Abstract
Cryptococcosis is a fungal disease affecting more than one million people per year worldwide. The main etiological agents of cryptococcosis are the two sibling species Cryptococcus neoformans and Cryptococcus gattii that present numerous differences in geographical distribution, ecological niches, epidemiology, pathobiology, clinical presentation and molecular characters. Genotyping of the two Cryptococcus species at subspecies level supplies relevant information to understand how this fungus has spread worldwide, the nature of its population structure, and how it evolved to be a deadly pathogen. At present, nine major molecular types have been recognized: VNI, VNII, VNB, VNIII, and VNIV among C. neoformans isolates, and VGI, VGII, VGIII, and VGIV among C. gattii isolates. In this paper all the information available in the literature concerning the isolation of the two Cryptococcus species has been collected and analyzed on the basis of their geographical origin, source of isolation, level of identification, species, and molecular type. A detailed analysis of the geographical distribution of the major molecular types in each continent has been described and represented on thematic maps. This study represents a useful tool to start new epidemiological surveys on the basis of the present knowledge.
1. Introduction
According to the last report (December 2010) from the Joint United Nations Program on HIV/AIDS and the World Health Organization (http://www.unaids.org/), 34 million people worldwide suffer from HIV infection/AIDS, 2.7 million people are newly infected every year from this disease, and 1.8 million people die from AIDS-related causes. Neuropathological conditions are present in approximately 70% to 90% of AIDS patients [1]. Cryptococcal meningitis is considered an AIDS-defining condition [2–4], and it is the most common fungal infection of the central nervous system and the third most frequent neurological complication in AIDS patients [1]. The main etiological agents of cryptococcosis are the basidiomycetous yeasts Cryptococcus neoformans and Cryptococcus gattii which can also infect, although with a significantly lower incidence, people with decreased immunity such as individuals with sarcoidosis, lymphoproliferative disorders, those undergoing immunosuppressive therapies [4–6], and more rarely immunocompetent people [7, 8]. Worldwide, C. neoformans and C. gattii infections cause an estimated one million cases of cryptococcal meningitis per year among people with HIV/AIDS, resulting in nearly 625,000 deaths (Centers for Disease Control and Prevention, CDC, Atlanta, USA, http://www.cdc.gov/). The greatest burden of disease occurs in sub-Saharan Africa, where mortality is estimated to be 50% to 70% [9, 10]. In the United States and other developed countries, cryptococcosis is decreasing among persons with HIV/AIDS due to the availability of high active antiretroviral therapy, and, at present, the mortality is around 12% (CDC, Atlanta, USA).
Although in the past the etiological agent of cryptococcosis was considered a homogeneous anamorphic species (C. neoformans), now the two species C. neoformans and C. gattii have been separated on the basis of numerous differences such as geographical distribution, ecological niches, epidemiology, pathobiology, clinical presentation, and molecular characters. C. neoformans species is further classified in two varieties, C. neoformans var. grubii (serotype A) and C. neoformans var. neoformans (serotype D), which are also able to recombine and to produce diploid or aneuploid intervarietal AD hybrids [78, 131, 132]. C. neoformans has been widely associated to avian excreta [44, 97, 98, 133–136] although it has been isolated also from other sources [11, 28, 60, 99, 100, 137].
C. gattii species is classified in two different serotypes, B and C, which have not yet elevated to the variety status. This species was thought to be restricted to tropical and subtropical regions but a recent outbreak due to C. gattii infection, which occurred in Vancouver Island and North West Pacific Coast of America [121, 138], has expanded the geographical area of this pathogen also to temperate regions. In addition, interspecies hybrids between C. neoformans and C. gattii have been found in Colombia, Brazil, India, and The Netherlands [29, 122, 139, 140].
Besides a prevalent asexual life cycle, both species have also a bipolar sexual cycle with two mating types, MATa and MATα, the latter being the most prevalently isolated from both patients and environment. Filobasidiella neoformans and Filobasidiella bacillispora are the sexual states of C. neoformans and C. gattii, respectively [141, 142]. During sexual recombination, either filaments with clamp connections and basidiospores are produced [143]. Recombinant basidiospores are also produced via same-sex mating [144] and are thought to be the propagules responsible for the infection of the host [143].
The availability of whole genome sequences from C. neoformans and C. gattii strains and the recent progresses in the molecular biology have greatly advanced our understanding of this pathogenic yeast [145, 146]. The disease aspects of cryptococcal infection are becoming better defined, while the life cycle of this fungus in the environment remains less well established. How this fungus has spread worldwide, the nature of its population structure, and how it evolved to be a deadly pathogen are ongoing research subjects that are key to our understanding of this environmental pathogen.
Due to the importance of the C. neoformans/C. gattii species complex as human fungal pathogens, several research groups are currently focusing on the molecular determination of the number of genetically diverged subgroups within each species. Several molecular typing techniques have been applied: multilocus enzyme electrophoresis (MLEE) [147]; DNA fingerprinting [148]; random amplification of polymorphic DNA (RAPD) [12, 149]; PCR fingerprinting [150]; amplified fragment length polymorphism (AFLP) [11]; restriction fragment length polymorphism (RFLP) of PLB1 [13], GEF1 [37], or URA5 genes [64]; sequencing of ITS1-5.8S-ITS2 rDNA region [38] or intergenic spacer region (IGS) [151] and, more recently, multilocus sequence typing (MLST) [14, 45, 53]; multilocus microsatellite typing (MLMT) [89, 140]; matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method (MALDI-TOF) analysis [79, 152, 153].
The multitude of data obtained with different typing methods has raised the problem to compare the results and the need to standardize genotypes nomenclature among comparable methods. The first steps towards this direction have been recently achieved by the comparison of the results obtained using the most common Cryptococcus typing methods [15] and by the identification of eight major molecular types among the two Cryptococcus species. A second step was to standardize a gold standard typing method able to produce unambiguous and comparable results [154]. Finally, a global database was implemented in order to collect the different genotypic profile and to make the data available for the research community [154]. This task is the main aim of the activity of the ISHAM Cryptococcus working Group for “Genotyping of Cryptococcus neoformans and Cryptococcus gattii” which promotes the genotyping of the two Cryptococcus species to elucidate the global epidemiology of this life-threatening pathogen.
2. Standardization of Cryptococcus Molecular Typing Methods
Although numerous molecular techniques have been applied to subtype C. neoformans and C. gattii strains, only three methods were proved to produce comparable results: PCR fingerprinting, AFLP, and MLST.
PCR fingerprinting is based on the amplification of DNA sequences flanked by simple DNA repeats which are used as single primers in the PCR. The amplification produces a banding profile that discriminates the strains at subspecies level. The primers employed in PCR fingerprinting include the minisatellite-specific core sequence of wild-type phage M13 (5′-GAGGGTGGCGGTTCT-3′) and the microsatellite-specific primer (GACA)4. The technique was first applied to Cryptococcus typing in 1993 [150] to study a set of cryptococcal strains. A high polymorphism was detected among the investigated strains which could be separated in two groups corresponding to serotype A and serotype D and a third one including both serotypes B and C. In a different study, a variety of genotyping clusters was identified during the investigation of some Italian C. neoformans isolates by (GACA)4 PCR fingerprinting [155]. The results showed a strong correlation between genotypes and serotypes. The more prevalent genotype was named VN1 and corresponded to C. neoformans var. neoformans, serotype D, a second genotype was identified as VN6 and corresponding to C. neoformans var. grubii, serotype A, and further two genotypes (VN3 and VN4) included isolates with a banding pattern intermediate between VN1 and VN6 suggesting that these strains were AD hybrids. Therefore, this study provided, for the first time, a tool to identify unambiguously intervarietal AD hybrids. In a first attempt to standardize the technique, PCR fingerprinting, using either M13 (GACA)4 primer, and RAPD were applied to genotype 356 C. neoformans global isolates [15]. Both typing methods were able to identify four different molecular types: VNI and VNII corresponding to C. neoformans var. grubii, serotype A, VNIV corresponded to C. neoformans var. neoformans, serotype D, and VNIII including all AD hybrids. Later, a collaborative network between Spanish and Latin American researchers was established [64], and the 340 isolates collected were investigated by M13 PCR fingerprinting and URA5 RFLP. The results showed, for the first time, the distribution of eight molecular types in the studied countries. The molecular types VNI, VNII, VNIII and VNIV were recognized among the C. neoformans isolates, as previously reported, and further four molecular types, VGI, VGII, VGIII, and VGIV, were found among C. gattii isolates. PCR fingerprinting was then applied in several studies, and, at present, thousands of strains from different countries of the world have been characterized using this typing method [14, 30, 37, 39, 56, 78, 80, 93, 97, 101, 120, 156–164].
