimensional texture feature vectors. SVRrbf was found to yield the best overall classification performance, as shown in Fig. 4. It must also be noted here that the classification performance of the fkNN classifier, when texture features were extracted from the first post-contrast image alone, was comparable to that of SVRrbf. This suggests that SVRrbf better uses the supplementary information provided by texture features extracted from later post-contrast images in distinguishing between benign and malignant lesions.
To gain further insight into which post-contrast images contribute the most relevant information to the classification task, different post-contrast images were ranked during the supervised learning task; the results are shown in Fig. 5 for texture features f4 and f6 and reflect the general trend for most features analyzed in this study. The most important contribution to the reported classification performance comes from the inclusion of texture features extracted from the third and fourth post-contrast images. When selecting the three best features using MI criteria, the best combination of features, as shown in Fig. 5, includes texture features extracted from the third and fourth post-contrast images, as well as one of the earlier post-contrast images (first or second). This is further illustrated in Fig. 6 (second column), which shows the texture feature curves for f4 and f6 over the first, third, and fourth post-contrast images for the lesions in Fig. 1. The curves in Fig. 6 are similar to characteristic benign and malignant dynamic time curve signatures (Fig. 1), which suggests that such dynamic texture quantification does in fact incorporate dynamic characteristics of these lesions in the classification task. In conventional dynamic analysis, evaluating the dynamic behavior of a lesion involves extracting the time series of every lesion pixel. Once extracted, these pixel time series are used to generate a single time series that represents the behavior of the lesion as a whole. While there are several approaches for accomplishing this (taking the mean, using different clustering approaches, etc.), the proposed approach represents the entire lesion enhancement pattern at each time point by a single texture feature. The results obtained here are in agreement with those in a previous work that suggested that such texture feature curves may perform better than signal intensity time curves as they are more robust to bias field and intensity non-standardness [34].
This work revealed certain limitations of using Haralick texture features for characterizing the enhancement pattern in small lesions from DCE-MRI. While 14 texture features are described in [17], feature f14 (Maximal Correlation Coefficient) was undefined for the lesions used in this study. This is a consequence of re-binning the gray levels found in small lesions to 32 bins, which results in certain bins remaining empty; under such conditions, f14 is undefined. Another problem was encountered with the smallest two lesions in the dataset; re-binning to 32 gray levels resulted in a constant image for the ROIs for certain post-contrast images. Since the variance for constant images is zero, texture features f3 (Correlation) and f12 (Information Measure of Correlation I) were undefined for these lesions. In future research, statistical texture features can be replaced with more recent texture analysis techniques that characterize the underlying gray-level pattern through geometric information.
One limitation of this study was that only exams with negligible motion artifacts over time were included in the texture analysis and classification tasks. Future studies with less stringent inclusion criteria for exams could incorporate sophisticated nonlinear image registration methods as part of the pre-processing to compensate for motion artifacts over time [35]. Another limitation of this study regards the use of 2D lesion ROIs in texture analysis rather than 3D lesion volumes; volumetric analysis could not be performed with the image datasets used in this study owing to the anisotropy of the pixels involved. Although previous research has shown that volumetric analysis of lesions improves classification performance [8], arguments have been made against acquiring breast images with isotropic voxels owing to the longer imaging time involved as well as the smaller coverage of the area being imaged [9]. However, even with these limitations, this work demonstrates the applicability of dynamic texture quantification of the lesion enhancement pattern for automated character classification using DCE-MRI.
5. Conclusion
This study evaluated the performance of automated character classification of diagnostically challenging lesions, specifically those considered small (mean lesion diameter of 1.05 cm), and found that it can be significantly improved through dynamic texture quantification of the lesion enhancement pattern, i.e., extracting texture features from the lesion enhancement pattern on all five post-contrast images. The results suggest that such an approach to automated lesion character classification for DCE-MRI could be helpful in clinical practice. Larger controlled trials need to be conducted in order to validate the clinical applicability of this approach.
Acknoledgements
This research was funded in part by the National Institutes of Health (NIH) Award R01-DA-034977, the Clinical and Translational Science Award 5-28527 within the Upstate New York Translational Research Network (UNYTRN) of the Clinical and Translational Science Institute (CTSI), University of Rochester, and by the Center for Emerging and Innovative Sciences (CEIS), a NYSTAR-designated Center for Advanced Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Benjamin Mintz for his assistance with lesion annotation, and Prof. M.F. Reiser, FACR, FRCR, from the Department of Radiology, University of Munich, Germany, for his support.
