Abstract
The exact sequence of events in biosyntheses of natural products is essential not only to understand and learn from nature's strategies and tricks to assemble complex natural products, but also for yield optimization of desired natural products, and for pathway engineering and muta-synthetic preparation of analogues of bioactive natural products. Biosyntheses of natural products were classically studied applying in vivo experiments, usually by combining incorporation experiments with stable-isotope labeled precursors with cross-feeding experiments of putative intermediates. Later genetic studies were dominant, which consist of gene cluster determination and analysis of gene inactivation experiments. From such studies various biosynthetic pathways were proposed, to a large extent just through in silico analyses of the biosynthetic gene clusters after DNA sequencing. Investigations of the complex biosyntheses of the angucycline group anticancer drugs landomycin, jadomycin and gilvocarcin revealed that in vivo and in silico studies were insufficient to delineate the true biosynthetic sequence of events. Neither was it possible to unambiguously assign enzyme activities, especially where multiple functional enzymes were involved. However, many of the intriguing ambiguities could be solved after in vitro reconstitution of major segments of these pathways, and subsequent systematic variations of the used enzyme mixtures. This method has been recently termed ‘combinatorial biosynthetic enzymology’.
Introduction
Landomycins and gilvocarcins belong to the angucycline group of natural products, the largest group of polyketide-derived natural products, rich in biological activities and intriguing chemical scaffolds. The unusual repetitive saccharide decoration pattern and high degree of deoxygenation in the aglycone moiety stimulated biosynthetic inquisitiveness for the landomycins, while particularly the oxidative framework rearrangements by post-polyketide synthase tailoring oxidoreductases triggered extensive biosynthetic studies of the kinamycin, jadomycin and gilvocarcin biosyntheses [1•].
With so far 26 members, the landomycins [2–5], produced by Streptomyces cyanogenus S-136 and Streptomyces globisporus 1912, with the principal products landomycin A (1, Figure 1) and landomycin E (2), respectively, are one of the largest and most studied families among the typical angucyclines. Their structures differ from each other in their saccharidal length and composition as well as oxygenation pattern of the aglycone moiety, for example, landomycin Z (3) [1,6,7]. The landomycins have received much attention for their structures [8–14,15••] and anticancer activities, partly because of the fact that they are not substrates of multi-drug resistance efflux pumps. They interfere with DNA synthesis, but do not bind directly to DNA. Their exact cell target and mechanisms-of-action remain elusive [16–21].
Figure 1.
Representative chemical structures of the landomycin and gilvocarcin groups of anticancer drugs.
Although already discovered in the mid-1950s [22], the chemical structures of the gilvocarcin group of potent anticancer agents remained elusive until 1981, when an X-ray structure [23] revealed the relative configuration of gilvocarcin M (4), which in turn led to the structure determination of various other members of this group of [24–26]. Gilvocarcin V (5 = toromycin [27,28], anandimycin [24,29]), the major and most active metabolite of Streptomyces griseoflavus Gö 3592 as well as of various other Streptomyces species, is usually produced along with the minor congeners gilvocarcin M (4) and E (6) that vary with respect to their 8-substitution [24,27,30,31]. Several gilvocarcin analogues (e.g. 7–9, Figure 1), now collectively called the gilvocarcin group of natural products, have been isolated from different Streptomyces species all containing the characteristic polyketide-derived benzo[d]naphtho[1,2-b]pyran-6-one chromophore and different C-glycosidically linked sugar units [32–36]. The group is known for their strong antitumor activities, unique mode of action and low toxicity [25,34,37]. The 8-vinyl side chain of the benzo[d]naphtho[1,2-b]pyran-6-one moiety undergoes photoactivated [2+2]-cycloaddition with thymine residues of DNA under irradiating conditions with low energy UV or visible light [29,38,39], and the sugar moiety appears to be essential for the observed gilvocarcin-mediated cross-linking of histone H3 or heat shock protein GRP78 with DNA resulting in the disruption of DNA replication and transcription [40••]. Gilvocarcins also exhibit strong antibacterial [22] and antiviral properties [41]; however, the inherent poor solubility of these molecules appeared to be a major obstacle toward their development as therapeutics [1,42–44].
Incorporation experiments with stable-isotope labeled precursors
The landomycin pathway was investigated by incorporation studies and genetic experiments. The carbon backbone of landomycinone (10) is derived from 10 acetate and malonate units. Experiments involving 18O-labeled molecular oxygen (18O2) and CH3C18O18OH indicated that only two of the six oxygen atoms of 1 (Figure 2), namely those 1-position and 8-position, originate from the polyketide building blocks [3], however, the 18O-incorporation experiments failed to further solve the intriguing biosynthesis of the aglycone [45].
Figure 2.
Incorporation experiments with stable-isotope labeled precursors.
Incorporation studies with isotope-labeled precursors [46–49] suggested that the unique benzo[d]naphtho[1,2-b]pyran-6-one chromophore of the gilvocarcins emerges from a polyketide-derived angucyclinone intermediate through a complex oxidative rearrangement process; however, the details and exact sequence of events and involvement of enzymes remained elusive (Figure 2) [50].
Gene cluster analysis and conclusions from gene inactivation and gene complementation experiments
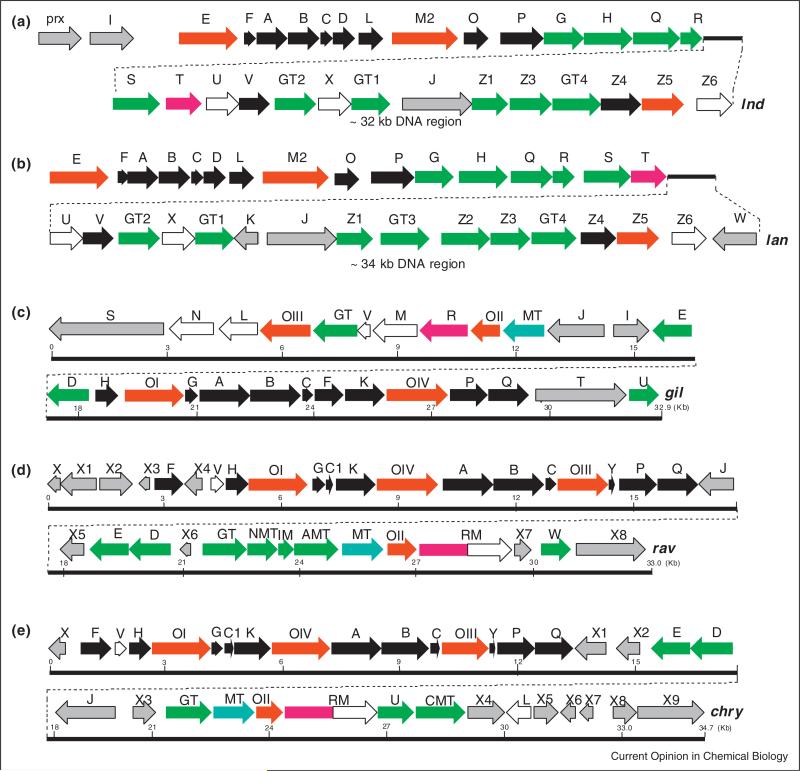
Two gene clusters of landomycin producers (lan from the 1-producer S. cyanogenus, and lnd from the 2-producer S. globisporus) were cloned (Figure 3) [51–55,56•,57•]. The clusters are almost identically organized, with only three biosynthetic genes missing in the lnd cluster, namely equivalents of lanK, lanGT3, and lanZ2. Many of the functions of the gene-products could be unambiguously assigned thorough gene inactivation/complementation studies. The entire glycosylation sequence was solved, many of the regulatory aspects of the landomycin bio-synthesis could be deduced [4,53–55,56•,57•,58•,59–67], and a couple of new genetically engineered landomycins were generated [19,68–71]. However, the deduction of the post-PKS tailoring oxidoreductase catalyzed reactions of the aglycone biosynthesis remained ambiguous, although some information was gained from accumulated products upon gene inactivation (see Figure 4 for examples) [69,72,73]. However, the exact substrates of the involved enzymes and the exact sequence of their actions remained ambiguous.
