Abstract
A quantitative polymerase chain reaction assay with melt curve analysis (qPCR-MCA) was applied for the detection of protozoan oocysts in 501 human fecal samples collected in Dominican Republic. Samples were subjected to qPCR using universal coccidia primers targeting 18S rDNA to detect oocysts followed by MCA to identify oocyst species based on amplicon melting temperature. Putative positive samples were also tested by conventional PCR and microscopy. Cystoisospora belli (×3), Cryptosporidium parvum (×3), Cryptosporidium hominis (×5), Cryptosporidium meleagridis (×1), Cryptosporidium canis (×1), and Cyclospora cayetanensis (×9) were detected by qPCR-MCA and confirmed by sequencing. This assay consistently detected 10 copies of the cloned target fragment and can be considered more efficient and sensitive than microscopy flotation methods for detecting multiple species of oocysts in human feces. The qPCR-MCA is a reliable protozoan oocyst screening assay for use on clinical and environmental samples in public health, food safety and veterinary programs.
Introduction
Oocyst-producing protozoa known as coccidia are a large and relatively diverse subclass of the Apicomplexan phylum of parasites that infects virtually all vertebrate hosts. Some members of this group have significant veterinary and public health implications, especially related to agriculture, food safety, and economics. Diagnosis of the infection is typically by detection of oocysts in fecal samples of the definitive host using traditional microscopy techniques, which are labor-intensive, lack sensitivity and specificity, and require parasitology expertise to identify pathogenic coccidian species. In humans, gastrointestinal illnesses caused by Cryptosporidium spp., Cyclospora cayetanensis, and Cystoisospora belli are of significant concern, particularly for the elderly, young children, and immunocompromised individuals.1 There are several diagnostic limitations for the detection of C. cayetanensis. It is not included in routine ova-parasite fecal examinations, oocysts are shed intermittently in low numbers, and they have poor uptake of many histological stains.2 Both C. cayetanensis and C. belli are probably underreported because of, in part, these factors.3
Existing molecular methods for detecting Cyclospora can overcome some of the limitations of traditional methods; however, most cannot distinguish between Cyclospora and closely related non-zoonotic Eimeria spp., which may lead to false-positive results.4 Therefore, a reliable and specific assay for detection of Cyclospora and other common protozoan oocysts of human health importance, such as Cryptosporidium spp. and C. belli, would be a valuable diagnostic tool. We recently reported on the development of a real-time quantitative polymerase chain reaction assay with melting curve analysis (qPCR-MCA) using a universal coccidia primer cocktail for use in detection and differentiation of a wide range of coccidia species of public and veterinary health concern.5 Using genomic DNA (gDNA) from pure isolates of coccidia, we showed that C. cayetanensis, Cryptosporidium parvum, Cryptosporidium muris, Toxoplasma gondii, Eimeria bovis, Eimeria acervulina, Cystoisospora suis, and Sarcocystis cruzi could be identified by unique melting curves and differentiated based on a specific melting temperature (Tm). However, the assay had not been validated for use on field matrices, such as food, water, or feces, where PCR inhibitors and background DNA are commonly present and could interfere with detection and identification of species. The objectives of the present study were to assess the suitability of the qPCR-MCA assay for use in screening human fecal samples for a variety of protozoan oocysts of public health importance and determine the occurrence of these parasites in the Santiago region of Dominican Republic.
Materials and Methods
Fecal samples.
A total of 501 human fecal samples was collected from patients ages 20 days to 89 years (age information not available for all samples) at nine different hospitals, clinics, or medical centers in the north central region of Dominican Republic. The samples were mixed with two volumes of 2.5% potassium dichromate and stored at 4°C until used. As part of a training initiative in Dominican Republic, a portion of each sample was initially screened by flotation microscopy (as below) for parasites before being shipped to Canada for additional analysis. The first 103 samples were analyzed by flotation microscopy followed by qPCR-MCA. Because of the laborious requirement of microscopic examination, the remaining 398 samples were initially analyzed by qPCR-MCA, and any samples found positive were then examined by flotation microscopy for visualization of the parasites. Oocysts of E. papillata were used routinely as controls for the DNA extraction procedure and preparation of the qPCR standard curve. E. papillata oocysts had been propagated by passage in mice and isolated from feces by washing and straining with water followed by flotation with sucrose. The oocysts were sporulated (72%) and stored in 2.5% potassium dichromate at 4°C until use.
Microscopy and parasite recovery.