References
- [1].Wiberg MK, Aspelin P, Sylvan M, Bone B. Comparison of lesion size estimated by dynamic MR imaging, mammography and histopathology in breast neoplasms. Eur. Radiol. 2003;13:1207–1212. doi: 10.1007/s00330-002-1718-2. [DOI] [PubMed] [Google Scholar]
- [2].Boetes C, Mus RDM, Holland R, Barentsz JO, Strijk SP, Wobbes T, Hendriks JHCL, Ruys SHJ. Breast-tumors-comparative accuracy of MR-imaging relative to mammography and US for demonstrating extent. Radiology. 1995;197:743–747. doi: 10.1148/radiology.197.3.7480749. [DOI] [PubMed] [Google Scholar]
- [3].Kuhl CK, Mielcareck P, Klaschik S, Leutner C, Wardelmann E, Gieseke J, Schild HH. Dynamic breast MR imaging: Are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology. 1999;211:101–110. doi: 10.1148/radiology.211.1.r99ap38101. [DOI] [PubMed] [Google Scholar]
- [4].Chen W, Giger ML, Lan L, Bick U. Computerized interpretation of breast MRI: Investigation of enhancement-variance dynamics. Med. Phys. 2004;31:1076–1082. doi: 10.1118/1.1695652. [DOI] [PubMed] [Google Scholar]
- [5].Wismüller A, Meyer-Base A, Lange O, Schlossbauer T, Kallergi M, Reiser MF, Leinsinger G. Segmentation and classification of dynamic breast magnetic resonance image data. J. Electron. Imaging. 2006;15:0130201–01302013. [Google Scholar]
- [6].Meinel LA, Stolpen AH, Berbaum KS, Fajardo LL, Reinhardt JM. Breast MRI lesion classification: Improved performance of human readers with a backpropagation neural network computer-aided diagnosis (CAD) system. J. Magn. Reson. Imaging. 2007;25:89–95. doi: 10.1002/jmri.20794. [DOI] [PubMed] [Google Scholar]
- [7].Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn. Reson. Med. 2003;50:92–98. doi: 10.1002/mrm.10496. [DOI] [PubMed] [Google Scholar]
- [8].Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn. Reson. Med. 2007;58:562–571. doi: 10.1002/mrm.21347. [DOI] [PubMed] [Google Scholar]
- [9].Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad. Radiol. 2008;15:1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wedegärtner U, Bick U, Wörtler K, Rummeny E, Bongartz G. Differentiation between benign and malignant findings on MR-mammography: usefulness of morphological criteria. Eur. Radiol. 2001;11:1645–1650. doi: 10.1007/s003300100885. [DOI] [PubMed] [Google Scholar]
- [11].Baum F, Fischer U, Vosshenrich R, Grabbe E. Classification of hypervascularized lesions in CE MR imaging of the breast. Eur. Radiol. 2002;12:1087–1092. doi: 10.1007/s00330-001-1213-1. [DOI] [PubMed] [Google Scholar]
- [12].Szabo BK, Aspelin P, Wiberg MK, Bone B. Dynamic MR imaging of the breast. Analysis of kinetic and morphologic diagnostic criteria. Acta Radiol. 2003;44:379–386. doi: 10.1080/j.1600-0455.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- [13].Fischer DR, Wurdinger S, Boettcher J, Malich A, Kaiser WA. Further signs in the evaluation of magnetic resonance mammography: a retrospective study. Invest. Radiol. 2005;40:430–435. doi: 10.1097/01.rli.0000167138.52283.aa. [DOI] [PubMed] [Google Scholar]
- [14].Leinsinger G, Schlossbauer T, Scherr M, Lange O, Reiser M, Wismüller A. Cluster analysis of signal-intensity time course in dynamic breast MRI: does unsupervised vector quantization help to evaluate small mammographic lesions? Eur. Radiol. 2006;16:1138–1146. doi: 10.1007/s00330-005-0053-9. [DOI] [PubMed] [Google Scholar]