The AFLP typing method is based on digestion of DNA samples with a frequent and a rare cutting endonuclease enzyme combined with amplification using an adaptor that creates specificity at the restriction sites. Subsequent rounds of PCR are able to select a unique profile depending on the number of nucleotides added to the primers. Fluorescently labeled fragments are separated by an automated capillary sequencer and visualized as a virtual banding profile [165]. Application of this technique to Cryptococcus typing requires a digestion with MseI and EcoRI restriction enzymes and an amplification with the two selective primers MseI-G and EcoRI-AC [11]. The analysis of 207 global C. neoformans and C. gattii isolates led to identify three AFLP genotypes (AFLP1, APLP2, and AFLP 3) among C. neoformans strains and three (AFLP4, AFLP5, and AFLP6) among C. gattii. In addition, two further subtypes of the genotype AFLP1 were identified as AFLP1A and AFLB1B [11]. Since AFLP is a technique with a high discriminatory power, being able to assign a unique profile to each strain, it contributed to elucidate the cause of the outbreak of C. gattii in Vancouver Island [157]. In the study, two AFLP6 subtypes were clearly identified as the cause of the outbreak: AFLP6A and AFLP6B. Since AFLP6A was isolated in 75% of the cases in Vancouver Island environment and AFLP6B isolates were less frequent, it was hypothesized that the former genotype was more virulent than the latter. The higher virulence of AFLP6A than AFLP6B was actually shown later in murine model [14]. Finally, AFLP application to Cryptococcus typing contributed to discover new interspecies hybrids between C. neoformans and C. gattii. Both AFLP1/AFLP4 (AFLP9) and AFLP3/AFLP4 (AFLP8) hybrid isolates were identified during some studies carried out in The Netherlands [122, 139, 140].
MLST is a typing technique based on sequence analysis of a set of polymorphic loci. The combination of the different allele types of the selected loci determines the MLST genotype [166]. One hundred and two global C. neoformans var. grubii isolates were analyzed by MLST in a first study which employed the following set of 12 loci: CAP10, CAP59, GPD1, LAC1, MPD1, MP88, SOD1, TEF1α, TOP1, URE1, and IGS1 [53]. The results showed two major clades among the studied isolates, corresponding to PCR fingerprinting molecular types VNI and VNII and a third new clade, VNB, including only isolates from Botswana.
A second MLST study investigated 202 global C. gattii isolates in order to elucidate the origin of Vancouver Island outbreak isolates [14]. MLST analysis, using seven loci (CAP10, GPD1, IGS1, LAC1, MPD1, PLB1, and TEF1α) and the two mating-type specific loci SXIα and SXIa, was able to differentiate all the four PCR fingerprinting molecular types, VGI-VGIV, as well as both Vancouver Island outbreak subtypes, VGIIa and VGIIb.
Although others studies have been carried out using alternative MLST schemes [31, 73, 167], the research community involved in C. neoformans and C. gattii genotyping has reached an agreement to adopt a common MLST scheme based on the two main studies reported earlier. During a meeting in 2007, the ISHAM Cryptococcus working group members established to adopt MLST as the gold standard technique for C. neoformans and C. gattii molecular typing [154]. The standard MLST scheme includes the sequencing of seven loci, GPD1, IGS1, CAP59, LAC1, SOD1, PLB1, and URA5, which combined represent the minimum number of genes giving the maximum discrimination power. In addition, genotype nomenclature between the three main typing methods (PCR fingerprinting, AFLP, and MLST) was compared and standardized as reported in Table 1, where reference strains are also indicated.
Table 1.
Genotype nomenclature adopted by the main molecular typing techniques and correspondence to standard nomenclature.
| Species/variety | Standard nomenclature |
PCR-fingerprinting M13 [15] | PCR-fingerprinting (GACA)4[155] | AFLP [11] |
MLST [14, 53] |
Reference strains |
|---|---|---|---|---|---|---|
|
C. neoformans
var. grubii |
VNI | VNI | VN6 | AFLP1 | VNI | ATCC MYA-4564 [15] |
| VNII | VNII | VN6 | AFLP1A or AFLP1B | VNII | ATCC MYA-4565 [15] |
|
| VNB | VNI or VNII | VN6 | AFLP1A or AFLP1B | VNB | bt1, bt131, bt100 [53] | |
|
C. neoformans
var. neoformans |
VNIV | VNIV | VN1 | AFLP2 | — | ATCC MYA-4567 [15] |
| Inter-varietal AD hybrids |
VNIII | VNIII | VN3 or VN4 | AFLP3 | — | ATCC 32045 [168] |
| C. gattii | VGI | VGI | — | AFLP4 | VGI | ATCC MYA-4560 [64] |
| VGII | VGII | — | AFLP6 | VGII | ATCC MYA-4561 [64] | |
| VGIII | VGIII | — | AFLP5 | VGIII | ATCC MYA-4562 [64] | |
| VGIV | VGIV | — | AFLP7 | VGIV | ATCC MYA-4563 [64] | |
| Inter-species hybrids |
VNI/VGI | VNI/VGI | — | AFLP9 | — | CBS 10496 [122] |
| VNIV/VGI | VNIV/VGI | — | AFLP8 | — | CBS 10488 [122] | |
| VNI/VGII | VNI/VGII | — | — | — | WM 05-272 [29] |
ATCC: American Type Culture Collection (http://www.atcc.org/); CBS: Centraalbureau voor Schimmelcultures (http://www.cbs.knaw.nl/); WM: Westmead Millennium Institute, Sydney, Australia; bt: isolate from Botswana.
The standard MLST scheme was applied in some recent studies contributing to identify new MLST genotypes. The investigation of 13 Korean C. neoformans var. grubii isolates led to the identification of a clonal population, designated genotype VNIc, which was prevalently isolated from non-AIDS patients [99]. The same finding seems to be confirmed by other authors who analyzed 35 isolates from Japanese non-HIV patients with cryptococcosis [100]. More recently, a large MLST study [121], carried out on 183 C. neoformans var. grubii Thai clinical isolates, revealed a low diversity of this population compared to that found in Africa and the Americas. The analysis showed also that the MLST data were consistent with a proposed ancestral African origin of C. neoformans var. grubii. MLST profiles of 107 Ugandan C. neoformans var. grubii clinical isolates were shown to be associated with the host immunological response providing a new tool to predict virulence [57]. Finally, four serotype-C VGIV C. gattii clinical isolates were identified in India, and their MLST profiles were found to be strictly correlated to those from South African VGIV isolates [34]. In order to include all C. neoformans var. grubii sequences and genotypes obtained using the standard MLST scheme, a preliminary MLST database was constructed at the Imperial College of London (London, UK, http://www.mlst.net/). Unfortunately, this database has the limit to require a long time for sequence check before the sequences could be included and assigned with the right sequence type code. To overcome these limits, a new MLST database has been established at the Molecular Mycology Research Laboratory (University of Sydney, Sydney, Australia, http://www.mycologylab.org/). The system assigns the sequence code automatically when a sequence is compared with the database and a progressive sequence code to new sequences. Subsequently, the users are required to send all the data necessary for quality control as well as the clinical data to complete the database. At present, the C. neoformans var. grubii database contains 355 strains with 110 sequence types, and the C. gattii database contains 400 strains with 160 sequence types [169].