Figure 3.
Gene clusters of the landomycin and gilvocarcin groups: (a,b): landomycin E (lnd) and landomycin A (lan) gene clusters in comparison. (c–e) Gilvocarcin (gil), ravidomycin (rav) and chrysomycin (chry) gene clusters in comparison. PKS and other genes encoding the polyketide frame = black, sugar biosynthesis and glycosyltransfer = green, methyltransfer = cyan, regulatory and resistance = gray, oxygenases/oxidoreductases = red, unknown = white.
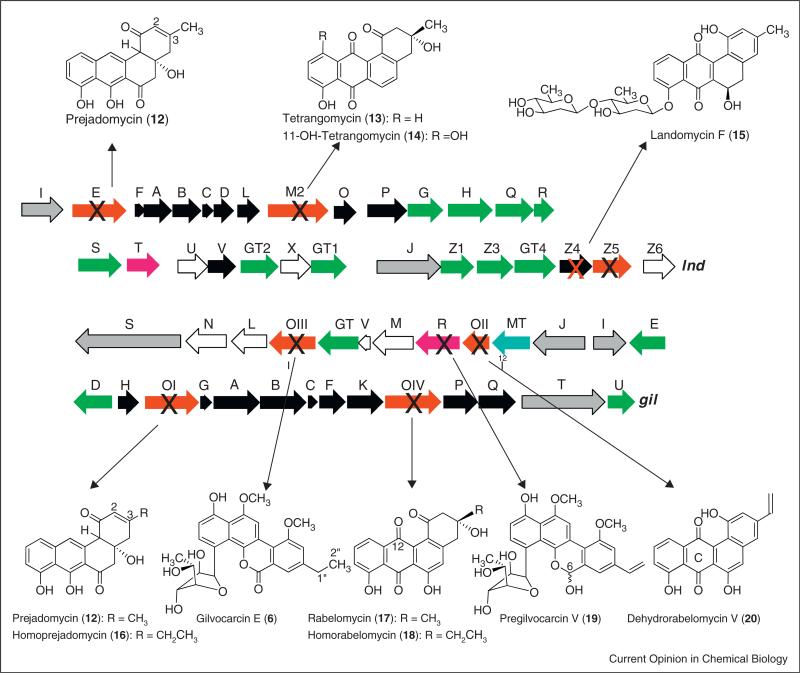
Figure 4.
Inactivation of genes of the landomycin (above) and gilvocarcin (below) biosynthesis. The major accumulation product/s of the corresponding mutant strains is/are shown.
The gilvocarcin biosynthetic gene cluster (gil) was cloned and heterologously expressed [30], and the clusters of chrysomycin (chry) and ravidomycin (rav) were cloned and analyzed (Figure 3) [74•,75•,76,77•,78•]. The functions of the post-PKS gene products were assigned after gene inactivation, complementation, cross-feeding experiments along with few in vitro and in vivo studies of activity of individual enzymes or enzyme mixtures [75•,79–82,83•]. However, many of the inactivation mutants accumulated biosynthetic shunt products, and left ambiguity over post-PKS biosynthetic steps (for some examples, see Figure 4) [49,80,82]. The exact sequence of events and the intriguing oxidative rearrangement mechanism remained ambiguous. It was not even possible to assign the enzyme responsible for the crucial C–C-bond cleavage. Instead, the production of the angucyclinones 12/16, 17/18 and 20, respectively (Figure 4), by three different oxygenase-deficient mutants led to the hypothesis that all three oxygenases (GilOI, GilOII and GilOIV) might form a multi-enzyme complex that catalyzes a concerted pathway which includes a C–C bond cleavage that eventually leads to the formation of the unique gilvocarcin scaffold [80]. The inactivation of oxidoreductase gene gilR led to the accumulation of mainly the intermediate pregilvocarcin V (19) [81,83•], and GilR has been shown to catalyze the very last step of the gilvocarcin biosynthesis by converting pregilvocarcin V (19) to the final lactone containing product gilvocarcin V 5 (Figures 4 and 6).
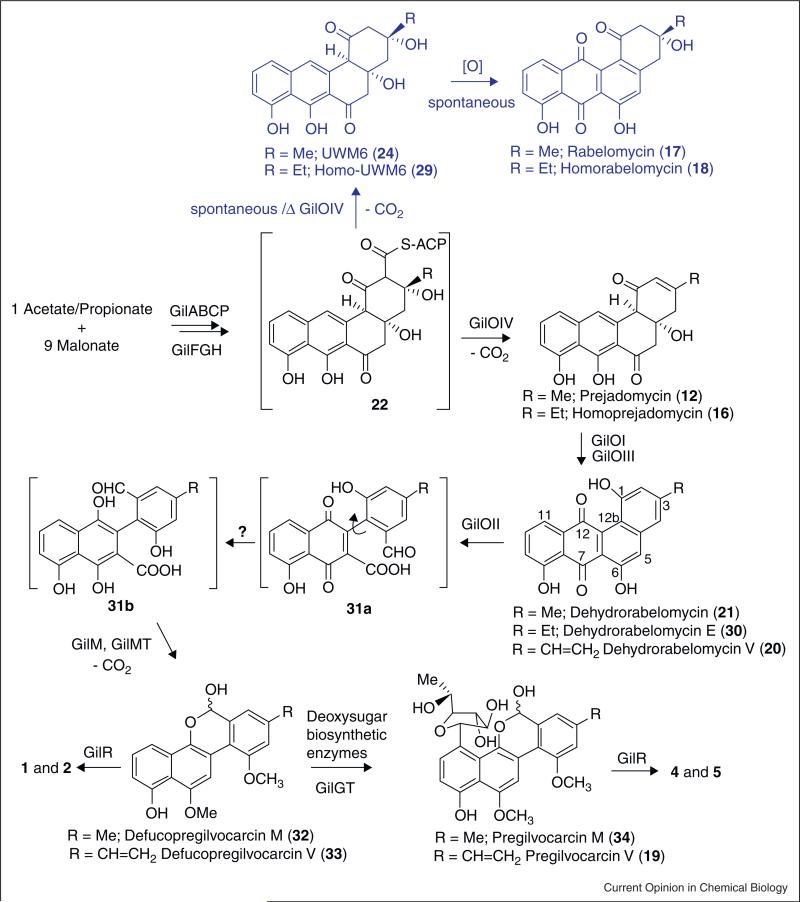
Figure 6.
Gilvocarcin biosynthesis. The pathway leading to the different gilvocarcins is depicted in black, while a shunt pathway branch is shown in blue.
Studies of single enzymes
All enzymes postulated to be involved in the biosynthesis of the landomycin aglycone (LanM2, LanE, LanV, and LanZ4/Z5) were interrogated with the available angucyclinones 12, 13, 17, 21, 24–26 (for structures see Figures 4 and 5) [84,85•], many of which were previously proposed to be intermediates of the landomycin pathway [77•]. Interrogating the overexpressed enzymes LanM2 and LanV, respectively, with tetrangomycin (13), tetrangulol (25), UWM6 (24), rabelomycin (17), dehydrorabelomycin (21), 11-deoxylandomycinone (26), and prejadomycin (12) showed no conversions whatsoever, proving that neither of these angucyclinones was a substrate for either enzyme. When the same group of angucyclinones was interrogated with the co-expressed enzyme pair LanZ4/Z5, only tetrangomycin (13) and 11-deoxylandomycinone (26) were converted, the former into its 11-hydroxylated derivative 14, the latter into tetrangulol (25) as well as into 27. However, landomycinone (10) was not formed, suggesting that 10 is not an intermediate of the pathway. Nevertheless, the experiments suggested for the first time two different functions of lanZ4/Z4, acting either (i) as 11-hydroxylase or (ii) as 5,6-dehydratase, or both. Overexpressed LanE was able to convert prejadomycin (12) and UWM6 (24) into dehydrorabelomycin (21) and rabelomycin (17), respectively. However, the conversion of 24 into 17 was also known to occur non-enzymatically. Yet, the experiments showed LanE to posses dual functionality, namely as (i) 4a,12b-dehydratase and (ii) as 12-oxygenase. The experiments also determined the cofactors for each of these enzymes (Kharel MK, Pahari P, Shaaban KA, Wang G, Morris C, Rohr J: Elucidation of post-PKS tailoring steps involved in landomycin biosynthesis. Org Biomol Chem, unpublished data) [85•].
