Abstract
Polymerase chain reaction was used to detect Leishmania siamensis DNA from clinical samples collected from six leishmaniasis patients during 2011–2012. The samples used in this study came from bone marrow, blood, buffy coat, saliva, urine, and tissue biopsy specimens. Saliva was a good source for L. siamensis DNA by polymerase chain reaction. L. siamensis DNA was also found in saliva of an asymptomatic case-patient. Levels of L. siamensis DNA in saliva decreased until being undetectable after treatment. These levels could be used as a marker to evaluate efficacy of the treatment. A larger study is needed to evaluate this method as a screening and survey tool to study the silent background of Leishmania infection among the at-risk population.
Leishmaniasis is a neglected tropical diseases caused by an obligate intracellular protozoa belonging to the genus Leishmania. The disease is transmitted to vertebrate hosts by infected female sand flies taking a blood meal.1 Leishmaniasis presents in three clinical forms; visceral, cutaneous, and mucocutaneous.2 Clinical presentation of leishmaniasis depends on the species of Leishmania and the immunity of the host.3
Detection and species identification of the parasites is essential for prognostic and therapeutic reasons and surveys.4 Several laboratory techniques have been used for diagnosis of Leishmania infection. They are microscopy, culture, immunologic techniques (enzyme-linked immunosorbent assay, direct agglutination test, and recombinant protein K39 dipstick test), and molecular techniques (polymerase chain reaction [PCR] and quantitative PCR).5–7 In comparisons of microscopic examination, culture, and PCR in detecting Leishmania parasites, PCR has shown to have a significantly higher sensitivity than culture and microscopic examination (97%, 78%, and 76%8–10 sensitivity, respectively). New cases of leishmaniasis caused by L. siamensis, a novel species of Leishmania, have been documented in patients in Thailand11–17 and Myanmar (unpublished data). The infection was described in immunocompromised patients, mostly persons infected with human immunodeficiency virus (HIV).11–17 Clinical presentations of these patients have included visceral,11–13 diffuse cutaneous,15 and overlapping diffuse cutaneous and visceral forms.14
With low prevalence of leishmaniasis in Thailand and Myanmar, screening tests for leishmaniasis such as enzyme-linked immunosorbent assay, direct agglutination test, and recombinant protein K39 dipstick test are not readily available. Moreover, sensitivity and specificity of these serologic tests for detection of L. siamensis infection have never been fully documented. Diagnosis of L. siamensis infection relies on microscopic examination, culture, and detection of parasite DNA by PCR.11–15 Although microscopic examination and culture of Leishmania parasites have high specificity, they require experience and have low sensitivity. The PCR is commonly used to diagnose leishmaniasis caused by L. siamensis. Bone marrow, blood, buffy coat, tissue, saliva, and urine have been successfully used for detection of L. siamensis DNA by PCR.14
Saliva has shown to be a good source for L. siamensis DNA.14 There are also reports of using saliva to identify other Leishmania species.18–20 Collection of saliva is noninvasive and convenient for field studies. We describe a PCR to amplify the internal transcribed spacer1 (ITS1) gene of L. siamensis from six infected patients and compare it with specimens collected from patients and different clinical presentations. Details of clinical presentations and management of some patients enrolled in this study have been described.14
Bone marrow, blood or saliva was smeared on glass slides, and fixed with absolute methanol (Sigma-Aldrich, St. Louis, MO) for one minute. The slides were then stained with Giemsa (Sigma-Aldrich) in phosphate buffer, pH 7.2. Tissue biopsy specimens were stained with hematoxylin and eosin. Stained slides were then examined under a light microscope (Olympus, Tokyo, Japan).
Schneider's insect medium (Sigma-Aldrich) containing 20% fetal bovine serum, 100 U/mL of penicillin, and 100 mg/mL of streptomycin (Sigma-Aldrich) was used for culturing Leishmania parasites. One hundred microliters of bone marrow, blood, or saliva was inoculated into 5 mL of culture media in a 25-cm3 flask and incubated at 25 ± 2°C. Cultures were inspected for parasites every 24 hours by using an inverted microscope (Olympus).
Two hundred microliters of blood or bone marrow and 50 μL of buffy coat were used for DNA extraction by using a blood DNA extraction kit (Invisorb® Spin Blood Mini Kit (STRATEC Molecular, Berlin, Germany). Thirty milliliters of urine or 1 mL of saliva were centrifuged for 5 minutes at 5,000 × g, and the pellets were collected and used for further DNA extraction. Tissue specimens, urine pellets, and saliva pellets were used for DNA extraction by tissue DNA extraction (Invisorb® Spin Tissue Mini Kit) according to the manufacturer's instructions. Extracted DNA was eluted in 80 μL of elution buffer. Quantity and quality of the extracted DNA was determined by using a Nanodrop 2000c Apparatus (Thermo Scientific, Singapore). Extracted DNA samples were kept at −80°C for long-term storage. Blood, saliva, and urine were collected from three healthy uninfected persons and used for the PCR as negative controls.