3. Combined Epidemiological Analysis
A total of 68,811 C. neoformans and C. gattii isolates, reported by hundreds of global research studies, were analyzed. Data search was performed in PubMed database (http://www.ncbi.nih.gov/) using the keyword “cryptococcus” combined with a country name, that is, “cryptococcus italy.” Each reference from the resulting list of references was selected if it reported data concerning the isolation of one or more Cryptococcus species complex isolates. Isolates reported without an identification code or without a citation were considered new isolates and included in the analysis, whereas isolates reported from more than one paper were considered only once. Then, all the isolates were analyzed on the basis of their geographical origin, source of isolation, level of identification, species, and molecular type.
3.1. Oceania
A total of 2,518 Cryptococcus species complex isolates were reported from four countries of Oceania: Australia, New Zealand, Papua New Guinea, and Hawaii Islands. Most of the strains were isolated in Australia representing 85.4% of the isolates reported. Sixty-five percent of the isolates were from clinical source, whereas 35% were from environmental and veterinary sources (Figure 1). C. neoformans was isolated from cat, dog, horse, koala, ferret, Potorous gilbertii [16, 170–173], and from Eucalyptus camaldulensis and pine needles [11], while C. gattii was isolated from kiwi, cat, dog, horse, sheep, cow, koala, quokka, cockatoo, ferret, Potorous tridactylus, echidna, African grey parrot, and dolphin [17–19, 174, 175], and from Eucalyptus camaldulensis, Eucalyptus tereticornis, Syncarpia glomulifera, insect frass, olive seedlings, and plant debris [11, 20, 21, 176].
Figure 1.
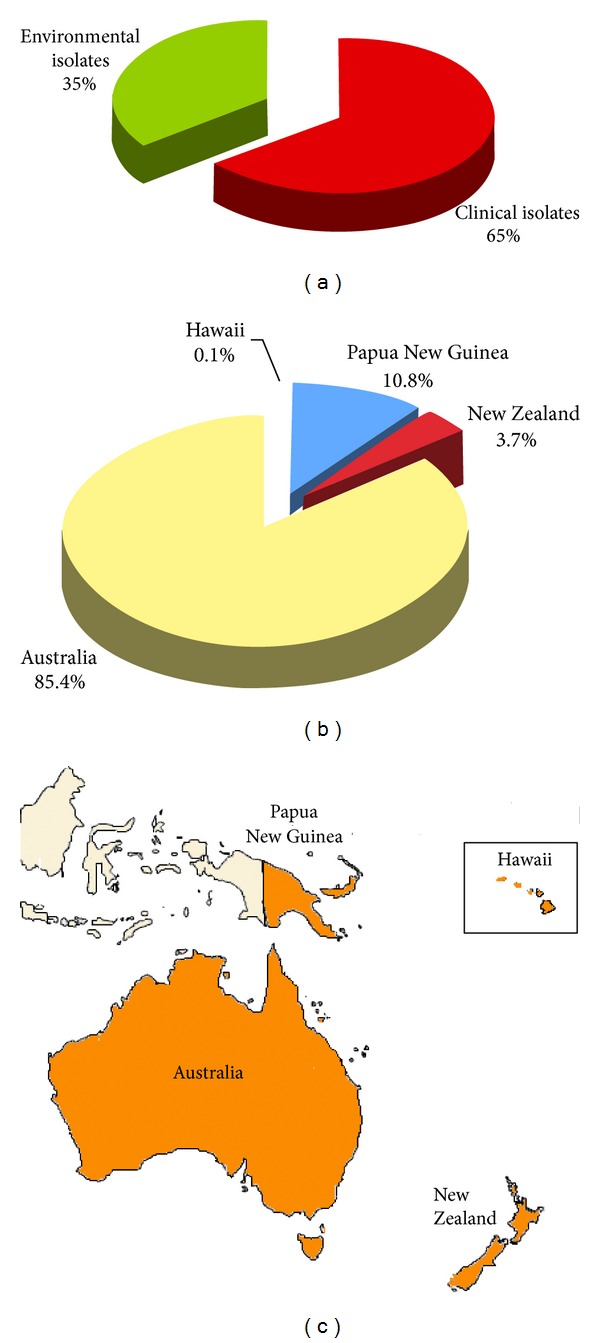
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 1,594) and environmental (n = 924) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different countries of Oceania (b). Map of the geographical distribution of the Cryptococcus neoformans and Cryptococcus gattii isolates in Oceania (c). Clinical isolates were reported from red-colored countries, whereas both clinical and environmental isolates were reported from orange-colored countries.
Identification at species level was performed for 62% of the isolates, variety or serotype was identified in 7%, and molecular type in 22% (n = 543). Only a small percentage of the isolates (9%) was identified as Cryptococcus species complex (Figure 2). A total of 1,328 C. gattii and 900 C. neoformans isolates were reported. Among the isolates identified at molecular type level, VGI represented the more frequently isolated molecular type (39%) followed by VNI (27%) and VGII (22%), the other molecular types were less frequent. No VGIV isolates have been yet isolated from Oceania.
Figure 2.
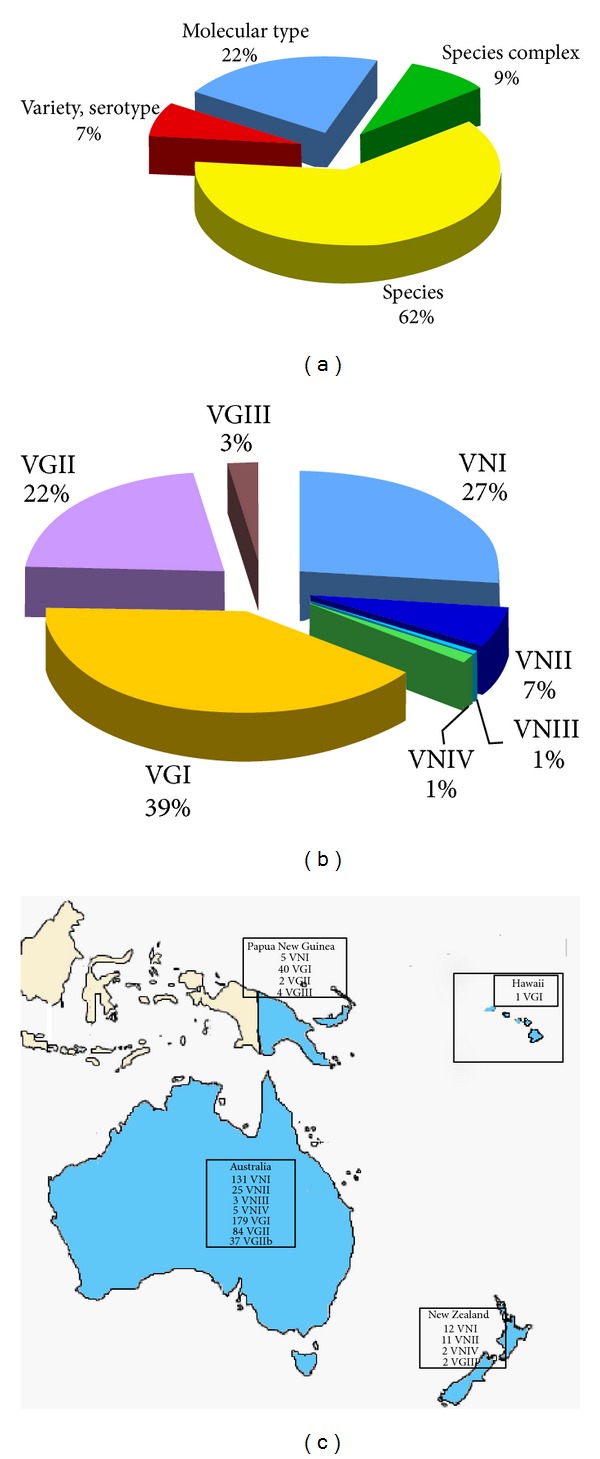
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 2,518) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 543) (b). Geographic distribution of the molecular types identified in Oceania (c). Molecular typing data have been combined from the following references: Australia [11–27], New Zealand [13, 15], and Papua New Guinea [13–15, 23, 25].