Fecal samples (140) were prepared for microscopy examination using a modified sucrose flotation method.6 Briefly, approximately 2 mL washed fecal suspension were mixed with a sucrose solution (specific gravity of 1.27) to fill a 16 × 100-mm glass tube and form a convex meniscus, which was covered with a coverslip to form a seal, and centrifuged at room temperature for 10 minutes at 300 × g using a swing-out rotor. The coverslip was removed onto a standard microscope slide and examined for helminth eggs at 100× magnification and protozoan oocysts at 200× magnification in a hash tag pattern. Material under the coverslip was then collected by placing the slide and coverslipin a 50-mL plastic centrifuge tube with 15 mL 0.1% Tween-H2O and soaking for at least 5 minutes at room temperature to dissolve the sucrose solution. After soaking, slides and coverslips were rinsed with an additional 35 mL 0.1% Tween-H2O, and then discarded. Any parasites were concentrated by centrifugation (2,060 × g for 15 minutes) and stored at 4°C. Some fecal samples (26) were selected for an additional examination using a ZnSO4 (specific gravity of 1.18) flotation method specifically for detection of non-oocyst protozoa, such as Giardia and Entamoeba,7 based on clinical information provided by sample providers in Dominican Republic. Enough ZnSO4 solution was mixed with 2 mL fecal suspension to fill a 16 × 100-mm glass tube and form a convex meniscus, which was covered with a coverslip to form a seal, and then, it was centrifuged at room temperature for 5 minutes at 260 × g using a swing-out rotor. Coverslips were removed 10 minutes after centrifugation was complete, placed on a slide with a droplet of iodine, and examined as above. Parasites observed were collected from glass coverslips and slides as above.
DNA extraction.
DNA was extracted from slide wash sediments and E. papillata controls using a QIAamp DNA Micro Kit (Qiagen, Mississaugua, Canada) with modifications as described previously.8 DNA extraction directly from fecal samples was performed using a QIAamp DNA Stool Mini Kit (Qiagen) with the following modifications to the manufacturer's protocols. (1) Approximately 1–2 mL fecal suspension were washed three times with milli-Q H2O by centrifugation (20,000 × g for 15 minutes) to remove potassium dichromate. The remaining 200 μL fecal pellet was mixed with 1.4 mL ASL buffer (Qiagen) and subjected to eight cycles of freezing in liquid nitrogen for 1 minute followed by thawing in a 95°C water bath for 1 minute. (2) The sample was then incubated with 20 μL proteinase K (20 mg/mL; Qiagen) for 18 hours overnight at 56°C. (3) The lysed suspension was centrifuged at 20,000 × g for 3 minutes to pellet stool particles, and 1.4 mL supernatant were transferred to a clean 2-mL tube and treated with the provided InhibitEX tablet to remove inhibitors according to the manufacturer's (Qiagen) protocol. (4) Resulting lysate was incubated with 200 μL Buffer AL (Qiagen) for 10 minutes at 70°C followed by purification through QIAamp microcolumns (Qiagen) according to the manufacturer's protocol and elution with 35 μL AE buffer (Qiagen). A negative control of only reagents and a positive control of 104 E. papillata oocysts were included in each batch of samples for DNA extraction. For the qPCR standard curve, DNA was extracted from 10-fold serial dilutions of purified E. papillata oocysts (105–101), which were enumerated by triplicate readings using a McMaster chamber.
Plasmid controls.
Plasmid DNA from a selection of coccidia species was run on each qPCR plate along with test samples. The melting curves of test samples were visually compared with the coccidia plasmid DNA controls to identify the species, which was then confirmed by sequencing of the qPCR products. To generate the plasmid DNA controls, gDNA from E. bovis, E. tenella, E. necatrix, C. cayetanensis, T. gondii, C. belli, C. parvum, C. meleagridis, and C. hominis was amplified with the universal coccidia primer cocktail, cloned, and purified as below for positive samples. Purified plasmid DNA was digested with Sca1 (New England Biolabs, Pickering, Canada) overnight at 37°C with supplied NEBuffer according to the manufacturer's instructions. The digested plasmid DNA was analyzed by gel electrophoresis, and linearized plasmid fragments were excised and purified using the QIAquick Gel Extraction Kit (Qiagen). The plasmid DNA was quantified by spectrophotometer (Nanodrop 2000c; Thermo Scientific, Wilmington, DE) and diluted to 0.05 ng/μL for use as routine controls in the qPCR assay; 10-fold dilutions of plasmid DNA from C. cayetanensis were also prepared to be equivalent to 105–100 copies of the small subunit ribosomal DNA (SSU rDNA) fragment. Six replicates of each copy number dilution were analyzed by qPCR-MCA to determine assay sensitivity as described below.
qPCR amplification.