The PCR was performed in a final volume of 25 μL containing approximately 100 ng of extracted DNA, 10 μM of each primer, 25 mM of MgCl2, 2 mM of dNTPs, and 1 unit of Taq DNA polymerase (Fermentas, Pittsburgh, PA). The primers were designed to anneal specifically to the ITS1 regions of ribosomal RNA (rRNA) of Leishmania parasites described by Spanakos and others:21 (LeF: 5′-TCC GCC CGA AAG TTC ACC GAT A-3′ and LeR: 5′-CCA AGT CAT CCA TCG CGA CAC G-3′). The PCRs were performed in a PCR Mastercycler® Pro (Eppendorf, Hamburg, Germany) with conditions as follows: denaturation at 94°C for 4 minutes; followed by 40 cycles at 94°C for 1 minute, 65°C for 1 minute, and 72°C for 1 minute; and a final extension at 72°C for 7 minutes. Aliquots of the PCR amplicons were analyzed by electrophoresis on 1.5% agarose gels, stained with ethidium bromide, and visualized with Quantity One Quantification Analysis Software version 4.5.2 (Gel Doc EQ System; Bio-Rad, Hercules, CA). The extracted DNA samples from an uninfected persons and a no DNA template (double-distilled water) were used as negative controls in every PCR. Reactions in which either or both negative controls contained bands were discarded.
Confirmatory testing for PCR was performed by using another set of primers to amplify the partial small subunit (SSU) rRNA genes of the Leishmania parasite. The primer sequences (R221: 5′-GGT TCC TTT CCT GAT TTA CG-3′ and R332: 5′-GGC CGG TAA AGG CCG AAT AG-3′) and PCR conditions were described by Van Eys and others.22
The PCR amplicons were ligated into the pGEM-T Easy Vector (Promega, Madison, WI). Ligation reactions mixture was composed of 5 μL of 2× Rapid ligation buffer, 3 μL of PCR products, 1 μL pGEM-T Easy Vector, and 1 μL double-distilled water. The ligated vectors were transformed into DH5α competent cells and chimeric plasmids were screened by blue–white colony selection system. The suspected positive colonies were cultured and used for further plasmid DNA extraction by using the Invisorb® Spin Plasmid Mini Kit following the manufacturer's instructions. Purified plasmids were sequenced by 1st BASE DNA sequencing services (1st Base Laboratories, Kuala Lumpur, Malaysia) by using universal forward T7 primer. Nucleotide sequences were analyzed by using BioEdit Sequence Alignment Editor Version 7.0.9.0 (www.mbio.ncsu.edu/bioedit/bioedit.html), and consensus sequences were compared with available sequence data in a GenBank by BLAST search (www.ncbi.nlm.gov/BLAST).
The study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB no. 385/55).
Patient 1 was a 46-year-old Thai man who was a rubber planter from southern Thailand. He has been given a diagnosis with HIV infection in 2003 and received boosted lopinavir and lamivudine. His CD4+ T cell count was 175 cells/mm3, and serum virus levels were < 40 copies/mL. He also had Evans syndrome (an autoimmune disorder with destruction of erythrocytes, platelets, and leukocytes), a left knee ulcer, and hepatosplenomegaly. A bone marrow study showed Leishmania amastigotes within macrophages. Bone marrow, blood, discharge from the ulcer, saliva, and urine were cultured and DNA was extracted for PCR.
He improved after two weeks of intravenous amphotericin B deoxycholate (1 mg/kg/day), followed by 400 mg of itraconazole/day. A recurrence after two months of itraconazole therapy was re-treated with three weeks of intravenous liposomal amphotericin B (3 mg/kg/day), followed by 400 mg of itraconazole per day. Blood and saliva were collected for PCR three months after re-treatment and were negative. Details of this patient were reported by Chusri and others.14
Patient 2 was 30-year-old pet store owner from southern Thailand who had been given a diagnosis of HIV infection in 1999. The patient received tenofovir, lamivudine, and nevirapine. His CD4+ T-cell count was 111 cells/mm3 and viral RNA was not detectable (< 40 copies/mL). He had multiple papules and plaques with ulceration and discharges. He also had anemia, thrombocytopenia, and hepatosplenomegaly. Bone marrow, papule, and ulcer biopsy specimens showed Leishmania amastigotes within macrophages. Bone marrow, blood, tissue biopsy specimens, and saliva and urine samples were used for culture and PCR. The patient received intravenous amphotericin B deoxycholate (1 mg/kg/day) for two weeks, followed by 400 mg of itraconazole per day. Blood and saliva were collected three months later and were negative for Leishmania and no recurrence was observed. This patient was reported by Chusri and others.14
Patient 3 was a 60-year-old man with diabetes who lived in Yangon, Myanmar and had not previously traveled abroad. He showed development of fever, multiple infiltrative skin lesions, and oral ulcers. A skin biopsy specimen indicated Sweet's syndrome or acute febrile neutrophilic dermatosis, which is a skin disease with fever and painful skin lesions that is commonly present on arms, neck, face, and back. He was treated with systemic corticosteroids for two months without improvement. He came to Thailand for further evaluation. Multiple erythematous, shiny, infiltrative plaques; nodules on face, trunk, and extremities; and oral ulcers and white patches on the buccal mucosa were observed. Lymph nodes, liver, and spleen were not palpable. Complete blood counts, blood urea nitrogen levels, creatinine levels, and liver function test results were within normal limits. Results for antinuclear antibodies, antibodies against HIV, and C-reactive protein levels were unremarkable.