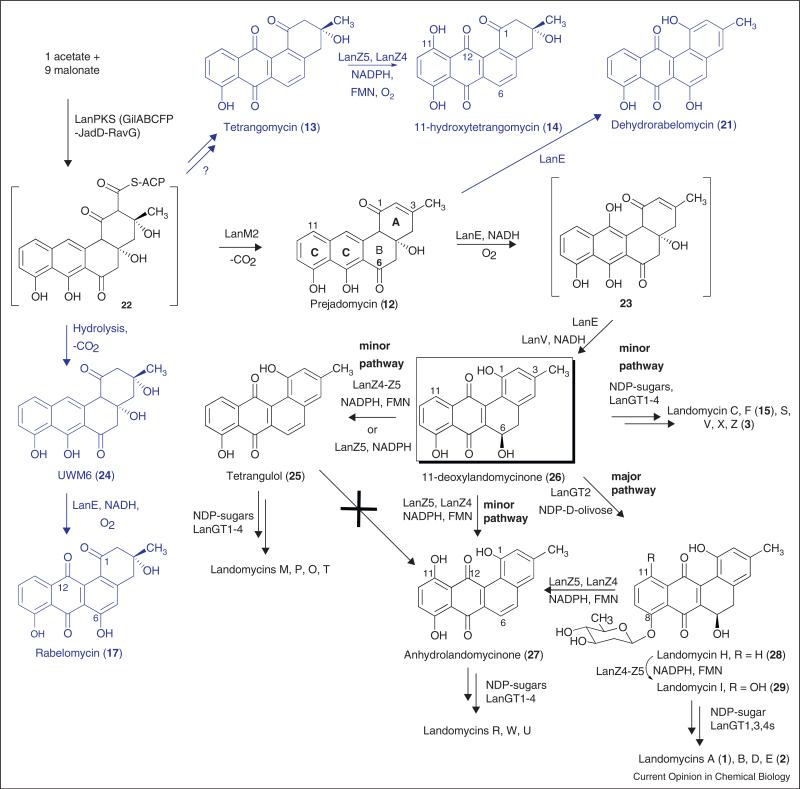
Figure 5.
Landomycin biosynthesis. The pathway leading to the three different series of landomycins, with different aglycones, is depicted in black, while shunt pathways are shown in blue.
Work with overexpressed oxidoreductase enzymes of the gilvocarcin pathway also revealed little, since in most cases the substrates remained obscure. From all the oxygenases, only GilOI reacted, and converted UWM6 (24) into rabelomycin (17). This confirmed previous genetic studies on the related jadomycin bio-synthetic pathway [86], in which it was found that oxygenases JadH, the equivalent of GilOI, and JadF, the equivalent of GilOIV, both seem to possess dual functionality (i.e. also catalyzed dehydration reactions in ring A of the benz[a]anthracene derived tetracyclic angucyclinone — JadF/GilOIV a 2,3-dehydration; JadH/GilOI a 4a,12b-dehydration, Figure 6). Since rabelomycin (17) was not an intermediate of the gilvocarcin biosynthesis, it was deduced that most, if not all, of the observed reactions might also occur spontaneously.
Overexpressed oxidoreductase GilR was studied, and was shown to catalyze the very last step of gilvocarcin biosynthesis, namely the conversion of pregilvocarcin to gilvocarcin [81], showing that this enzyme establishes the central lactone moiety from a hemiacetal. The latter was the first hint that the hypothetical 5,6-bond cleavage product of the (unknown) proposed angucyclinone intermediate is an aldehyde-acid rather than the originally proposed di-acid shown in Figure 2. The enzyme contains covalently bound FAD, which needs to be regenerated by dissolved oxygen. The 1.65 Å-resolution crystal structure of GilR showed this enzyme as dimer with bi-covalently linked FAD at the 8a-position and 6-position mediated through His-65 and Cys-125, respectively.
Combinatorial biosynthetic enzymology
It is necessary to have genes from several related pathways available to be able to overcome problems with the overexpression of soluble enzymes. For the studies of angucycline pathways, we had genes available from six different pathways: landomycin (lan and lnd), urdamycin (urd), jadomycin (jad), gilvocarcin (gil), ravidomycin (rav) and chrysomycin (chry). An angucyclinone expression system was constructed, which contained the minimal PKS genes for the production of UWM6 (24), an unstable angucyclinone that was believed to be a key intermediate of many pathways, including the jadomycin, urdamycin, landomycin and gilvocarcin pathways. UWM6 is unstable and converts non-enzymatically into rabelomycin (17). This PKS cassette contained genes from the gilvocarcin, ravidomycin and jadomycin pathways, and expressed the following enzymes: GilAB (co-expressed ketosynthase/chain length factor), RavC (acyl carrier protein), GilF (PKS-associated ketoreductase), JadD, RavG (cyclases) and GilP (MCAT) [75•]. For the characterization of the oxidoreductases involved in the landomycinone biosyn-thesis, both, in vivo experiments utilizing minimal PKS enzymes and different combinations of post-PKS tailoring enzymes in a heterologous host, and in vitro studies utilizing individual isolated enzymes were applied (Kharel MK, Pahari P, Shaaban KA, Wang G, Morris C, Rohr J: Elucidation of post-PKS tailoring steps involved in landomycin biosynthesis. Org Biomol Chem, unpublished data) [85•]. While the in vivo studies (not described here) were complicated by secondary catalytic events and therefore led to little new knowledge, the in vitro experiments delineated the sequence of events toward the four different angucyclinones used as agly-cones in the landomycin group: landomycinone (10), 11-deoxy-landomycinone (26), anhydro-landomycinone (27) and tetrangulol (25). With an aim to elucidate the functional role of each of the Lan-post-PKS enzymes (LanM2, E, V, Z4 and Z5) and the timing of their actions, various expression cassettes were constructed, in which individual genes encoding these oxidoreductase enzymes were co-expressed with PKS genes alone, and then in groups. A heterologous expression host, Streptomyces lividans TK64, was used for the co-expression.
Using the above-mentioned PKS-cassette mixture of enzymes along with a mixture of 5 presumed post-PKS enzymes LanM2, LanE, LanV and LanZ4/Z5 led to the production of tetrangulol (25). An interrogation of just these 5 post-PKS enzymes with potential angucyclinone intermediates showed a conversion of prejadomycin (12) to tetrangulol (25) via 11-deoxylandomycinone (26). However, the same mixture of enzymes was unable to convert UWM6 (24) into any of the aglycones of the landomycin group, and led to the production of 17 (as with LanE alone, see above), which proved that neither 24 nor 17 is an intermediate of the landomycin pathway. This enzyme mixture was then systematically varied and reduced, which revealed the following results: prejadomycin (12) was converted by the mixture of LanE and LanV (and necessary cofactors) into 26. This confirmed LanE's role as 4a,12b-dehydratase/12-oxygenase, and also confirmed LanV's suggested role as 6-ketoreductase [87]. Adding LanZ4/Z5 to this mixture resulted in the formation of tetrangulol (25), while the addition of LanM2 did not lead to any change in the product profile. The experiments clearly proved that neither UWM6 (24), nor tetrangomycin (13) nor rabelomycin (17) are true intermediates of the landomycin pathway as suggested earlier after cross-feeding experiments with whole organisms, which apparently possess secondary enzymatic activities to ‘recycle’ these shunt products. Prejadomycin (12) is the first verified angucyclinone intermediate, and is converted into 11-deoxylandomyci-none (26) through the activities of two enzymes, LanE and LanV. Exclusive production of prejadomycin (12) instead of UWM6 (24, the product of the PKS enzymes alone) through co-expression of LanM2 with the PKS enzymes indicated that LanM2 is the first tailoring enzyme, responsible for catalyzing the 2,3-dehydration step — the same reaction found for JadF in jadomycin biosynthesis [86,88].