All qPCR-MCA assays were performed using the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules). The reaction mix was loaded into clear-well PCR plates (catalogue no. HSP9611; Bio-Rad) and contained 1× SsoFast EvaGreen Supermix (Bio-Rad), 400 nM Crypto-F, Crypto-R, Cyclo-F, Cyclo-R, and Sarco-R, 800 nM Toxo-F,5 100 ng/μL bovine serum albumin (Sigma-Aldrich, Oakville, Canada), 2 μL DNA template, and sterile dH2O to a final volume of 25 μL. Standard curves for all qPCR runs consisted of DNA from diluted E. papillata oocysts (105–101). The qPCR cycling conditions were as follows: 2 minutes at 98°C followed by 40 cycles of denaturing at 98°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 30 seconds. Data collection was enabled at the extension step. The melt curve protocol followed with 10 seconds at 95°C and then 10 seconds each at 0.2°C increments between 62°C and 95°C. Data collection was enabled at each increment of the melt curve. Melt and standard curves were generated by the CFX Manager Software (version 2.0; Bio-Rad) as described previously.5 Oocyst-positive samples were identified by comparison of the test sample melt curves with the melt curves of the protozoan controls included in each run. Previously published conventional PCR assays targeting internal transcribed spacer 2 (ITS-2), 5.8S + ITS-2, and Cryptosporidium oocyst wall protein (COWP) regions8–10 were used for additional confirmation of the positive samples of C. cayetanensis, C. belli, and Cryptosporidium spp., respectively.
Cloning and sequence analysis.
Test samples that were positive for protozoan oocysts by qPCR-MCA (i.e., melt curve shape and Tm matched a positive control in the run) were cloned and sequenced to confirm species identification. First, qPCR products were analyzed by gel electrophoresis and purified with the Qiaquick Gel Extraction Kit (Qiagen). The purified products were subjected to an A-tailing reaction, which consisted of 4 μL qPCR product, 5 units GoTaq Flexi DNA polymerase (Promega, Madison), 0.2 mM dATP, 5× GoTaq reaction buffer, and dH2O to a final volume of 10 μL incubated at 70°C for 15 minutes. The A-tailed products were then ligated into plasmid cloning vector pGEM-T Easy (Promega) according to the manufacturer's instructions. Plasmid DNA was purified using the QIAprep Miniprep Kit (Qiagen) and sequenced in both directions using the T7 and SP6 primers (Promega). Forward and reverse sequences were assembled using Clone Manager software (version 9.0; Sci-Ed Software, Cary), and the vector and primer sequences were removed. Nucleotide sequences were compared with the GenBank collection of sequences using BLASTN and identified based on the degree of sequence identity.
Results
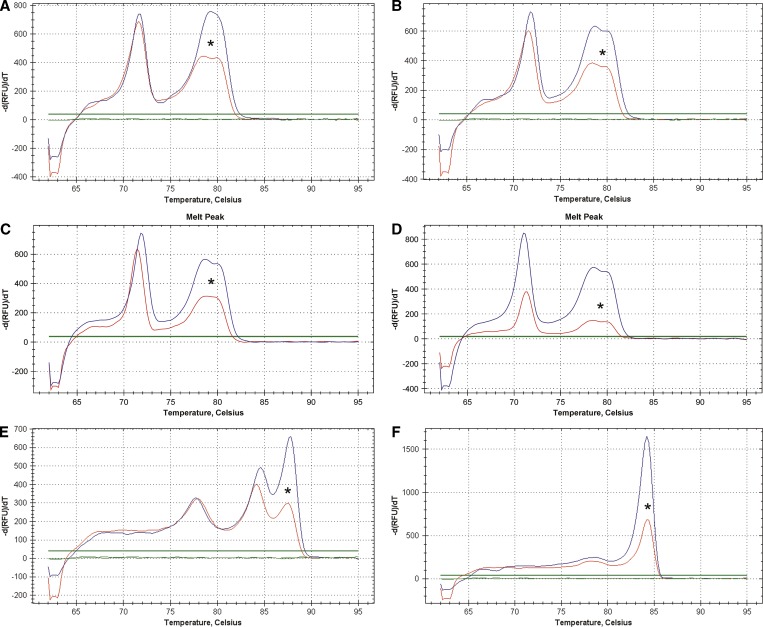
During the first phase of the project, human fecal samples were first examined microscopically for parasites, and the DNA extracted from the slide washes or directly from the feces was tested by qPCR-MCA. Of 103 samples examined, oocysts of Cystoisospora spp. (×2) and Cryptosporidium spp. (×1) and eggs of Trichuris spp. (×1) were observed by microscopy (Figure 1 and Table 1). Using qPCR-MCA, C. belli was detected and confirmed by sequencing and conventional PCR for both samples, which were detected by microscopy. The Cryptosporidium sample observed by microscopy could not be confirmed by qPCR-MCA. Another sample that was positive for C. hominis by qPCR was confirmed by sequencing and conventional PCR amplification of the COWP gene fragment (Table 1). Figure 2 shows representative melt curve graphs for both the protozoan oocyst plasmid controls and the isolates detected in this study. The qPCR-MCA could consistently detect 10 copies of the cloned C. cayetanensis SSU rDNA plasmid DNA fragment. When plotted along the standard curve consisting of serially diluted gDNA from 105 E. papillata oocysts, the quantification cycle (Cq) values for all coccidian-positive samples were equivalent to a range of 0.5–5 oocysts.