A new skin biopsy specimen of a trunk nodule showed diffuse histiocytic infiltrate and multinucleated giant cells in the upper and deep dermis. Many round-to-oval small organisms were present in histiocytes and fibrous stroma. They stained positive with Periodic Acid–Schiff (PAS) stain. Numerous small yeast-liked organisms were present in histiocytes and in the stroma. Some of these organisms were large and contained small basophilic dots in cytoplasm near nuclei stained positively with PAS stain but not with Gomori's methenamine silver stain and Giemsa. This finding suggested the presence of Leishmania parasites. Blood samples, tissue biopsy specimens, saliva and urine samples were used for culture and were negative. PCR testing of the ITS1 region of the rRNA gene in blood, skin biopsy specimen, urine, and saliva, and DNA sequencing identified L. siamensis. The patient was treated with intravenous amphotericin B for 40 days (total dose = 2.1 grams). After regression of the cutaneous lesions, he was discharged. When seen again two months later, he had gained weight but a few skin lesions were still present. He returned home and was lost to follow-up.
Patient 4 was the 22-year-old daughter of patient 3; she came to Thailand with her father. She was healthy and lived with her father in Yangon. Results of her physical examination were unremarkable. Saliva and urine samples were collected and used for culture; these samples were negative for L. siamensis. PCR testing of the ITS1 region of the rRNA gene in saliva DNA, followed by DNA sequencing, identified L. siamensis. Whole blood and buffy coat were then collected for Leishmania detection by culture, staining with Giemsa, and PCR. Only PCR identified L. siamensis in buffy coat. We planned to follow-up the patient for detection of Leishmania parasites without treatment of leishmaniasis but the patient was lost to follow-up.
Patient 5 was a 45-year-old Thai man living in Chiang Rai, Thailand. He has been given a diagnosis of HIV infection in 2005. He had a CD4+ T cell count of < 50 cells/mm3. He was treated with stavudine, lamivudine, and nevirapine. In 2007, He showed development of lumpy skin lesions that were first not investigated. He was later hospitalized with fever, oral candidiasis, pancytopenia, pancreatitis, type 2 diabetes mellitus, epistaxis, perianal abscess, urinary retention, and abnormal liver function test results. Skin biopsy specimens from lesions present for more than five years showed epidermal hyperplasia, diffuse fibrosis, dilated blood vessels, and mild perivascular lymphohistiocytic infiltrates in the dermis. There were a few clumps of small parasites in histiocytes and extracellularly between collagen fibers in the upper dermis. They stained with Giemsa. Blood, tissue biopsy specimen, and saliva and urine samples were used for culture but were negative. A PCR of ITS1 region of the rRNA gene in blood, tissue biopsy specimen, and saliva DNA, followed by DNA sequencing, confirmed the presence of L. siamensis. The patient was then confirmed as having cutaneous leishmaniasis. He was severely debilitated and died of systemic bacterial infection without treatment of leishmaniasis.
Patient 6 was a 34-year-old Burmese man from Yangon who was seropositive for HIV for six years. He was treated with truvada, legalon, isoniazid, rifampicin, ethambutal, and pyrazinamide. Five months later, he was hospitalized with high fever and diarrhea. He was empirically treated with clarithromycin and moxifloxacin. He showed development of multiple, umbilicated, erythematous papules on his neck, arms, and chest wall. Skin biopsy specimens showed a moderately dense superficial and deep perivascular and interstitial histiocytic infiltrate. Many small yeast-like organisms were present in the cytoplasm of histiocytes and between collagen fibers. They stained with PAS and Giemsa, leading to diagnosis of cutaneous leishmaniasis. Blood, tissue, saliva, and urine samples were cultured. A PCR of the ITS1 region of the rRNA gene in blood, skin biopsy specimen, and saliva DNA, followed by DNA sequencing, identified L. siamensis. The patient was treated with liposomal amphotericin B and anti-tuberculosis therapy was continued. Clinical recovery resulted within one month.