Figure 2 shows the molecular type geographical distribution in the different countries of Oceania. Although C. gattii, with molecular types VGI, VGII, and VGIII, is prevalent in Australia and in Papua New Guinea, only two VGIII isolates were reported from New Zealand where, on the contrary, C. neoformans (VNI, VNII, and VNIV) is prevalently isolated.
3.2. Asia
The combined analysis including all the Asian countries showed that a total of 19,651 C. neoformans and C. gattii isolates were reported. China, India, and Thailand, together, mainly contributed to the study reporting the 80% of the Asian isolates. Six percent of the isolates were recovered from the environment or from animals in Turkey, Israel, Iran, India, Nepal, China, Thailand, Malaysia, Taiwan, Republic of Korea, and Japan (Figure 3). In most of the environmental surveys, C. gattii was isolated from tree samples, namely, from Syzygium cumini, Mimusops elengi, Azadirachta indica, Acacia nilotica, Cassia fistola, Manikara hexandra, Polyalthia longifolia, Eucalyptus camaldulensis, Tamarindus indica, Cassia marginata, and Mangifera indica [32, 33, 177], while the only ten isolates from an animal source were recovered from koalas living in two different zoos in Japan [178, 179]. On the contrary, C. neoformans was prevalently isolated from pigeon and other birds excreta [180] and less frequently from trees such as Eucalyptus tree, Tamarindus arjuna, Tamarindus indica, Cassia fistola, Syzygium cumini, and Ficus religiosa [33, 177, 181, 182], as well as from some vegetables and fruit (tomato, carrot, banana, eggplant, papaya, apple, and guava) [183, 184]. Among animals, few C. neoformans isolates were isolated from cat and dog and one from a bandicoot [185, 186].
Figure 3.
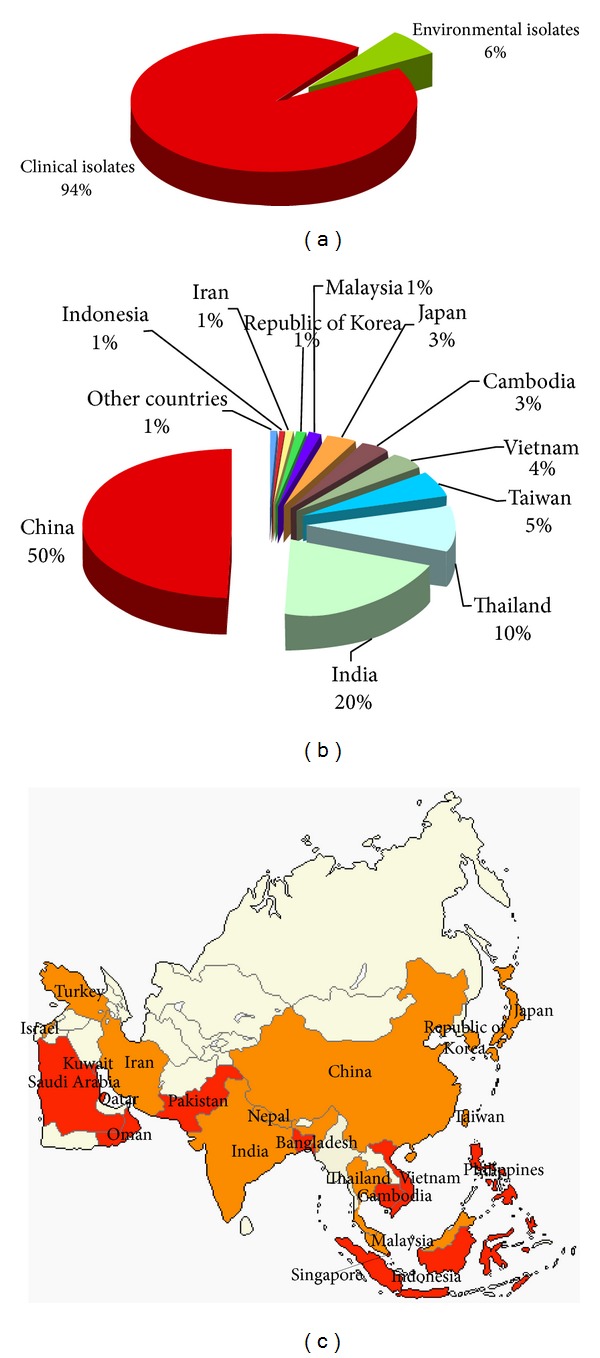
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 18,412) and environmental (n = 1,239) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different Asian countries (b). Map of the geographical distribution of the Cryptococcus neoformans and Cryptococcus gattii isolates in Asia (c). Clinical isolates were reported from red-colored countries, whereas both clinical and environmental isolates were reported from orange-colored countries.
The majority of the Asian isolates (74%) were identified just at species complex level, 7% at species level and 11% at variety/serotype level, while the molecular type was determined only in 8% (n = 1,708) (Figure 4). C. neoformans was the species prevalently isolated in Asia (n = 5,192), being C. gattii about tenfold less frequently (n = 682) isolated.
Figure 4.
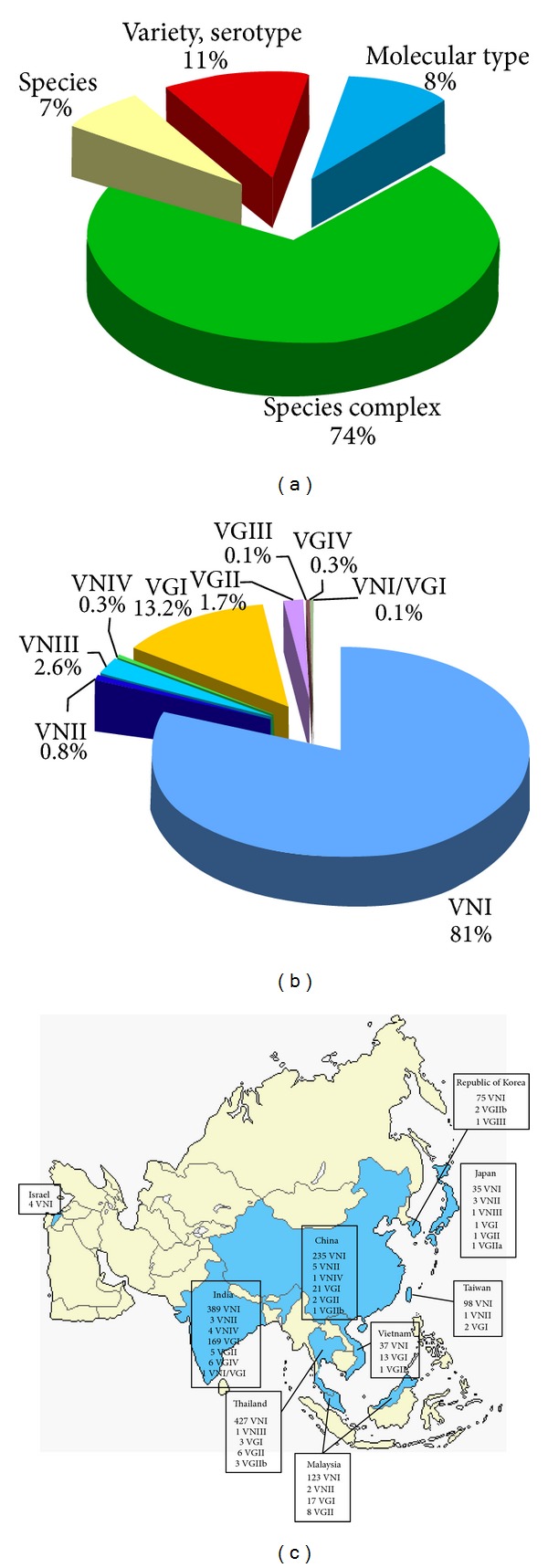
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 19,651) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 1,708) (b). Geographic distribution of the molecular types identified in Asia (c). Molecular typing data have been combined from the following references: India [11, 14, 15, 28–36], China [8, 37–43], Thailand [11, 15, 44–48], Malaysia [49, 50], Vietnam [51], Taiwan [38, 52], Japan [38, 53, 54], Republic of Korea [55], and Israel [56].