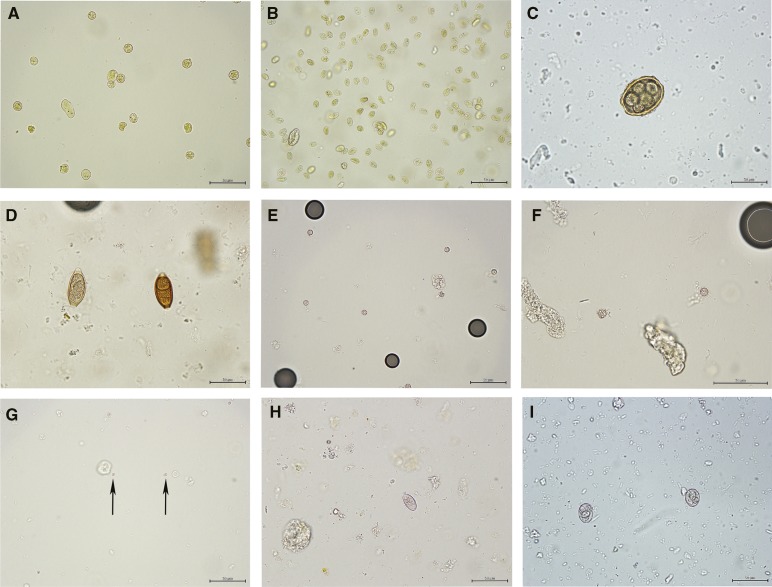
Figure 1.
Representative photos of parasites observed in human fecal samples collected in Dominican Republic: (A) Entamoeba cysts stained with iodine, (B) G. duodenalis cysts stained with iodine, (C) A. lumbricoides egg, (D) T. trichiuria egg, (E and F) C. cayetanensis oocysts, (G) Cryptosporidium oocysts, (H) C. belli oocysts, and (I) E. papillata oocysts (control sample). Scale bars: 50 μm. Crown Copyright c 2012. Published by AJTMH. All rights reserved.
Table 1.
Parasites detected by flotation microscopy and qPCR-MCA
| Parasite detected | Detection method | Species identified (no.) and confirmed by sequencing | Mean Tm (°C; SEM) | ||
|---|---|---|---|---|---|
| Flotation microscopy (N = 140) | qPCR-MCA (N = 501) | PCR (N = 22) | |||
| Cryptosporidium | 10 | 10 | 9 | C. hominis (5) | 78.38 (0.02) |
| C. parvum (3) | 78.53 (0.06) | ||||
| C. canis (1) | 78.80 (0) | ||||
| C. meleagridis (1) | 78.45 (0) | ||||
| Cyclospora | 2 | 9 | 5 | C. cayetanensis (9) | 84.19 (0.06) |
| 87.53 (0.08) | |||||
| Cystoisospora | 3 | 3 | 2 | C. belli (3) | 84.33 (0.07) |
Figure 2.
Representative melt curve graphs for (A) C. hominis, (B) C. parvum, (C) C. canis, (D) C. meleagridis, (E) C. cayetanensis, and (F) C. belli. Melt curve graphs from plasmid DNA controls are unmarked, and those graphs from field isolates identified in this study are marked with an asterisk. Temperature (°C) is displayed on the x axis, and −d(RFU)/dT is displayed on the y axis. Crown Copyright c 2012. Published by AJTMH. All rights reserved.