Specimens were obtained from patients with different clinical presentations of leishmaniasis (Table 1) . Five of 6 patients were immunocompromised, only patient 4 was immunocompetent. Amastigotes of L. siamensis were detected in bone marrow and blood of patients 1 and 2, and in tissue biopsy specimens of patients 2, 3, 5, and 6 by microscopic examinations. Cultures were positive only for patients 1 and 2 (Table 1). Patients 3, 5, and 6 were treated with antifungal drugs before blood was collected for culture. Leishmania parasites in saliva were not detected in any cases by microscopy.
Table 1.
Results of microscopic examination, culture, and polymerase chain reaction for detection of L. siamensis DNA from various clinical specimens collected under different conditions and different clinical presentations from six patients*
| Patient | Age (years) | Sex/nationality | Clinical presentation/host conditions | Microscopic examination/culture for Leishmania | PCR detection of L. siamensis DNA | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood smear | Skin biopsy | Blood/bone marrow culture | Bone marrow | Blood | Buffy coat | Saliva | Urine | Tissue | |||||
| 1 | 46 | M/Thai | CL and VL/HIV infection and prednisolone therapy | + | N/A | +/+ | + | + | + | + | + | N/A | Chusri and others14 |
| 2 | 30 | M/Thai | CL and VL/HIV infection | + | + | –/+ | + | + | + | + | + | + | Chusri and others14 |
| 3 | 60 | M/Burmese | CL/DM and Prednisolone therapy | – | + | –/N/A | N/A | + | + | + | + | + | Unpublished data |
| 4 | 22 | F/Burmese | Asymptomatic/normal | – | N/A | –/N/A | N/A | - | + | + | – | N/A | Unpublished data |
| 5 | 45 | M/Thai | CL/HIV infection | – | + | –/N/A | N/A | + | + | + | – | + | Unpublished data |
| 6 | 34 | M/Burmese | CL/HIV infection | – | + | –/N/A | N/A | + | + | + | – | + | Unpublished data |
PCR = polymerase chain reaction; CL = cutaneous leishmaniasis; VL = visceral leishmaniasis; HIV = human immunodeficiency virus; NA = not available.
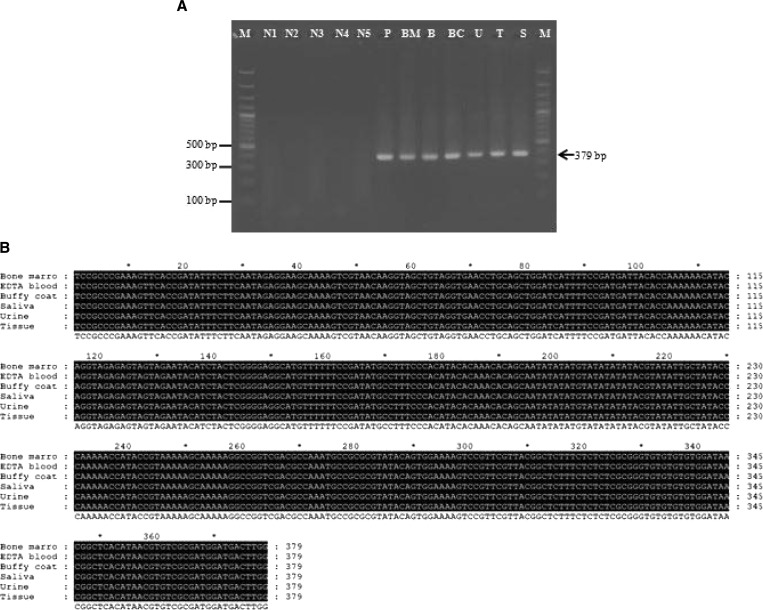
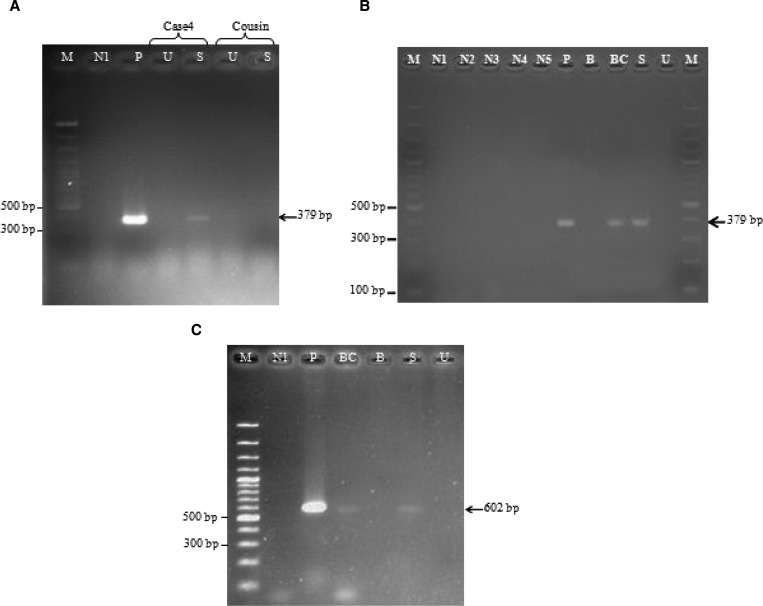
The PCR amplicon used in this study was 379 basepairs (Figure 1A ). Nucleotide sequence analysis of all samples identified L. siamensis (Figure 1B). In cases where bone marrow or tissue biopsy specimens were available, both types of specimens were also positive by PCR (Table 1 and Figure 1A). Saliva and buffy coats were positive by PCR in all cases. Saliva and urine from an asymptomatic patient (patient 4) and her cousin was used for screening by PCR, but only patient 4 had a positive result (Figure 2A ). Saliva and urine samples were collected from patient 4 again when she provided a blood sample, the PCR result was positive for saliva and buffy coat samples (Figure 2B). The PCR was also performed with another set of primers specific for the SSU rRNA gene for Leishmania parasite. DNA extracted from saliva and buffy coat of patient 4 was amplified by these primers (Figure 2C), and sequences of the amplified PCR products were 100% identical to the SSU rRNA gene of L. siamensis (GenBank accession no GQ226033).