Eighty-one percent of the isolates belong to VNI and 13.2% to VGI molecular type. VGII molecular type is also represented, although in low percentages, in all the Asian countries included in the analysis, except for Israel and Taiwan. VNIII and VNIV molecular types are present in China and in India, as well as one VNIII isolate was reported from Thailand, whereas they are absent in the other Asian countries. Isolates belonging to VGIV molecular type and one interspecies VN1/VG1 hybrid were reported only in India [29, 34]. VGIII seems to be very rare or absent in Asia since only one isolate was detected in Republic of Korea [55] (Figure 4).
3.3. Africa
Isolation of 19,753 C. neoformans and C. gattii strains was reported from 25 of the 58 African countries and mainly from South Africa (79%). Environmental surveys, carried out in eight countries (Tunisia, Egypt, Nigeria, Republic Democratic of Congo, Burundi, Zimbabwe, Botswana, and South Africa), recovered only 1% out the total reported isolates (Figure 5). C. neoformans was not only isolated from pigeon and other birds excreta but also from soil and house dust [60, 187, 188], as well as from trees such as Eucalyptus camaldulensis, mopane tree, and baobab [60, 189]. C. gattii was isolated from soil, Eucalyptus camaldulensis, and almond tree [60, 189]. Two veterinary isolates were also reported from two cases of cryptococcosis affecting South African cheetahs [38, 190]. The majority of the studies reported only the species of the isolates (68%), 19% were reported as Cryptococcus species complex, and 11% as variety or serotype (Figure 6). Molecular typing techniques were applied to identify 2% of the isolates (n = 505). Of these, 68% were molecular type VNI. VNII and VNIII represent 11% and 1% of the isolates, respectively, and have been reported only from Uganda and South Africa. Thirteen percent of the African isolates belongs to VNB molecular type, which was initially considered endemic of Botswana [53] but that is present also in South Africa, Rwanda, and Republic Democratic of Congo. In addition, a consistent population of the rare VGIV molecular type was isolated in Botswana and Malawi [58]. Only one isolate belonging to VGII and four belonging to VGI molecular type were reported from Senegal [14] and Republic Democratic of Congo [11], respectively, whereas VNIV was totally absent among the African isolates included in the present study (Figure 6).
Figure 5.
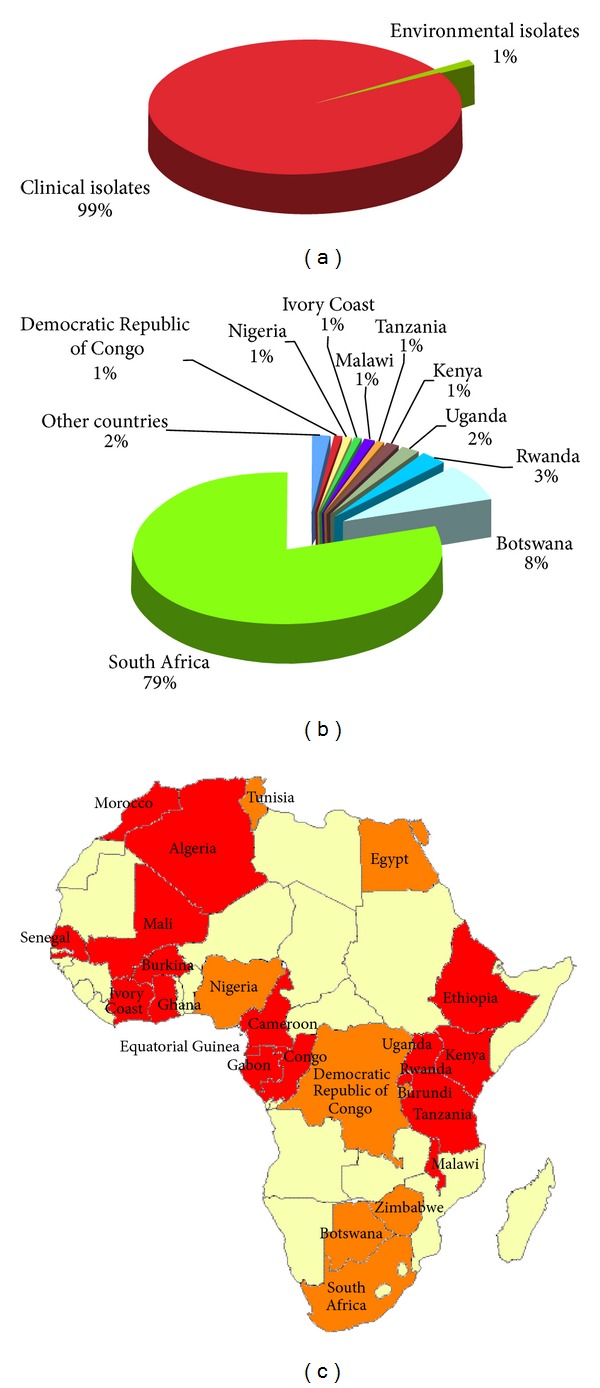
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 19,436) and environmental (n = 211) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different African countries (b). Map of the geographical distribution of the Cryptococcus neoformans and Cryptococcus gattii isolates in Africa (c). Clinical isolates were reported from red-colored countries, whereas both clinical and environmental isolates were reported from orange-colored countries.
Figure 6.
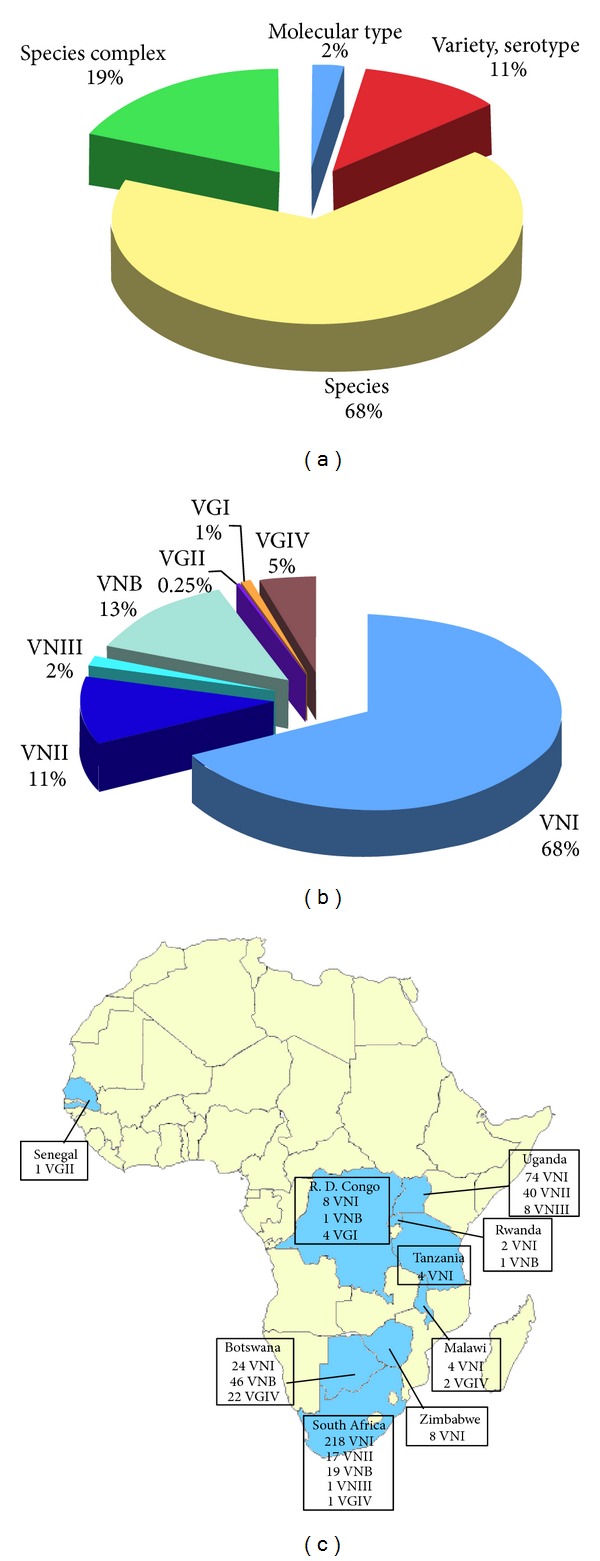
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 19,647) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 505) (b). Geographic distribution of the molecular types identified in Africa (c). Molecular typing data have been combined from the following references: Senegal [14], Republic Democratic of Congo [11, 53], Uganda [53, 57], Rwanda [11], Tanzania [53], Malawi [53, 58, 59], Zimbabwe [11], Botswana [53, 58, 60, 61], and South Africa [11, 15, 38, 60, 62, 63].