During the second phase of the project, DNA was extracted directly from washed human fecal samples and screened for oocysts using qPCR-MCA. Samples positive for protozoan oocysts by qPCR-MCA were then examined by microscopy for visualization of oocysts. Of 398 samples examined during this phase, Cryptosporidium (×9), Cyclospora (×9), Cystoisospora (×1), and three unknown protozoa (melting curves did not match any of the available controls; Tm = 81.6°C for two samples and Tm = 84°C for the third sample) were detected by qPCR-MCA (Table 1). Cryptosporidium oocysts were observed by microscopy in all nine samples identified by qPCR-MCA. Cyclospora oocysts were observed by microscopy in two of nine samples identified by qPCR-MCA. Sequencing confirmed the positive protozoan oocyst samples as C. hominis (×4), C. parvum (×3), C. canis (×1), C. meleagridis (×1), and C. cayetanensis (×9) (Figure 2 and Table 1). Conventional PCR assays confirmed eight of nine positive Cryptosporidium spp. and five of nine C. cayetanensis samples. Three samples with melt curves in the Tm range of protozoan oocysts were sequenced and identified as Adelina bambarooniae (two samples; 97% identity by BLAST analysis), which is a coccidian parasite that infects cane beetles, and Baldinia anauniensis (one sample; 96% identity), which is a dinoflagellate.
In addition to those samples examined by Sheather's flotation for visual confirmation of oocysts detected by the qPCR-MCA, 39 samples were selected for examination by microscopy (13 samples using Sheather's flotation for helminth eggs and 26 samples using ZnSO4 for other protozoa) based on clinical information provided by sample providers in Dominican Republic. From these selected samples, Trichuris (×2), Ascaris (×1), Giardia (×6), and Entameoba cysts (×10) were identified based on morphological features (Figure 1).
Discussion
The primary aim of this study was to show the application of the qPCR assay with MCA for detection and identification of protozoan oocysts in human fecal samples. We selected Latin America as a source for the samples, because some areas within this region are known to be endemic for protozoan parasites, such as Cryptosporidium and C. cayetanensis.2 The study was performed on samples from Dominican Republic, where there is little information about the occurrence of protozoan oocysts. In developing countries where molecular diagnostic instrumentation and expertise are limited, microscopy remains the gold standard for detection of protozoa in humans. Limitations of microscopic methods can include intermittent shedding and need to examine multiple fecal samples, inconsistent staining of oocysts, time required for oocysts to be sporulated for taxonomic identification, and need for parasitology experience to distinguish oocysts from fecal debris.2 Although several commercial immunofluorescent antibody (IFA) kits are available for Cryptosporidium spp., there are currently none for C. cayetanensis or C. belli. Because of the limited amount of fecal material provided in this study, we opted to perform a simple sucrose flotation to screen all fecal samples for parasites by microscopy followed by qPCR-MCA in the first phase of the project. In the second phase, we screened fecal samples first using qPCR-MCA followed by microscopy for oocyst visualization.
The most recent report on the occurrence of helminths and protozoa in Dominican Republic was published in 1981, and it indicated a high prevalence of A. lumbricoides (13.1%), T. trichiura (35.8%), and E. coli (24.8%) in rural sugar plantation employees.11 G. duodenalis was less frequently detected (0.7%). The present study showed that the prevalence of these helminth parasites was lower, possibly because of the inclusion of both urban and rural populations, improvements in parasite control programs, and improvements in sanitation and hygiene practices. However, Giardia and Entamoeba were detected more frequently in this study. Reports of Cyclospora, Cryptosporidium, and Cystoisospora from Dominican Republic relate primarily to travel-associated cases of infection12,13; thus little published information is available for the prevalence in the local population.