Figure 1.
Polymerase chain reaction amplification of various sources of specimen of patient case 2 (A) were analyzed by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. Lane BM, bone marrow; lane B, Blood; lane BC, buffy coat; lane S, saliva; lane U, urine; lane T, tissue; lane M,: molecular mass marker (100 basepairs [bp]; lane, P, positive control containing extracted DNA from cultured L. siamensis; lane N1, negative control (no DNA template: double-distilled water); lanes N2–N5: negative control (DNA template from non- infected saliva, urine, blood, and buffy coat, respectively). Comparison of internal transcribed spacer 1 (ITS1) gene sequences amplified from various sources of specimen of the patient 2 (B), Amplified sequences of L. siamensis ITS gene from bone marrow, blood, saliva, urine, and tissue biopsy of patient 2 were assigned GenBank numbers KF227887–KF227892, respectively.
Figure 2.
Polymerase chain reaction (PCR) amplification of the internal transcribed spacer 1 (ITS1) gene of Leishmania parasites of the first saliva and urine samples collected from case 4 and her cousin (A) and the second saliva, urine, blood, and buffy coat samples collected from case 4 (B), PCR amplification of case 4 by using primers annealed specifically to small subunit ribosomal RNA of Leishmania parasites (C). Lane B, blood; lane BC, buffy coat; lane S, saliva; lane U, urine; lane M, molecular mass marker (100 basepairs [bp]); lane P, positive control containing DNA from cultured L. siamensis; lane N1, negative control (no DNA template: double-distilled water); lanes N2–N5, negative control (DNA template from non-infected saliva, urine, blood, and buffy coat, respectively).
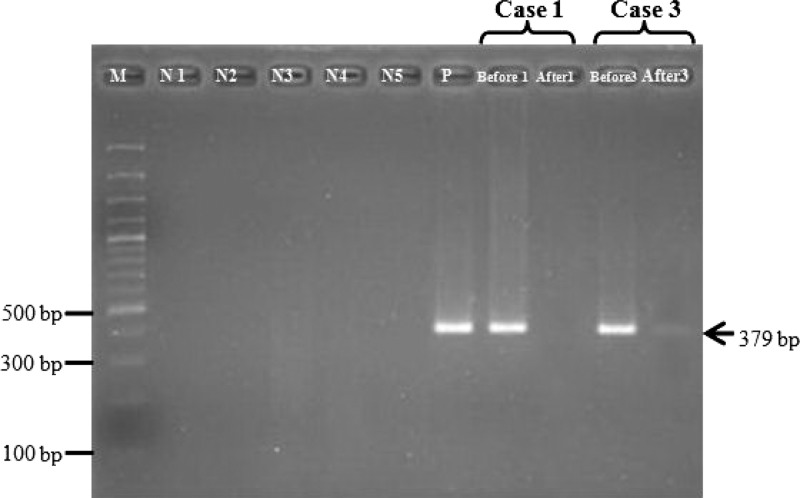
Detection of L. siamensis DNA in saliva pre-treatment and post-treatment was performed for patient 1. Blood and saliva were collected three months after treatment, but L. siamensis DNA was not detected in blood and saliva samples. Blood and saliva was collected two weeks after treatment from patient 3 and L. siamensis DNA was still detected (Figure 3).
Figure 3.