3.4. Europe
The map in Figure 7 shows the European countries reporting the isolation of at least one Cryptococcus species complex strain. Data were lacking from some Balkan and Eastern European countries. The majority of the isolates were reported from France, Spain, Italy, and United Kingdom representing 82% out of the total (n = 8,736). Nine percent of the isolates were detected from environmental and veterinary sources (Figure 7). C. neoformans not only isolated from pigeon and other birds excreta, but also from bat guano and red fox faeces [65, 191, 192]. Veterinary isolates include strains recovered from cat, dog, magpie, and some isolates from striped grass mouse and degu living in a zoo [193–196]. Few C. neoformans strains were isolated from trees, namely, from Eucalyptus camaldulensis and oak tree [74, 197]. Most of the C. gattii natural isolates were from Eucalyptus camaldulensis, Douglas tree, carob tree, and stone pine [66, 74], whereas C. gattii animal infections were reported in a ferret and in some goats [65, 67].
Figure 7.
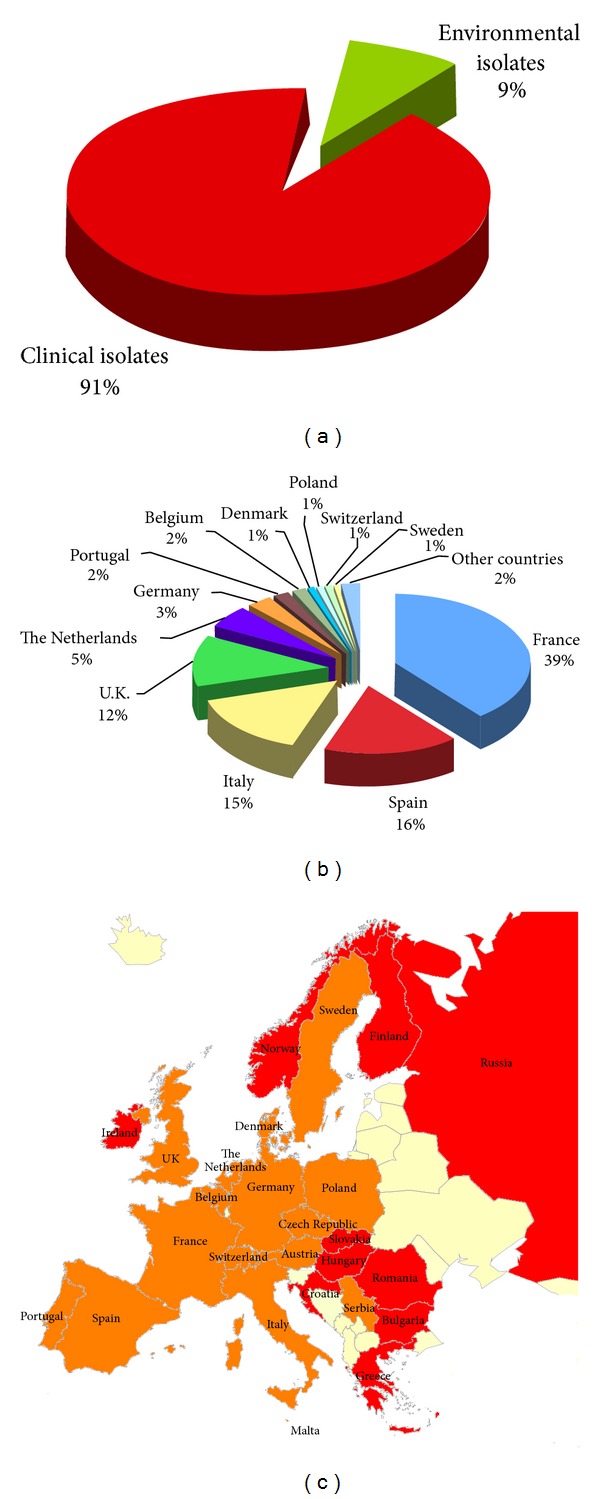
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 7,959) and environmental (n = 777) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different European countries (b). Map of the geographical distribution of the Cryptococcus neoformans and Cryptococcus gattii isolates in Europe (c). Clinical isolates were reported from red-colored countries, whereas both clinical and environmental isolates were reported from orange-colored countries.
Variety or serotype was determined in 34% of the European isolates, species in 25%, molecular type in 15%, while the 26% was reported as Cryptococcus species complex (Figure 8). A total of 6,371 isolates were identified as C. neoformans and 94 as C. gattii.
Figure 8.
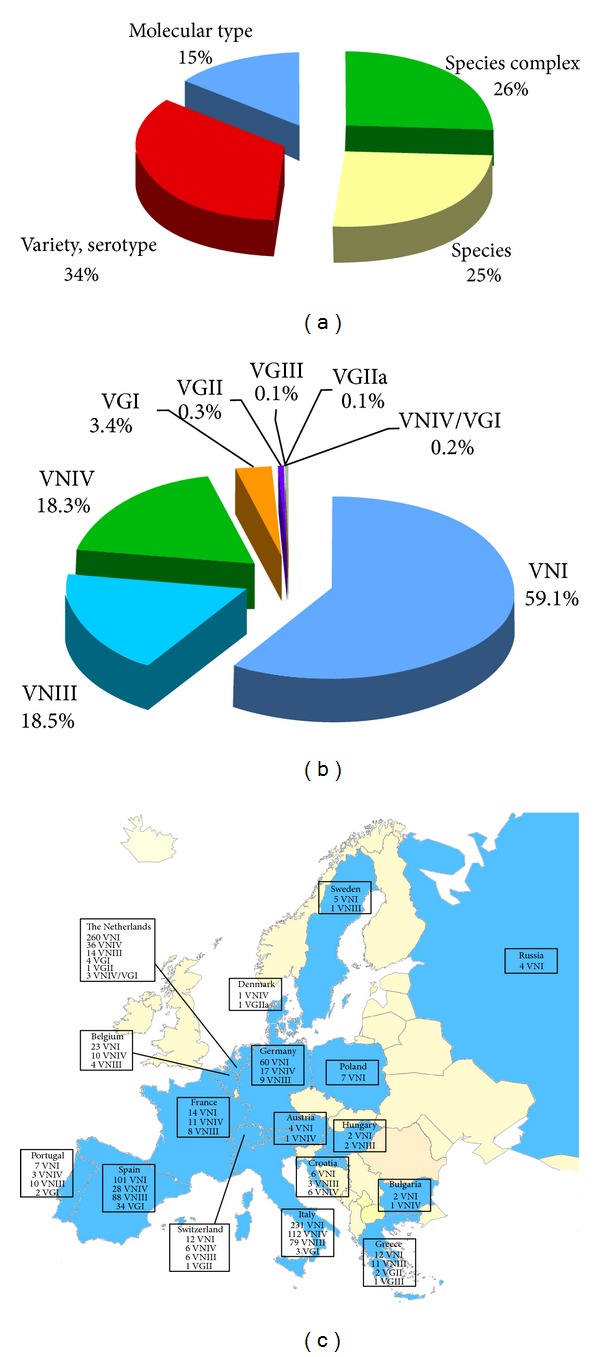
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 8,736) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 1,269) (b). Geographic distribution of the molecular types identified in Europe (c). Molecular typing data have been combined from the following references: Portugal [14, 56], Spain [56, 64–71], France [11, 72], Belgium [11, 56], The Netherlands [11, 73–75], Switzerland [73, 76, 77], Austria [56, 77], Italy [38, 53, 56, 78–85], Germany [13, 77], Denmark [11, 86], Sweden [56], Bulgaria [56], Russia [56], Greece [34, 56, 87], Croatia [88], Hungary [56], and Poland [56].