Furthermore, it was concluded that this elimination happens simultaneously with the cleavage of the thereby generated angucyclinone 12 from the ACP. This parallels the findings we recently established for GilOIV and JadF, respectively, in the gilvocarcin and jadomycin biosyntheses, respectively [89••], and is corroborated by the fact that LanM2 cannot convert 24 into 12 in vitro. Overall, this thorough study of function and substrates specifici-ties of landomycin post-PKS tailoring enzymes led to a revised pathway for landomycin biosynthesis, shown in Figure 5.
To gain a better understanding of the role of individual enzymes involved in the gilvocarcin biosynthesis and to identify the actual biosynthetic sequence of events, the focus of the studies was also shifted toward in vitro studies with enzymes mixtures. The first step was to establish the above mentioned PKS-enzyme cassette that enabled the production of UWM6 (24), also believed to be a gilvocarcin pathway intermediate. Then the scope of the investigations was expanded involving selected post-PKS enzymes. It was assumed that the minimal set of the enzymes GilOI, GilOII, GilOIV (oxygenases), GilM, GilMT (methyl transferases), and oxidoreductase GilR was necessary to establish both the oxidative rearrangement and follow-up reactions toward the simplest gilvocarcin skeleton, the defuco-gilvocarcin scaffold. Like many PKS enzymes, one of the gil post-PKS enzymes, GilOIV, resisted all efforts to express in soluble form, and was replaced by its homologue JadF from the jadomycin biosynthetic pathway [80]. Also, instead of the using the slowly acting enzyme GilH for the reductase necessary to regenerate cofactor FADH2, Escherichia coli flavin reductase Fre was used [90].
To test the activity of the PKS enzymes, all seven PKS enzymes (GilAB, RavC, GilF, GilP, JadD, RavG) were incubated with one equivalent of acetyl-CoA and nine equivalents of malonyl-CoA in the presence of cofactor NADPH. To ensure enough supply of NADPH for the reaction, the well established NADPH-regeneration system consisting of glucose-6-phosphate (G6P) and glucose-6-phosphate dehydrogenase (G6PDH) from E. coli was used. As expected, the products UWM6 (24) and rabelomycin (17) were formed. With fully functional PKS-enzymes in hand, a cocktail of 15 enzymes was mixed, consisting of the above listed PKS enzymes and of all of the anticipated post-PKS enzymes (GilOI, GilOII, JadF, GilM, GilMT, GilR, and E. coli Fre). This enzyme mixture was incubated with acetyl-CoA, malonyl-CoA, cofactors NADPH, FAD, SAM (S-adenosyl methionine) along with the NADPH regeneration system G6P and G6PDH, and was shown to produce defucogilvocarcin M (32, identical with the isolated 32 from the previously characterized GilGT-minus mutant [82]) along with shunt product rabelomycin (17, Figure 6).
Once the enzymes required for the production of 32 were established, the enzyme mixture was systematically reduced to deduce the events of the complex oxidative rearrangement cascade. Two of the previously proposed pathway intermediates UWM6 (24) and prejadomycin (12) were used as substrates instead of acetyl-CoA and malonyl-CoA in the multi-enzyme reaction described above. Prejadomycin 12 was completely converted into 32 both in the presence and absence of the PKS-enzymes. This confirmed that 12 is indeed a true intermediate of the gilvocarcin M (4) pathway, and that the designated PKS enzymes have no role in converting 12 to 32, while the same enzyme mixture failed to convert 24 to 32, in contradiction to previously proposed hypotheses [86,91]. Instead, rabelomycin (17) was exclusively produced. This led to the new hypothesis that one or more of the designated post-PKS enzymes may only act on angucyclinone substrates still tethered to the acyl carrier protein, and that UWM6 (24) is a shunt product formed by spontaneous hydrolysis and decarboxylation of the ACP-tethered intermediate 22 (Figure 6). Shunt product rabelomycin (17) in turn is produced spontaneously from 24 by aerial oxidation and spontaneous dehydration, like observed before [75•].
Further reducing the enzyme mixture eventually revealed the point at which a stable, non-ACP-tethered pathway intermediate emerges from the mixture. Removal of GilOI, GilOII, or JadF, respectively, from the enzyme cocktail produced prejadomycin (12), dehydrorabelomycin (21) and rabelomycin (17), respectively, whereas removal of GilM/GilMT led to unidentified products. Finally, addition of individual oxygenases to the PKS enzyme mixture showed that only one additional enzyme, namely JadF (soluble replacement of GilOIV), needed to be added to the PKS-enzymes to convert acetyl-CoA/malonyl-CoA to the pathway intermediate prejadomycin (12). GilOIV/JadF was previously suggested to be a bifunctional enzyme with an (unclear) oxygenase and a 2,3-dehydratase activity [86]. The enzyme mix reactions performed here confirmed that GilOIV/JadF indeed catalyzes the 2,3-dehydration, but moreover also serves as key enzyme bridging PKS and post-PKS reactions by catalyzing the hydrolysis and decarboxylation of the ACP-tethered angucycline 22 to 12 (Figure 6).
To determine the fate of prejadomycin (12), the study was reduced to experiments interrogating single or more of the remaining post-PKS enzymes, using 12 as the substrate. These studies identified GilOI as the enzyme responsible for the next reaction, the oxidation of 12 to dehydrorabelomycin (21), proving that GilOI is a bifunctional enzyme which performs a 4a,12b-dehydration and C-12 oxygenation. Identical observations and conclusions were reported previously for JadH, the homologous enzyme of the jadomycin pathway [86,91,92], and now also for LanE (see above).
Furthermore, an enzyme mixture consisting of the remaining enzymes GilOII, GilM, GilMT, GilR along with E. coli flavin reductase Fre [90] was able to convert 21 completely into 32 confirming 21 as a true intermediate of the gilvocarcin pathway. Since GilOII was the only oxygenase present in this reaction mixture, it also was concluded that GilOII is the enzyme responsible for the C–C bond cleavage reaction, which is a key reaction for the establishment of the unique dibenzochromen-6-one backbone of the gilvocarcin group of natural products. However, it still remains ambiguous whether GilM, GilMT or GilR may partake in this cleavage reaction. Overall, these results led to a major revision of gilvocarcin biosynthesis overwriting all earlier proposed hypotheses that suggested GilOI and/or GilOIV or both in complex with GilOII to be required for the C–C-bond cleavage [80]. Since earlier cross-complementation results have shown that GilOIV and GilOI are functionally equivalent to JadF and JadH of the jadomycin pathway, respectively, and that both pathways may share an identical mechanism for the C5–C6 bond cleavage of an angucyclinone intermediate, it is now evident the GilOII-equivalent of the jad pathway, JadG [80], is likely responsible for the C6–C6-bond cleavage in the jadomycin pathway. Replacing GilOII with JadG in the post-PKS enzyme cocktail and incubation of this enzyme mixture with substrate 21 confirmed this conclusion, since the substrate was converted into 34 demonstrating the functional equivalence of GilOII and JadG. Currently, investigations are in progress to seek clarification of the remaining ambiguous biosynthetic steps of the gilvocarcin pathway, beyond the C–C-bond cleavage. It also remains to be shown whether the reaction mechanism of the cofactor free oxygenase GilOII [30] involves sequential monooxygenase reactions (e.g. 5-hydroxylation followed by Baeyer–Villiger reaction [80]), or rather a dioxygenase mechanism, similar to the recently discussed mechanism discussed for the cofactor independent dioxygenase DpgC [93,94,95••] involved in the biosynthesis of the dihydroxyphenylglyoxylate building block of glycoprotein antibiotics. It also remains unclear how GilOII is regenerated, and which of the remaining enzymes performs the reduction of 31a to 31b. Possible candidates are GilR, GilM, or GilOII, some of these may act co-dependently.