Changes in L. siamensis DNA in saliva after treatment. Lane M, molecular mass marker (100 basepairs [bp])); lane N1, negative control (no-DNA template: double-distilled water), lanes N2–N5: negative control (DNA template from non-infected saliva, urine, blood, and buffy coat, respectively); lane P, positive control; lane Before1, 3: pre-treatment of case 1 and case 3; lane After 1, 3: post-treatment of case 1 and case 3, respectively.
Autochthonous leishmaniasis cases caused by L. siamensis have been reported in patients in Thailand and Myanmar. The prevalence of this disease has dramatically increased in past few years.12,14,15 Most cases from Thailand have been reported in the southern region of the country14, and cases in Myanmar patients were reported in Yangon; these cases were cutaneous, visceral, and asymptomatic cases (unpublished data). In this study, patients (except patient 4) were confirmed by demonstration of the parasites in blood smears, tissue biopsy specimens, or culture. The PCR was used to detect Leishmania DNA in bone marrow, blood, buffy coat, tissue, saliva, and urine. L. siamensis DNA was detected in the saliva of all 6 patients. Interestingly, in an asymptomatic leishmaniasis patient (patient 4), we were unable to detect Leishmania by microscopic examination and culture, but Leishmania DNA was detected in saliva and buffy coat. There was only one patient in whom the PCR result was positive. However, this woman was asymptomatic and we could not detect Leishmania by other means.
To avoid DNA contamination in the PCR, the PCR were performed with all precautions suggested by Kwok and Higuchi.23 There are several reports of viable L. donovani found in nasal, oral, and nasopharyngeal secretions,20,24 but L. siamensis in this study was not detected in saliva by either microscopic examination or culture. This finding led to inappropriate treatment with antifungal agents before it was confirmed by PCR in buffy coat and saliva. Urine is another source for detection of L. siamensis DNA. Although there are several reports of renal involvement in patients with leishmaniasis,12,25–27 the six patients had no evidence of renal disease. DNA extraction from urine requires 30 mL of urine, and we found that 50% of the patients in our series were negative for Leishmania DNA yet positive for DNA in saliva.
In regions in which the incidence of L. siamensis infection is low, immunologic diagnostic tests are not readily available. Demonstration of Leishmania by microscopic examination is the traditional test for diagnosis. It requires expertise to distinguish Leishmania from other pathogens such as Histoplasma capsulutum or Penicillium manefeii. Culture for Leishmania is available only in few laboratories. The current state of the art diagnosis of L. siamensis infection relies on PCR and nucleotide sequencing. These techniques are more sensitive than others,6,9,28–31 and they can now be performed in most provincial and university hospitals in Thailand. Although use of traditional screening tests for this disease is being investigated, PCR could be used for survey and surveillance studies, including asymptomatic persons. Our report demonstrates that saliva is a good source of L. siamensis DNA, and that parasite DNA can also be found in asymptomatic patients. Furthermore, in symptomatic patients in whom leishmaniasis is a possibility, multiple studies on different samples by using PCR with sequencing are indicated.
Footnotes
Financial support: This study was supported by Integrated Innovation Academic Center: IIAC Chulalongkorn University Centenary Academic Development Project; the Higher Education Research Promotion and National Research University Project of Thailand; the Office of the Higher Education Commission (HR1160A-56), a tuition fee scholarship from the Graduate School, Chulalongkorn University; the Thailand Research Fund to Wej Choochote (TRF Senior Research Scholar: RTA5480006); the Ratchadapisak Sompotch Fund, Faculty of Medicine, Chulalongkorn University (grant no. RA (MF) 02/56); and the National Science and Technology Development Agency (Thailand).
Authors' addresses: Atchara Phumee, Medical Science Programme, Faculty of Medicine, Chulalongkorn University, Bangkok 10330, Thailand, E-mail: amphumee@gmail.com. Kanyarat Kraivichian and Vivornpun Sanprasert, Department of Parasitology, Faculty of Medicine, Chulalongkorn University, Bangkok 10330, Thailand, E-mails: iamteaw@yahoo.com and vivornpun@gmail.com. Sarunyou Chusri, Division of Infectious Diseases, Department of Medicine, Faculty of Medicine, Prince of Songkla University, Songkhla 90110, Thailand, E-mail: sarunyouchusri@hotmail.com. Nopadon Noppakun, Division of Dermatology, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok 10330, Thailand and Bumrungrad International Hospital, Wattana, Bangkok, 10110 Thailand, E-mail: dr.nopadon@yahoo.com. Asda Vibhagool, Bumrungrad International Hospital, Wattana, Bangkok 10110, Thailand, E-mail: asdavi@gmail.com. Vich Tampanya, Chiangrai Prachanukroh Hospital, Chiang Rai, Chiang Rai 57000, Thailand, E-mail: aekcha@gmail.com. Henry Wilde, Division of Infectious Diseases, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok 10330 Thailand, E-mail: wildehenry@yahoo.com. Padet Siriyasatien, Department of Parasitology, Faculty of Medicine, Chulalongkorn University, Bangkok 10330, Thailand, and Excellence Center for Emerging Infectious Diseases, King Chulalongkorn Memorial Hospital, Thai Red Cross Society, Bangkok, Thailand, E-mail: padet.s@chula.ac.th.