The qPCR-MCA assay was as sensitive and specific as microscopic analysis for the detection of C. belli. One fecal sample that was presumed positive for Cryptosporidium oocysts by microscopy was not confirmed by qPCR-MCA. Because of the small amount of fecal material available, it was not possible to perform an additional microscopy method (such as IFA) to detect Cryptosporidium spp. in this study; therefore, it is possible the putative oocysts observed were, in fact, plant material or yeast and as a result, not amplified by the qPCR-MCA assay. Another sample that was negative by microscopy was positive by qPCR-MCA and confirmed by sequencing and conventional PCR amplification. The qPCR-MCA is likely more sensitive than flotation microscopy; the estimated recovery efficiency of the Sheather's flotation method is ∼60% for Cryptosporidium oocysts.14 Because the qPCR-MCA assay is performed on DNA extracted directly from a separate aliquot of feces, it is possible that, because of inadequate sample mixing or low oocyst number, the results do not concur. An additional seven cases of C. cayetanensis were identified by qPCR-MCA and confirmed by sequencing, but they were not seen on microscopy or all conventional PCR assays. In subclinical cases, C. cayetanensis is shed intermittently and in low numbers, and therefore, examination of only one sample by microscopy may not have been sensitive enough to detect the oocysts. It is also possible that the conventional PCR assay selected for confirmation was not sensitive enough to detect these C. cayetanensis-positive samples. All negative controls performed as expected during qPCR-MCA analysis, and therefore, the likelihood of false positives caused by cross-contamination is low.
We have shown that, by using a universal cocktail of seven primers, DNA from virtually any coccidia species can be amplified and then identified by melt curve shape and Tm.5 The SSU rDNA region targeted by the primer cocktail was selected, because it is highly variable within the subclass, which allows for many coccidia species to be differentiated based on their Tm, and the identification can be confirmed by sequencing of the PCR products. Primers to amplify this region, originally designed by Morgan and others,15 have also been modified by others and used in real-time PCR to detect and differentiate Cryptosporidium spp.16 The melt temperatures determined for the Cryptosporidium species detected in this study differ by a small margin of 0.27° to 0.42° and therefore, would require high-resolution melt curve analysis for reliable species identification. An additional benefit to using a broad-specificity universal primer set is that unknown, emerging, or opportunistic coccidian pathogens may be discovered if this assay was used for difficult-to-diagnose cases or with immunocompromised patients. This type of extended-range approach was used by Murphy and others3 to identify C. belli in an immunocompromised patient with persistent diarrhea of unknown cause.
The potential disadvantage of a broadly specific assay is that non-specific aberrant peaks are often observed when amplifying DNA from fecal or environmental samples. However, the qPCR-MCA allows these peaks to be distinguished from true positives by their Tm (typically less than 78°C) and shape (irregular) as well as comparison of melt peaks for test samples with well-characterized control samples included in each run. In this study, three samples displayed melt curves in the Tm range of coccidia but were not an exact match to any controls. The closest BLAST match for two of the samples indicated Adelina spp., which is a coccidian parasite of invertebrates that is not known to be zoonotic; it is likely a transient gut organism, or it may have been present as an opportunistic dweller if the host was immunocompromised. The DNA sequence from the other sample identified it as a dinoflagellate, a flagellate protist. The amplification of dinoflagellates and occasionally, fungi (another protist) suggests that the specificity of the primers used in the qPCR-MCA is not strictly coccidia-specific. This finding is supported by earlier rDNA studies showing a close phylogenetic relationship among some phyla of the diverse group of protists, including coccidia, dinoflagellates, piroplasms, and ciliates,17 However, when appropriate controls are used in each run, these non-specific amplicons are easily identified and may be ruled out by sequencing.