Conclusions
After all classical in vivo methods failed to reveal unambiguous results, in vitro pathway reconstitution and systematic recombination of its enzyme components (combinatorial biosynthetic enzymology) was critical to delineating the complex post-polyketide tailoring steps toward the landomycin aglycone mixture and the oxidative rearrangement cascade of the gilvocarcin and jadomycin biosyntheses. The studies also allowed for the first time to unambiguously assign many of the involved enzymes, which contradicted many of the earlier drawn hypotheses and conclusions. Although pathway reconstitution had been used before to understand the biosyn-thesis of vitamin B12 [96••], tetracenomycin C [97••], and enterocin [98], the highlighted examples here were the first, in which a systematic enzyme mix and match approach was used to delineate the sequence of biosynthetic events.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1•.Kharel MK, Pahari P, Shepherd MD, Tibrewal N, Nybo SE, Shaaban KA, Rohr J. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [This is the newest review on angucycline group natural products encompassing the period of 1997–2010, focusing predominantly on biosynthetic studies and synthetic strategies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henkel T, Rohr J, Beale JM, Schwenen L. Landomycins, new angucycline antibiotics from Streptomyces sp. I. Structural studies on landomycins A–D. J Antibiot. 1990;43:492–503. doi: 10.7164/antibiotics.43.492. [DOI] [PubMed] [Google Scholar]
- 3.Weber S, Zolke C, Rohr J, Beale JM. Investigations of the biosynthesis and structural revision of landomycin A. J Org Chem. 1994;59:4211–4214. [Google Scholar]
- 4.Gromyko O, Rebets Y, Ostash B, Luzhetskyy A, Fukuhara M, Bechthold A, Nakamura T, Fedorenko V. Generation of Streptomyces globisporus SMY622 strain with increased landomycin E production and its initial characterization. J Antibiot. 2004;57:383–389. doi: 10.7164/antibiotics.57.383. [DOI] [PubMed] [Google Scholar]
- 5.Matseliukh BP, Lavrinchuk V. The isolation and characteristics of mutant Streptomyces globisporus 1912 defective for landomycin E biosynthesis. Mikrobiol Z. 1999;61:22–27. [PubMed] [Google Scholar]
- 6.Shaaban KA, Srinivasan S, Kumar R, Damodaran C, Rohr J. Landomycins P-W, cytotoxic angucyclines from Streptomyces cyanogenus S-136. J Nat Prod. 2011;74:2–11. doi: 10.1021/np100469y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaaban KA, Stamatkin C, Damodaran C, Rohr J. 11-Deoxylandomycinone and landomycins X–Z, new cytotoxic angucyclin(on)es from a Streptomyces cyanogenus K62 mutant strain. J Antibiot. 2011;64:141–150. doi: 10.1038/ja.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Sulikowski GA. Synthesis of the hexasaccharide fragment of landomycin A: application of glycosyl tetrazoles and phosphites in the synthesis of a deoxyoligosaccharide. J Am Chem Soc. 1998;120:1392–1397. [Google Scholar]
- 9.Kirschning A, Chen G-W, Drager G, Schuberth I, Tietze LF. Syntheses and biological evaluation of new glyco-modified angucyclin-antibiotics. Bioorg Med Chem. 2000;8:2347–2354. doi: 10.1016/s0968-0896(00)00166-8. [DOI] [PubMed] [Google Scholar]
- 10.Roush WR, Bennett CE. A highly stereoselective synthesis of the landomycin A hexasaccharide unit. J Am Chem Soc. 2000;122:6124–6125. [Google Scholar]
- 11.Roush WR, Bennett CE, Roberts SE. Studies on the synthesis of landomycin A: synthesis and glycosidation reactions of l-rhodinosyl acetate derivatives. J Org Chem. 2001;66:6389–6393. doi: 10.1021/jo015756a. [DOI] [PubMed] [Google Scholar]
- 12.Roush WR, Neitz RJ. Studies on the synthesis of landomycin A. Synthesis of the originally assigned structure of the aglycone, landomycinone, and revision of the structure. J Org Chem. 2004;69:4906–4912. doi: 10.1021/jo049426c. [DOI] [PubMed] [Google Scholar]
- 13.Yu B, Wang P. Efficient synthesis of the hexasaccharide fragment of landomycin A: using phenyl 2,3-O-thionocarbonyl-1-thioglycosides as 2-deoxy-beta-glycoside precursors. Org Lett. 2002;4:1919–1922. doi: 10.1021/ol0259286. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M, O'Doherty GA. De novo synthesis of the trisaccharide subunit of landomycins A and E. Org Lett. 2008;10:2283–2286. doi: 10.1021/ol800697k. [DOI] [PubMed] [Google Scholar]
- 15••.Yang X, Fu B, Yu B. Total synthesis of landomycin A, a potent antitumor angucycline antibiotic. J Am Chem Soc. 2011;133:12433–12435. doi: 10.1021/ja205339p. [This is the first total synthesis of landomycin A, employing a clever strategy of mono-glycosylation followed by synthesizing the remaining pentasaccharide and combing these two synthons, thereby circumventing the problems of attaching a complex and instable hexasaccharide to the phenolic OH of the aglycone.] [DOI] [PubMed] [Google Scholar]
- 16.Crow RT, Rosenbaum B, Smith R, 3rd, Guo Y, Ramos KS, Sulikowski GA. Landomycin A inhibits DNA synthesis and G1/S cell cycle progression. Bioorg Med Chem Lett. 1999;9:1663–1666. doi: 10.1016/s0960-894x(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 17.Depenbrock H, Bornschlegl S, Peter R, Rohr J, Schmid P, Schweighart P, Block T, Rastetter J, Hanauske AR. Assessment of antitumor activity of landomycin A (NSC 6399187-A). Ann Hematol. 1996;73(Suppl. II):A80/316. [Google Scholar]
- 18.Korynevska A, Heffeter P, Matselyukh B, Elbling L, Micksche M, Stoika R, Berger W. Mechanisms underlying the anticancer activities of the angucycline landomycin E. Biochem Pharmacol. 2007;74:1713–1726. doi: 10.1016/j.bcp.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Ostash B, Korynevska A, Stoika R, Fedorenko V. Chemistry and biology of landomycins, an expanding family of polyketide natural products. Mini Rev Med Chem. 2009;9:1040–1051. doi: 10.2174/138955709788922593. [DOI] [PubMed] [Google Scholar]
- 20.Panchuk R, Korynevska A, Ostash B, Osyp Y, Fedorenko V, Stoika R. Study of the mechanism of landomycin E action on mammalian cells. Visn L’viv Univ Ser Biol. 2004;35:54–59. [Google Scholar]
- 21.Panchuk R, Korynevska A, Stoika R. Effect of landomycin E on expression of mRNA coding for transforming growth factor beta ligands and specific receptors in MCF-7 cells. Exp Oncol. 2005;27:330–332. [PubMed] [Google Scholar]
- 22.Strelitz F, Flon H, Ashenov IN. Chrysomycin: a new antibiotic substance for bacterial viruses. J Bacteriol. 1955;69:183–280. doi: 10.1128/jb.69.3.280-283.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirayama N, Takahashi K, Shirahata K, Ohashi Y, Sasada Y. Crystal and molecular structure of antibiotic gilvocarcin M. Bull Chem Soc Jpn. 1981;54:1338–1342. [Google Scholar]