References
- 1.Feasey N, Jones MW, Mabey DC, Solomon AW. Neglected tropical diseases. Br Med Bull. 2010;93:179–200. doi: 10.1093/bmb/ldp046. [DOI] [PubMed] [Google Scholar]
- 2.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 3.Roberts MT. Current understandings on the immunology of leishmaniasis and recent developments in prevention and treatment. Br Med Bull. 2006;75–76:115–130. doi: 10.1093/bmb/ldl003. doi:10.1093/bmb/ldl003. [DOI] [PubMed] [Google Scholar]
- 4.Berman JD. Human leishmaniasis: clinical, diagnostic and therapeutic developments in the last 10 years. Clin Infect Dis. 1997;24:684–703. doi: 10.1093/clind/24.4.684. [DOI] [PubMed] [Google Scholar]
- 5.Andresen K, Gaafar A, El-Hasan AM, Ismail A, Dafaila M, Theander TG, Kharazmi A. Evaluation of the polymerase chain reaction in the diagnosis of cutaneous leishmaniasis due to Leishmania major. A comparison with direct microscopy of smears and sections from lesions. Trans R Soc Trop Med Hyg. 1996;90:133–135. doi: 10.1016/s0035-9203(96)90112-1. [DOI] [PubMed] [Google Scholar]
- 6.Aviles H, Belli A, Armijos R, Monroy F, Harris E. PCR detection and identification of Leishmania parasites in clinical specimens in Ecuador: a comparison with classical diagnostic methods. J Parasitol. 1999;85:181–187. [PubMed] [Google Scholar]
- 7.Mary C, Faraut FO, Lascombe L, Dumon H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol. 2004;42:5249–5255. doi: 10.1128/JCM.42.11.5249-5255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahbazi F, Shahabi S, Kazemi B, Mohebali M, Abadi AR, Zare Z. Evaluation of PCR assay in diagnosis and identification of cutaneous leishmaniasis: a comparison with the parasitological methods. Parasitol Res. 2008;103:1159–1162. doi: 10.1007/s00436-008-1111-4. [DOI] [PubMed] [Google Scholar]
- 9.Osman OF, Oskam L, Zijlstra EE, Kroon NC, Schoone GJ, Khalil ET, EL-Hassan AM, Kager PA. Evaluation of PCR for diagnosis of visceral leishmaniasis. J Clin Microbiol. 1997;35:2454–2457. doi: 10.1128/jcm.35.10.2454-2457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lightner LK, Chulay JD, Bryceson AD. Comparison of microscopy and culture in the detection of Leishmania donovani from splenic aspirates. Am J Trop Med Hyg. 1983;32:296–299. doi: 10.4269/ajtmh.1983.32.296. [DOI] [PubMed] [Google Scholar]
- 11.Sukmee T, Siripattanapipong S, Mungthin M, Worapong J, Rangsin R, Samung Y, Kongkaew W, Bumrungsana K, Chanachai K, Apiwathanasorn C, Rujirojindakul P, Wattanasri S, Ungchusak K, Leelayoova S. A suspected new species of Leishmania, the causative agent of visceral leishmaniasis in a Thai patient. Int J Parasitol. 2008;38:617–622. doi: 10.1016/j.ijpara.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Suankratay C, Suwanpimolkul G, Wilde H, Siriyasatien P. Case report: autochthonous visceral leishmaniasis in a human immunodeficiency virus (HIV)-infected patient: the first in Thailand and review of the literature. Am J Trop Med Hyg. 2010;82:4–8. doi: 10.4269/ajtmh.2010.09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kongkaew W, Siriarayaporn P, Leelayoova S, Supparatpinyo K, Areechokchai D, Duang-ngern P, Chanachai K, Sukmee T, Samung Y, Sridurongkathum P. Autochthonous visceral leishmaniasis: a report of a second case in Thailand. Southeast Asian J Trop Med Public Health. 2007;38:8–12. [PubMed] [Google Scholar]
- 14.Chusri S, Hortiwakul T, Silpapojakul K, Siriyasatien P. Case report: consecutive cutaneous and visceral leishmaniasis manifestations involving a novel Leishmania species in two HIV patients in Thailand. Am J Trop Med Hyg. 2012;87:76–80. doi: 10.4269/ajtmh.2012.11-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bualert L, Charungkiattikul W, Thongsuksai P, Mungthin M, Siripattanapipong S, Khositnithikul R, Naaglor T, Ravel C, Baidouri FE, Leelayoova S. Case report: autochthonous disseminated dermal and visceral leishmaniasis in an AIDS patient, southern Thailand, caused by Leishmania siamensis. Am J Trop Med Hyg. 2012;86:821–824. doi: 10.4269/ajtmh.2012.11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viriyavejakul P, Viravan C, Riganti M, Punpoowong B. Imported cutaneous leishmaniasis in Thailand. Southeast Asian J Trop Med Public Health. 1997;28:558–562. [PubMed] [Google Scholar]