European molecular typing data are shown in Figure 8. The majority of the isolates belong to VNI molecular type (59%), although VNIII and VNIV molecular types were also reported in most of the countries representing 18.5% and 18.3%, respectively. C. gattii molecular types distribution in Europe is not yet well defined. VGI is the prevalent molecular type, being 43 isolates reported from Portugal, Spain, Italy, and The Netherlands. Few VGII isolates were reported from Greece, Switzerland, The Netherlands, and Denmark, and only one VGIII isolate was found in Greece. A relevant observation is the absence of typing results from the United Kingdom, despite the fact that 12% of the European isolates were reported from this country (Figure 8).
3.5. Central and South America
Among the 10,548 Cryptococcus species complex isolates reported from Central and South America, the 53% were reported from Brazil, 22% from Colombia, 15% from Argentina, and a lower percentage from other countries. A total of 8,590 (81%) strains were isolated from clinical sources and 1,958 (19%) from environmental and veterinary sources (Figure 9). Natural C. neoformans isolates were detected from pigeon and other birds excreta, soil, dust, and contaminated dwellings [94, 98, 198–200], as well as from Eucalyptus tree, almond tree, kassod tree, pink shower tree, Caesalpinia peltophoroides, and Anadenanthera peregrine [90, 99, 102, 201, 202]. Some isolates were also recovered from insects, bull, and sheep [99, 203, 204]. C. gattii was isolated from soil, dust, and psittaciformes bird excreta [94, 103, 199], and from Eucalyptus camaldulensis, almond tree, kassod tree, pottery tree, jungle tree, Corymbia ficifolia, and Cephalocereus royenii [92, 95, 102, 205–208]. Animal infection due to C. gattii was reported in a cheetah, a goat, and some psittacine birds [11, 91, 209].
Figure 9.
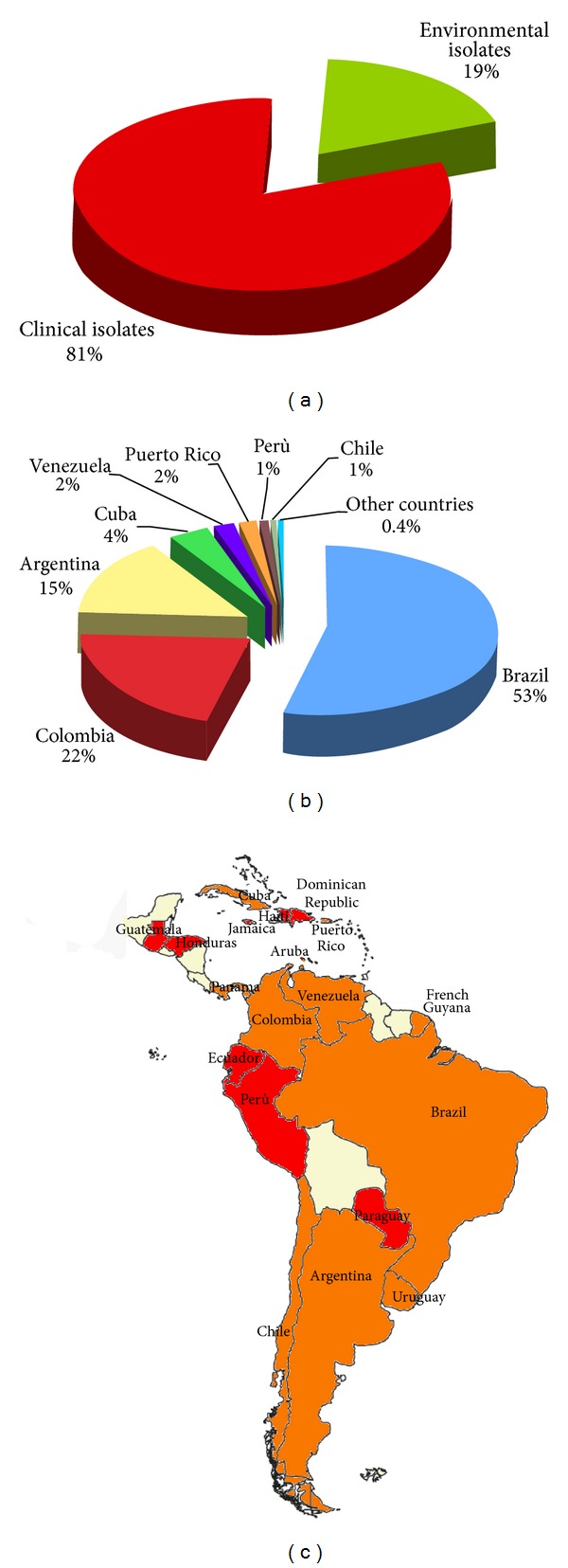
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 8,590) and environmental (n = 1,958) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different countries of Central and South America (b). Map of the geographical distribution of the Cryptoccocus neoformans and Cryptococcus gattii isolates in Central and South America (c). Clinical isolates were reported from red-coloredcountries, whereas both clinical and environmental isolates were reported from orange-colored countries.
Seventy-seven percent of the isolates were identified at least at species level (32% as variety or serotype, 23% as species, and 22% as molecular type), and 23% were reported as Cryptococcus species complex (Figure 10). C. neoformans was recognized in 6,665 and C. gattii in 1,464 isolates. The combined analysis of the molecular typing data reported from Brazil (1,439 isolates) showed that all the molecular types, except for VGIV, are represented in this country. The majority of the isolates in Brazil belong to VNI (n = 1033) molecular type followed by VGII (n = 266), while VNII, VNIV, VGI, and VGIII occurred in a lower but similar percentage. Two isolates of VNIII as well as one VNI/VGII hybrid were also reported. In Colombia (542 isolates), the prevalence of molecular types was similar to that observed in Brazil, except for VGIII, which occurred in a higher percentage than VNII, VGI, and VGIV, as well as VNIV, that was recognized only in one isolate. VNIII AD hybrids seem to be absent in Colombia. Data from Argentina (94 isolates) showed that the VGI molecular type is the prevalent genotype among C. gattii isolates, in contrast to that observed in Brazil and Colombia, where it is the VGII. In Cuba, 198 VNI isolates were detected, whereas only one isolate was VGI. On the contrary, in the near Puerto Rico, only C. gattii isolates (16 VGII and one VGIV) were reported. Finally, all the four C. neoformans molecular types were reported from Chile, but no C. gattii isolates were found (Figure 10).
Figure 10.