- 24.Balitz DM, O'Herron FA, Bush J, Vyas DM, Nettleton DE, Grulich RE, Bradner WT, Doyle TW, Arnold E, Clardy J. Antitumor agents from Streptomyces anandii: gilvocarcins V, M and E. J Antibiot. 1981;34:1544–1555. doi: 10.7164/antibiotics.34.1544. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto M, Okubo S, Tomita F, Marumo H. Gilvocarcins, new antitumor antibiotics. 3. Antitumor activity. J Antibiot. 1981;34:701–707. doi: 10.7164/antibiotics.34.701. [DOI] [PubMed] [Google Scholar]
- 26.Findlay JA, Liu J-S, Radics L, Rakhit S. The structure of ravidomycin. Can J Chem. 1981;59:3018–3020. [Google Scholar]
- 27.Hatano K, Hagashide E, Shibata M, Kameda Y, Horii SKM. Toromycin: a new antibiotic produced by Streptomyces collinas subsp. albescens subsp. nov. Agric Biol Chem. 1980;44:1157–1163. [Google Scholar]
- 28.Jain TC, Simolike GC, Jackman LM. Structure and stereochemistry of toromycin: studies of its acid-catalyzed rearrangement. Tetrahedron. 1983;39:599–605. [Google Scholar]
- 29.McGee LR, Misra R. Gilvocarcin photobiology. isolation and characterization of the DNA photoadduct. J Am Chem Soc. 1990;112:2386–2389. [Google Scholar]
- 30.Fischer C, Lipata F, Rohr J. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J Am Chem Soc. 2003;125:7818–7819. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Yoshida M, Tomita F, Shirahata K. Gilvocarcins, new antitumor antibiotics. 2. Structural elucidation. J Antibiot. 1981;34:271–275. doi: 10.7164/antibiotics.34.271. [DOI] [PubMed] [Google Scholar]
- 32.Sehgal SN, Czerkawski H, Kudelski A, Pandev K, Saucier R, Vezina C. Ravidomycin (AY-25,545), a new antitumor antibiotic. J Antibiot. 1983;36:355–361. doi: 10.7164/antibiotics.36.355. [DOI] [PubMed] [Google Scholar]
- 33.Weiss U, Yoshihira K, Highet RJ, White RJ, Wei TT. The chemistry of the antibiotics chrysomycin A and B. Antitumor activity of chrysomycin A. J Antibiot. 1982;35:1194–1201. doi: 10.7164/antibiotics.35.1194. [DOI] [PubMed] [Google Scholar]
- 34.Li YQ, Huang XS, Ishida K, Maier A, Kelter G, Jiang Y, Peschel G, Menzel KD, Li MG, Wen ML, et al. Plasticity in gilvocarcin-type C-glycoside pathways: discovery and antitumoral evaluation of polycarcin V from Streptomyces polyformus. Org Biomol Chem. 2008;6:3601–3605. doi: 10.1039/b808633h. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima S, Kojiri K, Suda H, Okanishi M. New antitumor substances, BE-12406A and BE-12406B, produced by a streptomycete. II. Structure determination. J Antibiot. 1991;44:1061–1064. doi: 10.7164/antibiotics.44.1061. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita N, Shin-ya K, Furihata K, Hayakawa Y, Seto H. New ravidomycin analogues, FE35A and FE35B, apoptosis inducers produced by Streptomyces rochei. J Antibiot. 1998;51:1105–1108. doi: 10.7164/antibiotics.51.1105. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto A, Fujiwara Y, Elespuru RK, Hanawalt PC. Photoactivated gilvocarcin V induces DNA-protein crosslinking in genes for human ribosomal RNA and dihydrofolate reductase. Photochem Photobiol. 1994;60:225–230. doi: 10.1111/j.1751-1097.1994.tb05095.x. [DOI] [PubMed] [Google Scholar]
- 38.Elespuru RK, Gonda SK. Activation of antitumor agent gilvocarcins by visible light. Science. 1984;223:69–71. doi: 10.1126/science.6229029. [DOI] [PubMed] [Google Scholar]
- 39.Tse-Dinh Y-C, McGee LR. Light-induced modifications of DNA by gilvocarcin V and its aglycone. Biochem Biophys Res Commun. 1987;143:808–812. doi: 10.1016/0006-291x(87)90320-2. [DOI] [PubMed] [Google Scholar]
- 40••.Matsumoto A, Hanawalt PC. Histone H3 and heat shock protein GRP78 are selectively cross-linked to DNA by photoactivated gilvocarcin V in human fibroblasts. Cancer Res. 2000;60:3921–3926. [This paper revealed a novel mechanism of action unique for the gilvocarcin group of anticancer agents.] [PubMed] [Google Scholar]
- 41.Lytle CD, Wagner SJ, Prodouz KN. Antiviral activity of gilvocarcin V plus UVA radiation. Photochem Photobiol. 1993;58:818–821. doi: 10.1111/j.1751-1097.1993.tb04976.x. [DOI] [PubMed] [Google Scholar]
- 42.Hua DH, Saha S. Gilvocarcins. J R Netherl Chem Soc. 1995;114:341–355. [Google Scholar]
- 43.Hosoya T, Takashiro E, Matsumoto T, Suzuki K. Total synthesis of the gilvocarcins. J Am Chem Soc. 1994;116:1004–1015. [Google Scholar]
- 44.Futagami S, Ohashi Y, Imura K, Hosoya T, Ohmori K, Matsumoto T, Suzuki K. Total synthesis of ravidomycin: revision of absolute and relative stereochemistry. Tetrahedron Lett. 2000;41:1063–1067. [Google Scholar]
- 45.Wohlert S-E, Bechthold A, Beninga C, Henkel T, Holzenkämpfer M, Kirschning A, Oelkers C, Ries M, Weber S, Weissbach U, et al. Investigations on the biosynthesis of landomycin A. In: Diederichsen U, Lindhorst TK, Westermann B, Wessjohann LA, editors. Bioorganic Chemistry. Wiley-VCH; 1999. pp. 305–312. [Google Scholar]
- 46.Takahashi K, Tomita F. Gilvocarcins, new antitumor antibiotics. 5. Biosynthesis of gilvocarcins: incorporation of 13C-labeled compounds into gilvocarcin aglycones. J Antibiot. 1983;36:1531–1535. doi: 10.7164/antibiotics.36.1531. [DOI] [PubMed] [Google Scholar]
- 47.Carter GT, Fantini AA, James JC, Borders DB, White RJ. Biosynthesis of ravidomycin. Use of 13C–13C double quantum NMR to follow precursor incorporation. Tetrahedron Lett. 1984;25:255–258. [Google Scholar]
- 48.Carter GT, Fantini AA, James JC, Borders DB, White RJ. Biosynthesis of chrysomycins A and B. Origin of the chromophore. J Antibiot. 1985;38:242–248. doi: 10.7164/antibiotics.38.242. [DOI] [PubMed] [Google Scholar]
- 49.Liu T, Fischer C, Beninga C, Rohr J. Oxidative rearrangement processes in the biosynthesis of gilvocarcin V. J Am Chem Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 50.Keyes RF, Kingston DGI. Stereochemistry of hydrogen loss on formation of the vinyl group in the biosynthesis of ravidomycin. J Org Chem. 1989;54:6127–6129. [Google Scholar]
- 51.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 52.Von Mulert U, Luzhetskyy A, Hofmann C, Mayer A, Bechthold A. Expression of the landomycin biosynthetic gene cluster in a PKS mutant of Streptomyces fradiae is dependent on the coexpression of a putative transcriptional activator gene. FEMS Microbiol Lett. 2004;230:91–97. doi: 10.1016/S0378-1097(03)00861-9. [DOI] [PubMed] [Google Scholar]
- 53.Ostash B, Rebets Y, Yuskevich V, Luzhetskyy A, Tkachenko V, Fedorenko V. Targeted disruption of Streptomyces globisporus lndF and lndL cyclase genes involved in landomycin E biosynthesis. Folia Microbiol (Praha) 2003;48:484–488. doi: 10.1007/BF02931329. [DOI] [PubMed] [Google Scholar]
- 54.Pankevych K, Kruegel H, Fedorenko VA. Cloning and sequencing of a putative positive transcription regulator gene of landomycin E biosynthetic gene cluster of Streptomyces globisporus 1912. Visn L’viv Univ Ser Biol. 2001;27:97–105. [Google Scholar]