An estimate of the number of oocysts detected per sample was based on a standard curve consisting of serially diluted DNA from 105 E. papillata oocysts. Sporulated Eimeria oocysts have eight haploid sporozoites and are thought to have approximately 140 copies of the rRNA gene,18 whereas Cryptosporidium has four sporozoites and may have as few as 5 copies of the gene.19 Thus, the amount of target DNA available per oocyst is much greater for Eimeria and other species that have eight sporozoites, such as C. belli. We could consistently detect 10 copies of the C. cayetanensis SSU rDNA plasmid DNA fragment, which may be equivalent to less than one oocyst, depending on the number of gene copies per organism and the sporulation stage of the oocyst. The Cryptosporidium and Cyclospora oocyst numbers extrapolated from the Eimeria standard curve will be underestimated, and the assay sensitivity for these species may be reduced, because they inherently have less target 18S rDNA. For instance, using a standard curve consisting of serially diluted DNA from 105 C. parvum oocysts, the Cq values ranged from 22 to 36 for 105–101 oocysts (data not shown). A similarly constructed standard curve using E. papillata would have a Cq range of 17–30 for 105–101 oocysts, a difference of about five to six cycles (about 2 logs) compared with C. parvum. Thus, using an Eimeria standard curve to estimate numbers of Cryptosporidium and Cyclospora oocysts is not ideal. However, it is not practical to run more than one standard curve on every plate, and therefore, E. papillata was selected for this study, because it can be easily propagated in mice, and a fresh source of oocysts may be available. As a result, the oocyst quantification aspect of this assay is not entirely accurate; however, because it was designed to target multiple coccidia species, its power is not in quantification but species differentiation and identification.5
There are several multiplex real-time PCR assays targeting diarrhea-causing parasitic protozoa, including G. duodenalis, Cryptosporidium spp., and E. histolytica20–22; however, this qPCR-MCA assay is the only published method for detection, differentiation, and identification of a wide range of protozoan oocyst species that are important for human and animal health. The current method requires only a single channel real-time instrument and does not use probes, and therefore, it is simpler and may be more cost-effective to run than other multiplex real-time PCR assays used for diarrhea-causing protozoa. Although the qPCR-MCA assay can differentiate to the genus and species level by Tm alone in many cases,5 distinguishing some species (such as Cryptosporidium) may require equipment and software capable of high-resolution melt curve analysis or sequencing to confirm results. The Tm values reported here using a qPCR mix containing EvaGreen dye differ from those values that we reported previously using SYBR Green qPCR chemistry.5 However, the unique double-peak melt curve profile previously observed for C. cayetanensis was retained, enabling effective differentiation between Cyclospora, Cystoisospora, and Eimeria species. The distinctive melt curve profiles observed for all species detected here, including the dual peak for Cyclospora, are the result of differences in nucleotide sequence composition, length, and G-C composition. It is important that gDNA or plasmid DNA from suitable controls be included in each qPCR run for comparison with test samples, because Tm values can also vary based on qPCR reagent chemistry, amplification instrument, and DNA concentration in addition to the expected variation because of nucleic acid composition.
We have shown the qPCR-MCA assay's use for simultaneous detection and identification of multiple protozoan oocysts species in human feces. The extraction of DNA directly from washed fecal samples followed by qPCR-MCA is more efficient than and at least as sensitive as standard microscopy flotation methods, which are laborious and lack specificity. This study shows the use of the qPCR-MCA as a reliable screening tool for oocyst detection and identification for use in clinical and environmental samples in public health, food safety, and veterinary programs.
ACKNOWLEDGMENTS
This work was supported and funded by the Canadian Food Inspection Agency. Crown Copyright © 2013 Published by AJTMH. All rights reserved. Excellent and valuable technical and sample collection services and coordination in Dominican Republic were provided by Ana Rodriguez, Maribel Sosa, Arnaldo Cruz, Jose Sanchez, and Dahiana Peña, Pontificia Universidad Católica Madre y Maestra. The authors thank Jenna Oakley for significant technical contributions and the other staff for assistance at the Centre for Food-Borne and Animal Parasitology, Canadian Food Inspection Agency Saskatoon Laboratory. Special thanks to Dr. Batol Al-Adhami for her helpful comments to improve the manuscript.
Footnotes
Authors' addresses: Laura F. Lalonde and Alvin A. Gajadhar, Centre for Food-Borne and Animal Parasitology, Canadian Food Inspection Agency Saskatoon Laboratory, Saskatoon, Saskatchewan, Canada, E-mails: laura.lalonde@inspection.gc.ca and alvin.gajadhar@inspection.gc.ca. Julissa Reyes, Facultad de Ciencias de la Salud, Pontificia Universidad Catolica Madre y Maestra, Santiago, Dominican Republic, E-mail: julareyes@hotmail.com.