- 17.Maharom P, Siripattanapipong S, Mungthin M, Naaglor T, Sukkawee R, Pudkorn R, Wattana W, Wanachiwanawin D, Areechokchai D, Leelayoova S. Visceral leishmaniasis caused by Leishmania infantum in Thailand. Southeast Asian J Trop Med Public Health. 2008;39:988–990. [PubMed] [Google Scholar]
- 18.Corvalan FH, Sampaio RN, Brustoloni YM, Andreotti R, Lima Júnior MS. DNA identification of Leishmania (Viannia) braziliensis in human saliva from a patient with American cutaneous leishmaniasis. J Venom Anim Toxins Incl Trop Dis. 2011;17:98–102. [Google Scholar]
- 19.Galaï Y, Chabchoub N, Abid MB, Abda IB, Bouafif NB, Amri F, Aoun K, Bouratbine A. Diagnosis of Mediterranean visceral leishmaniasis by detection of Leishmania antibodies and Leishmania DNA in oral fluid samples collected using an Oracol device. J Clin Microbiol. 2011;49:3150–3153. doi: 10.1128/JCM.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forkner CE, Zia LS. Viable Leishmania donovani in nasal and oral secretions of patients with kala-azar and the bearing of this finding on the transmission of the disease. J Exp Med. 1934;59:491–499. doi: 10.1084/jem.59.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spanakos G, Piperaki ET, Menounos PG, Tegos N, Flemetakis A, Vakalis NC. Detection and species identification of Old World Leishmania in clinical samples using a PCR-based method. Trans R Soc Trop Med Hyg. 2007;102:46–53. doi: 10.1016/j.trstmh.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Van Eys GJ, Schoone GJ, Kroon NC, Ebeling SB. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol. 1992;51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- 23.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 24.Mebrahtu YB, Hendricks LD, Oster CN, Lawyer PG, Perkins PV, Pamba H. Leishmania donovani parasites in the nasal secretions, tonsillopharyngeal mucosa, and urine centrifugates of visceral leishmaniasis patients in Kenya. Am J Trop Med Hyg. 1993;48:530–535. doi: 10.4269/ajtmh.1993.48.530. [DOI] [PubMed] [Google Scholar]
- 25.Efstratiadis G, Boura E, Giamalis P, Mandala E, Leontsini M, Tsiaousis G, Memmos D. Renal involvement in a patient with visceral leishmaniasis. Nephrol Dial Transplant. 2006;21:235–236. doi: 10.1093/ndt/gfi157. [DOI] [PubMed] [Google Scholar]
- 26.Dutra M, Martinelli R, de Carvalho EM, Rodrigues LE, Brito E, Rocha H. Renal involvement in visceral leishmaniasis. Am J Kidney Dis. 1985;6:22–27. doi: 10.1016/s0272-6386(85)80034-2. [DOI] [PubMed] [Google Scholar]
- 27.Amann K, Bogdan C, Harrer T, Rech J. Renal leishmaniasis as unusual cause of nephrotic syndrome in an HIV patient. J Am Soc Nephrol. 2012;23:586–590. doi: 10.1681/ASN.2011050472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero GA, Guerra MV, Paes MG, Cupolillo E, Bentin Toaldo C, Macedo VO, Fernandes O. Sensitivity of the polymerase chain reaction for the diagnosis of cutaneous leishmaniasis due to Leishmania (Viannia) guyanensis. Acta Trop. 2001;79:225–229. doi: 10.1016/s0001-706x(01)00140-1. [DOI] [PubMed] [Google Scholar]
- 29.Mathis A, Deplazes P. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J Clin Microbiol. 1995;33:1145–1149. doi: 10.1128/jcm.33.5.1145-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smits HL, Hartskeerl RA. PCR amplification reactions in parasitology. J Microbiol Methods. 1995;23:41–54. [Google Scholar]
- 31.Piarroux R, Azaiez R, Lossi AM, Reynier P, Muscatelli F, Gambarelli F, Fontes M, Dumon H, Quilici M. Isolation and characterization of a repetitive DNA sequence from Leishmania infantum: development of a visceral leishmaniasis polymerase chain reaction. Am J Trop Med Hyg. 1993;49:364–369. doi: 10.4269/ajtmh.1993.49.364. [DOI] [PubMed] [Google Scholar]