- 55.Rebets Y, Ostash B, Luzhetskyy A, Hoffmeister D, Braña AF, Méndez C, Salas JA, Bechthold A, Fedorenko V. Production of landomycins in Streptomyces globisporus 1912 and Streptomyces cyanogenus S136 is regulated by genes encoding putative transcriptional activators. FEMS Microbiol Lett. 2003;222:149–153. doi: 10.1016/S0378-1097(03)00258-1. [DOI] [PubMed] [Google Scholar]
- 56•.Rebets Y, Ostash B, Luzhetskyy A, Kushnir S, Fukuhara M, Bechthold A, Nashimoto M, Nakamura T, Fedorenko V. DNA-binding activity of LndI protein and temporal expression of the gene that upregulates landomycin E production in Streptomyces globisporus 1912. Microbiology. 2005;151:281–290. doi: 10.1099/mic.0.27244-0. [See annotation to Ref. [57•].] [DOI] [PubMed] [Google Scholar]
- 57•.Rebets YV, Ostash BO, Fukuhara M, Nakamura T, Fedorenko VO. Expression of the regulatory protein LndI for landomycin E production in Streptomyces globisporus 1912 is controlled by the availability of tRNA for the rare UUA codon. FEMS Microbiol Lett. 2006;256:30–37. doi: 10.1111/j.1574-6968.2005.00087.x. [Ref. [56•] and this paper as well as Ref. [66] describe a novel, temporary upregulation process, which is interesting in context with new hypotheses of secondary metabolites as signal molecules.] [DOI] [PubMed] [Google Scholar]
- 58•.Luzhetskyy A, Fedoryshyn M, Dürr C, Taguchi T, Novikov V, Bechthold A. Iteratively acting glycosyltransferases involved in the hexasaccharide biosynthesis of landomycin A. Chem Biol. 2005;12:725–729. doi: 10.1016/j.chembiol.2005.05.008. [Here, two iteratively acting glycosyltransferases are key to understand how a hexasaccharide can be biosynthesized by only four glycosyltransferases, see also Ref. [67].] [DOI] [PubMed] [Google Scholar]
- 59.Luzhetskyy A, Liu T, Fedoryshyn M, Ostash B, Fedorenko V, Rohr J, Bechthold A. Function of lanGT3, a glycosyltransferase gene involved in landomycin A biosynthesis. ChemBioChem. 2004;5:1567–1570. doi: 10.1002/cbic.200400123. [DOI] [PubMed] [Google Scholar]
- 60.Luzhetskyy A, Taguchi T, Fedoryshyn M, Dürr C, Wohlert SE, Novikov V, Bechthold A. LanGT2 catalyzes the first glycosylation step during landomycin A biosynthesis. ChemBioChem. 2005;6:1406–1410. doi: 10.1002/cbic.200500018. [DOI] [PubMed] [Google Scholar]
- 61.Luzhetskyy A, Vente A, Bechthold A. Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides. Mol Biol Syst. 2005;1:117–126. doi: 10.1039/b503215f. [DOI] [PubMed] [Google Scholar]
- 62.Ostash B, Ostash I, Zhu L, Kharel MK, Luzhetskyy A, Bechthold A, Walker S, Rohr J, Fedorenko V. Properties of lanK-based regulatory circuit involved in landomycin biosynthesis in Streptomyces cyanognes S136. Russ J Genet. 2010;46:530–535. [PMC free article] [PubMed] [Google Scholar]
- 63.Ostash I, Ostash B, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V. Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol Lett. 2008;285:195–202. doi: 10.1111/j.1574-6968.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 64.Ostash I, Ostash B, Walker S, Fedorenko V. Proton-dependent transporter gene lndJ confers resistance to landomycin E in Streptomyces globisporus. Genetika. 2007;43:1032–1037. [PubMed] [Google Scholar]
- 65.Ostash I, Rebets Y, Ostash B, Kobylyanskyy A, Myronovskyy M, Nakamura T, Walker S, Fedorenko V. An ABC transporter encoding gene lndW confers resistance to landomycin E. Arch Microbiol. 2008;190:105–109. doi: 10.1007/s00203-008-0367-5. [DOI] [PubMed] [Google Scholar]
- 66.Rebets Y, Dutko L, Ostash B, Luzhetskyy A, Kulachkovskyy O, Yamaguchi T, Nakamura T, Bechthold A, Fedorenko V. Function of lanI in regulation of landomycin A biosynthesis in Streptomyces cyanogenus S136 and cross-complementation studies with Streptomyces antibiotic regulatory proteins encoding genes. Arch Microbiol. 2008;189:111–120. doi: 10.1007/s00203-007-0299-5. [DOI] [PubMed] [Google Scholar]
- 67.Trefzer A, Fischer C, Stockert S, Westrich L, Künzel E, Girreser U, Rohr J, Bechthold A. Elucidation of the function of two glycosyltransferase genes (lanGT1 and lanGT4) involved in landomycin biosynthesis and generation of new oligosaccharide antibiotics. Chem Biol. 2001;8:1239–1252. doi: 10.1016/s1074-5521(01)00091-6. [DOI] [PubMed] [Google Scholar]
- 68.Luzhetskyy A, Zhu L, Gibson M, Fedoryshyn M, Dürr C, Hofmann C, Hoffmeister D, Ostash B, Mattingly C, Adams V, et al. Generation of novel landomycins M and O through targeted gene disruption. ChemBioChem. 2005;6:675–678. doi: 10.1002/cbic.200400316. [DOI] [PubMed] [Google Scholar]
- 69.Ostash B, Rix U, Remsing Rix LL, Liu T, Lombó F, Luzhetskyy A, Gromyko O, Wang C, Braña AF, Méndez C, et al. Generation of new landomycins by combinatorial biosynthetic manipulation of the LndGT4 gene of the landomycin E cluster in S. globisporus. Chem Biol. 2004;11:547–555. doi: 10.1016/j.chembiol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 70.Zhu L, Luzhetskyy A, Luzhetska M, Mattingly C, Adams V, Bechthold A, Rohr J. Generation of new landomycins with altered saccharide patterns through over-expression of the glycosyltransferase gene lanGT3 in the biosynthetic gene cluster of landomycin A in Streptomyces cyanogenus S-136. ChemBiochem. 2007;8:83–88. doi: 10.1002/cbic.200600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffmeister D, Weber M, Drager G, Ichinose K, Dürr C, Bechthold A. Rational saccharide extension by using the natural product glycosyltransferase LanGT4. ChemBioChem. 2004;5:369–371. doi: 10.1002/cbic.200300793. [DOI] [PubMed] [Google Scholar]
- 72.Zhu L, Ostash B, Rix U, Nur-e-Alam M, Mayers A, Luzhetskyy A, Méndez C, Salas JA, Bechthold A, Fedorenko V, et al. Identification of the function of gene lndM2 encoding a bifunctional oxygenase-reductase involved in the biosynthesis of the antitumor antibiotic landomycin E by Streptomyces globisporus 1912 supports the originally assigned structure for landomycinone. J Org Chem. 2005;70:631–638. doi: 10.1021/jo0483623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baig I, Kharel M, Kobylyanskyy A, Zhu LL, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J. On the acceptor substrate of C-glycosyltransferase UrdGT2: three prejadomycin C-glycosides from an engineered mutant of Streptomyces globisporus 1912 Delta lndE(urdGT2). Angew Chem Int Ed. 2006;45:7842–7846. doi: 10.1002/anie.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Kharel MK, Nybo SE, Shepherd MD, Rohr J. Cloning and characterization of the ravidomycin and chrysomycin biosynthetic gene clusters. ChemBioChem. 2010;11:523–532. doi: 10.1002/cbic.200900673. [Ravidomycin and chrysomycin biosynthetic gene clusters cloned in this study provided a ground for the comparative functional assignment of the closely related gilvocarcin biosynthetic genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Kharel MK, Pahari P, Lian H, Rohr J. Enzymatic total synthesis of rabelomycin, an angucycline group antibiotic. Org Lett. 2010;12:2814–2817. doi: 10.1021/ol1009009. [Angucycline PKS enzymes were used to generate rabelomycin utilizing the building blocks malonyl-CoA and acetyl-CoA. This work provided framework for the in vitro studies of landomycin, gilvocarcin and jadomycin biosyntheses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kulowski K, Wendt-Pienkowski E, Han L, Yang KQ, Vining LC, Hutchinson CR. Functional characterization of the jadI gene as a cyclase forming angucyclinones. J Am Chem Soc. 1999;121:1786–1794. [Google Scholar]