- [15].Schlossbauer T, Leinsinger G, Wismüller A, Lange O, Scherr M, Meyer-Baese A, Reiser M. Classification of small contrast enhancing breast lesions in dynamic magnetic resonance imaging using a combination of morphological criteria and dynamic analysis based on unsupervised vector-quantization. Invest. Radiol. 2008;43:56–64. doi: 10.1097/RLI.0b013e3181559932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lerski RA, Straughan K, Schad LR, Boyce D, Blüml S, Zuna I. Tissue characterization by magnetic-resonance spectroscopy and imaging-results of a concerted research-project of the European-economic-community. Introduction, objectives, and activities. Magn. Reson. Imaging. 1993;11:809–815. doi: 10.1016/0730-725x(93)90198-m. [DOI] [PubMed] [Google Scholar]
- [17].Haralick RM, Shanmuga K, Dinstein I. Textural features for image classification. IEEE T. Sys. Man Cyb. 1973;Smc3:610–621. [Google Scholar]
- [18].Haralick RM. Statistical and structural approaches to texture. Proc. IEEE. 1979;67:786–804. [Google Scholar]
- [19].Ikeda DM, Hylton NM, Kinkel K, Hochman MG, Kuhl CK, Kaiser WA, Weinreb JC, Smazal SF, Degani H, Viehweg P, Barclay J, Schnall MD. Development, standardization, and testing of a lexicon for reporting contrast-enhanced breast magnetic resonance imaging studies. J. Magn. Reson. Imaging. 2001;13:889–895. doi: 10.1002/jmri.1127. [DOI] [PubMed] [Google Scholar]
- [20].D'Orsi CJ, Bassett LW, Berg WA, Feig SA, Jackson JA, Kopans D. Breast Imaging Reporting and Data System Breast Imaging Atlas. American College of Radiology; Reston: 2003. [Google Scholar]
- [21].Drucker H, Burges CJC, Kaufman L, Smola A, Vapnik V. Support vector regression machines. Adv. Neur. In. 1996;9:155–161. [Google Scholar]
- [22].Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- [23].Chen W, Giger ML, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad. Radiol. 2006;13:63–72. doi: 10.1016/j.acra.2005.08.035. [DOI] [PubMed] [Google Scholar]
- [24].Kjaer L, Ring P, Thomsen C, Henriksen O. Texture analysis in quantitative MR imaging-tissue characterization of normal brain and intracranial tumors at 1.5-T. Acta Radiol. 1995;36:127–135. [PubMed] [Google Scholar]
- [25].Nagarajan MB, Huber MB, Schlossbauer T, Leinsinger G, Wismüller A. Analysis of breast lesions on contrast-enhanced magnetic resonance images using high-dimensional texture features. Proc. Int. Soc. for Opt. Engi Med. Imag. 2010;7624:1G1–1G8. [Google Scholar]
- [26].Tourassi GD, Frederick ED, Markey ML, Floyd CE., Jr. Application of the mutual information criterion for feature selection in computer-aided diagnosis. Med. Phys. 2001;28:2394–2402. doi: 10.1118/1.1418724. [DOI] [PubMed] [Google Scholar]
- [27].Duda RO, Hart PE, Stork DG. Pattern Classification. Wiley-Interscience Publication; 2000. [Google Scholar]
- [28].Keller JM, Gray MR, Givens JA. A fuzzy K-nearest neighbor algorithm. IEEE T. Sys. Man Cyb. 1985;15:580–585. [Google Scholar]
- [29].Wright SP. Adjusted P-values for simultaneous inference. Biometrics. 1992;48:1005–1013. [Google Scholar]
- [30].Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- [31].Gilhuijs KGA, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic- resonance imaging. Med. Phys. 1998;25:1647–1654. doi: 10.1118/1.598345. [DOI] [PubMed] [Google Scholar]
- [32].Zheng Y, Englander S, Baloch S, Zacharaki EI, Fan Y, Schnall MD, Shen D. STEP: Spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Med. Phys. 2009;36:3192–3204. doi: 10.1118/1.3151811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Buelow T, Saalbach A, Bergtholdt M, Wiemker R, Buurman H, Meinel LA, Newstead G. Heterogeneity of kinetic curve parameters as indicator for the malignancy of breast lesions in DCE MRI. Proc. Int. Soc. for Opt. Engi Med. Imag. 2010;7624:1H1–1H12. [Google Scholar]
- [34].Agner S, Soman S, Libfield E, McDonald M, Thomas S, Englander K, Rosen M, Chin D, Nosher J, Madabhushi A. Textural kinetics: A novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J. Digit. Imaging. 2011;24:446–463. doi: 10.1007/s10278-010-9298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]