References
- 1.Gerba CP, Rose JB, Haas CN. Sensitive populations: who is at the greatest risk? Int J Food Microbiol. 1996;30:113–123. doi: 10.1016/0168-1605(96)00996-8. [DOI] [PubMed] [Google Scholar]
- 2.Mansfield LS, Gajadhar AA. Cyclospora cayetanensis, a food- and waterborne coccidian parasite. Vet Parasitol. 2004;126:73–90. doi: 10.1016/j.vetpar.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Murphy SC, Hoogestraat DR, SenGupta DJ, Prentice J, Chakrapani A, Cookson BT. Molecular diagnosis of cystoisosporiasis using extended-range PCR screening. J Mol Diagn. 2011;13:359–362. doi: 10.1016/j.jmoldx.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields JM, Olson BH. PCR-restriction fragment length polymorphism method for detection of Cyclospora cayetanensis in environmental waters without microscopic confirmation. Appl Environ Microbiol. 2003;69:4662–4669. doi: 10.1128/AEM.69.8.4662-4669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalonde LF, Gajadhar AA. Detection and differentiation of coccidian oocysts by real-time PCR and melting curve analysis. J Parasitol. 2011;97:725–730. doi: 10.1645/GE-2706.1. [DOI] [PubMed] [Google Scholar]
- 6.Gajadhar AA. Host specificity studies and oocyst description of a Cryptosporidium sp. isolated from ostriches. Parasitol Res. 1994;80:316–319. doi: 10.1007/BF02351873. [DOI] [PubMed] [Google Scholar]
- 7.Levine ND. Laboratory diagnosis of protozoan infections. In: Levine ND, editor. Veterinary Protozoology. Ames, IA: Iowa State University Press; 1985. pp. 365–386. [Google Scholar]
- 8.Lalonde LF, Gajadhar AA. Highly sensitive and specific PCR assay for reliable detection of Cyclospora cayetanensis oocysts. Appl Environ Microbiol. 2008;74:4354–4358. doi: 10.1128/AEM.00032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 10.Taniuchi M, Verweij JJ, Sethabutr O, Bodhidatta L, Garcia L, Maro A, Kumburu H, Gratz J, Kibiki G, Houpt ER. Multiplex polymerase chain reaction method to detect Cyclospora, Cystoisospora, and Microsporidia in stool samples. Diagn Microbiol Infect Dis. 2011;71:386–390. doi: 10.1016/j.diagmicrobio.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins RF, Edwards LD. Prevalence of intestinal helminths and protozoans in a rural population segment of the Dominican Republic. Trans R Soc Trop Med Hyg. 1981;75:549–551. doi: 10.1016/0035-9203(81)90196-6. [DOI] [PubMed] [Google Scholar]
- 12.Green ST, McKendrick MW, Mohsen AH, Schmid ML, Prakasam SFR. Two simultaneous cases of Cyclospora cayetanensis enteritis returning from the Dominican Republic. J Travel Med. 2000;7:41–42. doi: 10.2310/7060.2000.00014. [DOI] [PubMed] [Google Scholar]
- 13.Weitzel T, Wichmann O, Mühlberger N, Reuter B, Hoof HD, Jelinek T. Epidemiological and clinical features of travel-associated cryptosporidiosis. Clin Microbiol Infect. 2006;12:921–924. doi: 10.1111/j.1469-0691.2006.01487.x. [DOI] [PubMed] [Google Scholar]
- 14.Kar S, Gawlowska S, Daugschies A, Bangoura B. Quantitative comparison of different purification and detection methods for Cryptosporidium parvum oocysts. Vet Parasitol. 2011;177:366–370. doi: 10.1016/j.vetpar.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Morgan UM, Constantine CC, Forbes DA, Thompson RC. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 16.Hadfield SJ, Robinson G, Elwin K, Chalmers RM. Detection and differentiation of Cryptosporidium spp. in human clinical samples by use of real-time PCR. J Clin Microbiol. 2011;49:918–924. doi: 10.1128/JCM.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gajadhar AA, Marquardt WC, Hall R, Gunderson J, Ariztia-Carmona EV, Sogin ML. Ribosomal RNA sequences of Sarcocystis muris, Theileria annulata and Crypthecodinium cohnii reveal evolutionary relationships among apicomplexans, dinoflagellates, and ciliates. Mol Biochem Parasitol. 1991;45:147–154. doi: 10.1016/0166-6851(91)90036-6. [DOI] [PubMed] [Google Scholar]
- 18.Morgan JAT, Morris GM, Wlodek BM, Byrnes R, Jenner M, Constantinoiu CC, Anderson GR, Lew-Tabor AE, Molloy JB, Gasser RB, Jorgensen WK. Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Mol Cell Probes. 2009;23:83–89. doi: 10.1016/j.mcp.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Le Blancq SM, Khramtsov NV, Zamani F, Upton SJ, Wu TW. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 20.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EAT, van Rooyen MAA, van Lieshout L, Polderman AM. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haque R, Roy S, Siddique A, Mondal U, Rahman SMM, Mondal D, Houpt E, Petri WA. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg. 2007;76:713–717. [PubMed] [Google Scholar]
- 22.Stark D, Al-Qassab SE, Barratt JLN, Stanley K, Roberts T, Marriott D, Harkness J, Ellis JT. Evaluation of multiplex tandem real-time PCR for detection of Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis in clinical stool samples. J Clin Microbiol. 2011;49:257–262. doi: 10.1128/JCM.01796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]