- 77•.Rohr J, Hertweck C. Structural diversity. I: type II PKS. In: Mander L, Liu H-W, editors. Comprehensive Natural Products II — Chemistry and Biology. Elsevier; 2010. pp. 227–303. (Townsend CA (Series Editor), vol. 1). [This review is the newest overview of type II PKSs.] [Google Scholar]
- 78•.Shepherd MD, Kharel MK, Zhu LL, Van Lanen SG, Rohr J. Delineating the earliest steps of gilvocarcin biosynthesis: role of GilP and GilQ in starter unit specificity. Here, an unusual mechanism of type II-PKS starter unit usage control is analyzed. Org Biomol Chem. 2010;8:3851–3856. doi: 10.1039/c0ob00036a. [DOI] [PubMed] [Google Scholar]
- 79.Liu T, Kharel MK, Zhu L, Bright SA, Mattingly C, Adams VR, Rohr J. Inactivation of the ketoreductase gilU gene of the gilvocarcin biosynthetic gene cluster yields new analogues with partly improved biological activity. ChemBioChem. 2009;10:278–286. doi: 10.1002/cbic.200800348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kharel MK, Zhu L, Liu T, Rohr J. Multi-oxygenase complexes of the gilvocarcin and jadomycin biosyntheses. J Am Chem Soc. 2007;129:3780–3781. doi: 10.1021/ja0680515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kharel MK, Pahari P, Lian H, Rohr J. GilR, an unusual lactone-forming enzyme involved in gilvocarcin biosynthesis. ChemBioChem. 2009;10:1305–1308. doi: 10.1002/cbic.200900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu T, Kharel MK, Fischer C, McCormick A, Rohr J. Inactivation of gilGT, encoding a C-glycosyltransferase, and gilOIII, encoding a P450 enzyme, allows the details of the late biosynthetic pathway to gilvocarcin V to be delineated. ChemBioChem. 2006;7:1070–1077. doi: 10.1002/cbic.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83•.Noinaj N, Bosserman MA, Schickli MA, Piszczek G, Kharel MK, Pahari P, Buchanan SK, Rohr J. The crystal structure and mechanism of an unusual oxidoreductase, GilR, involved in gilvocarcin V biosynthesis. J Biol Chem. 2011;286:23533–23543. doi: 10.1074/jbc.M111.247833. [Crystal structure of the hemiacetal dehydrogenase GilR revealed the key amino acid residues involved in the catalysis. It is considered as the bottleneck enzyme of the gilvocarcin biosynthetic pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kharel MK, Pahari P, Shaaban KA, Wang G, Morris C, Rohr J. Elucidation of post-PKS tailoring steps involved in landomycin biosynthesis. Org Biomol Chem. 2012 doi: 10.1039/c2ob07171a. doi:10.1039/C2OB07171A, in press. [DOI] [PubMed] [Google Scholar]
- 85.This work utilized combinatorial biosynthetic enzymology approach to generate 11-deoxylandomycinone in vitro. Biosynthetic route for other landomycin aglycones tetrangulol and anhydrolandomycinone were also determined.
- 86.Rix U, Wang C, Chen Y, Lipata FM, Remsing Rix LL, Greenwell LM, Vining LC, Yang K, Rohr J. The oxidative ring cleavage in jadomycin biosynthesis: a multistep oxygenation cascade in a biosynthetic black box. ChemBioChem. 2005;6:838–845. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 87.Mayer A, Taguchi T, Linnenbrink A, Hofmann C, Luzhetskyy A. Bechthold A: LanV, a bifunctional enzyme: aromatase and ketoreductase during landomycin A biosynthesis. ChemBioChem. 2005;6:2312–2315. doi: 10.1002/cbic.200500205. [DOI] [PubMed] [Google Scholar]
- 88.Kallio P, Liu Z, Mäntsälä P, Niemi J, Metsa-Ketelä M. Sequential action of two flavoenzymes, PgaE and PgaM, in angucycline biosynthesis: chemoenzymatic synthesis of gaudimycin C. Chem Biol. 2008;15:157–166. doi: 10.1016/j.chembiol.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 89••.Pahari P, Kharel MK, Shepherd MD, Van Lanen SG, Rohr J. Enzymatic total synthesis of defucogilvocarcin M and its implications on gilvocarcin biosynthesis. Angew Chem Int Ed. 2012;51:1216–1220. doi: 10.1002/anie.201105882. [This work utilizes a combinatorial biosynthetic enzymology approach to address complex biosynthetic questions. Minimum number of enzymes necessary for Defuco-gilvocarcin M was determined through systematic combination of enzymes in vitro.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin S, Van Lanen SG, Shen B. Regiospecific chlorination of (S)-beta-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Am Chem Soc. 2007;129:12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen YH, Wang CC, Greenwell L, Rix U, Hoffmeister D, Vining LC, Rohr J, Yang KQ. Functional analyses of oxygenases in jadomycin biosynthesis and identification of JadH as a bifunctional oxygenase/dehydrase. J Biol Chem. 2005;280:22508–22514. doi: 10.1074/jbc.M414229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y, Fan K, He Y, Xu X, Peng Y, Yu T, Jia C, Yang K. Characterization of JadH as an FAD- and NAD(P)H-dependent bifunctional hydroxylase/dehydrase in jadomycin biosynthesis. ChemBioChem. 2010;11:1055–1060. doi: 10.1002/cbic.201000178. [DOI] [PubMed] [Google Scholar]
- 93.Chen HW, Tseng CC, Hubbard BK, Walsh CT. Glycopeptide antibiotic biosynthesis: enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc Natl Acad Sci U S A. 2001;98:14901–14906. doi: 10.1073/pnas.221582098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fielding EN, Widboom PF, Bruner SD. Substrate recognition and catalysis by the cofactor-independent dioxygenase DpgC. Biochemistry. 2007;46:13994–14000. doi: 10.1021/bi701148b. [DOI] [PubMed] [Google Scholar]
- 95••.Widboom PF, Fielding EN, Liu Y, Bruner SD. Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis. Nature. 2007;447:342–345. doi: 10.1038/nature05702. [The work lays the foundation to understand cofactor free oxygenation events in secondary metabolism.] [DOI] [PubMed] [Google Scholar]
- 96••.Roessner CA, Scott AI. Achieving natural product synthesis and diversity via catalytic networking ex vivo. Chem Biol. 1996;3:325–330. doi: 10.1016/s1074-5521(96)90114-3. [This work marked a milestone for the understanding of the B12 biosynthetic pathway.] [DOI] [PubMed] [Google Scholar]
- 97••.Shen B, Hutchinson CR. Enzymatic synthesis of a bacterial polyketide from acetyl and malonyl coenzyme A. Science. 1993;262:1535–1540. doi: 10.1126/science.8248801. [This was the first enzymatic total synthesis of a polyketide antibiotic.] [DOI] [PubMed] [Google Scholar]
- 98.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Enzymatic total synthesis of enterocin polyketides. Nat Chem Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]