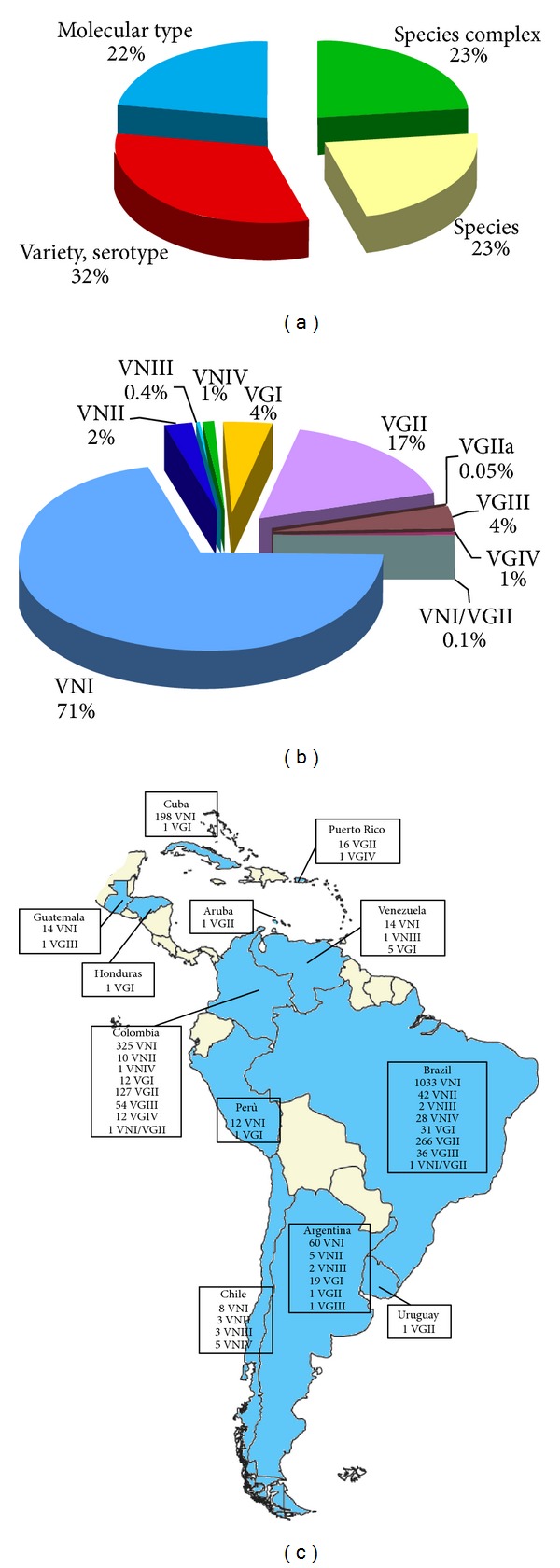
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 10,548) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 2,345) (b). Geographic distribution of the molecular types identified in Central and South America (c). Molecular typing data have been combined from the following references: Guatemala [64], Honduras [11], Cuba [89–91], Puerto Rico [92], Aruba [11, 17], Venezuela [64], Colombia [29, 64, 93–96], Perù [64], Uruguay [11, 17], Brazil [11, 17, 29, 38, 46, 64, 97–118], Argentina [15, 64, 119], and Chile [64].
3.6. North America
A total of 7,922 C. neoformans and C. gattii isolates were reported from the USA (79%), Canada (15%), and Mexico (6%). Eighty percent of the isolates were from clinical sources, whereas 20% were recovered from the environment and animals (Figure 11). Pigeon droppings were the main source for C. neoformans isolation [210, 211], although, in Mexico, it was also isolated from fruit and vegetables [212]. Three C. neoformans isolates (two in Canada and one in the United States) caused infection in ferrets [170, 213]. Isolation of C. gattii from the environment and from animals was widely reported from Canada during the monitoring of the Vancouver Island C. gattii outbreak. Soil, trees, and animals living in Vancouver Island (dogs, cats, horses, ferrets, and birds) resulted colonized or infected with this pathogen [121]. Outside Canada, VGIIa isolates were found in the environment and animals (air, water, soil, tree, cats, dogs, alpacas, and parrots) in Oregon and Washington State [123, 124], and one VGI strain was isolated from Eucalyptus camaldulensis in Mexico [214].
Figure 11.
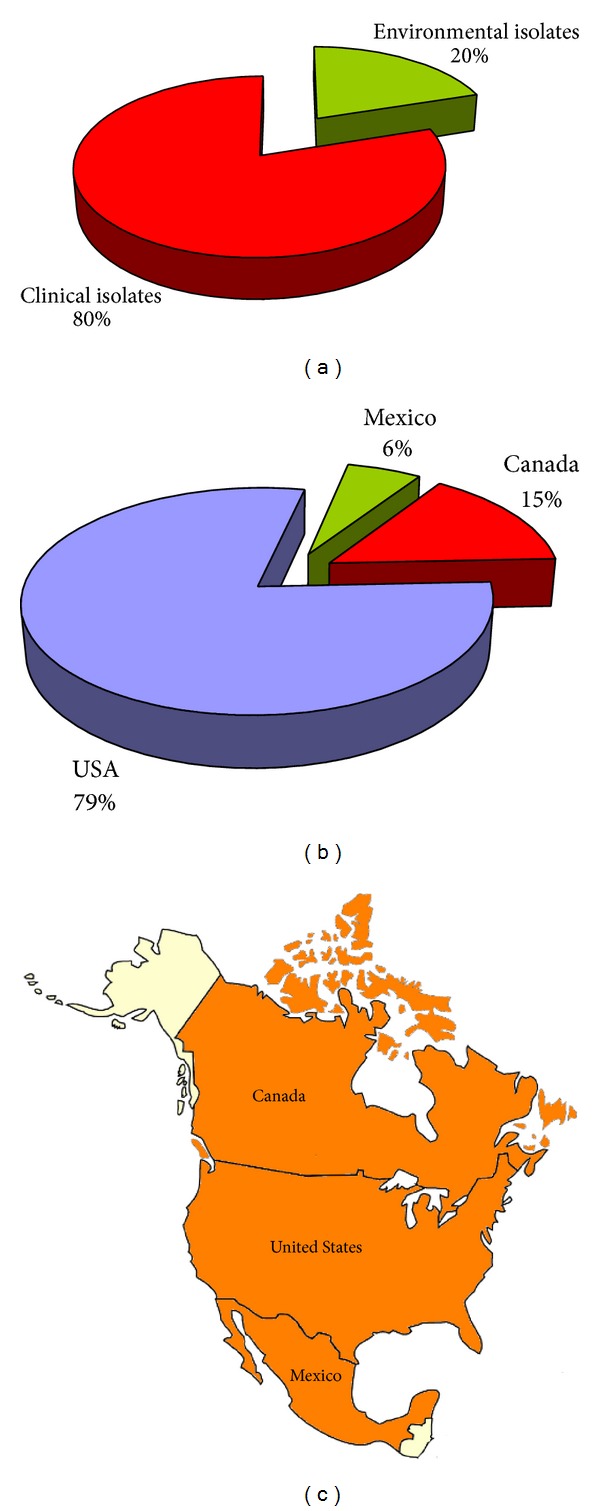
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates from clinical (n = 6,248) and environmental (n = 1,674) sources (a). Distribution of Cryptococcus neoformans and Cryptococcus gattii isolates in the different countries of North America (b). Map of the geographical distribution of the Cryptococcus neoformans and Cryptococcus gattii isolates in North America (c). Clinical isolates were reported from red-colored countries, whereas both clinical and environmental isolates were reported from orange-colored countries.
Almost half of the isolates (49%) reported from North America were identified only as Cryptococcus species complex, 10% were identified at species level, while variety or serotype was reported for 21%. Molecular type was determined in 20% of the isolates (n = 1,707) (Figure 12). Despite the fact that C. neoformans was more frequently isolated in North America than C. gattii (3,148 versus 885 isolates, resp.), 39% of the isolates identified by molecular techniques belong to VGIIa molecular type. This is due to the extensive effort produced to discover the cause of Vancouver Island outbreak which, at present, includes 473 VGIIa, 57 VGIIb, and 70 VGI isolates [121]. In addition, a recent study has reported the infection of a Canadian patient with an interspecies VNI/VGI AB hybrid strain [122]. VNI was the prevalent molecular type in both the United States and Mexico, where VNII, VNIII, VNIV, and VGI are also present in lower percentages. VGII and VGIIa C. gattii molecular types were reported from the Northwest Pacific Coast of United States, while VGIII was reported more frequently from Mexico and Southern California [64, 120, 125]. Five VGIV isolates were reported from Mexico [64, 120], although this molecular type is absent in Canada and in the United States.
Figure 12.
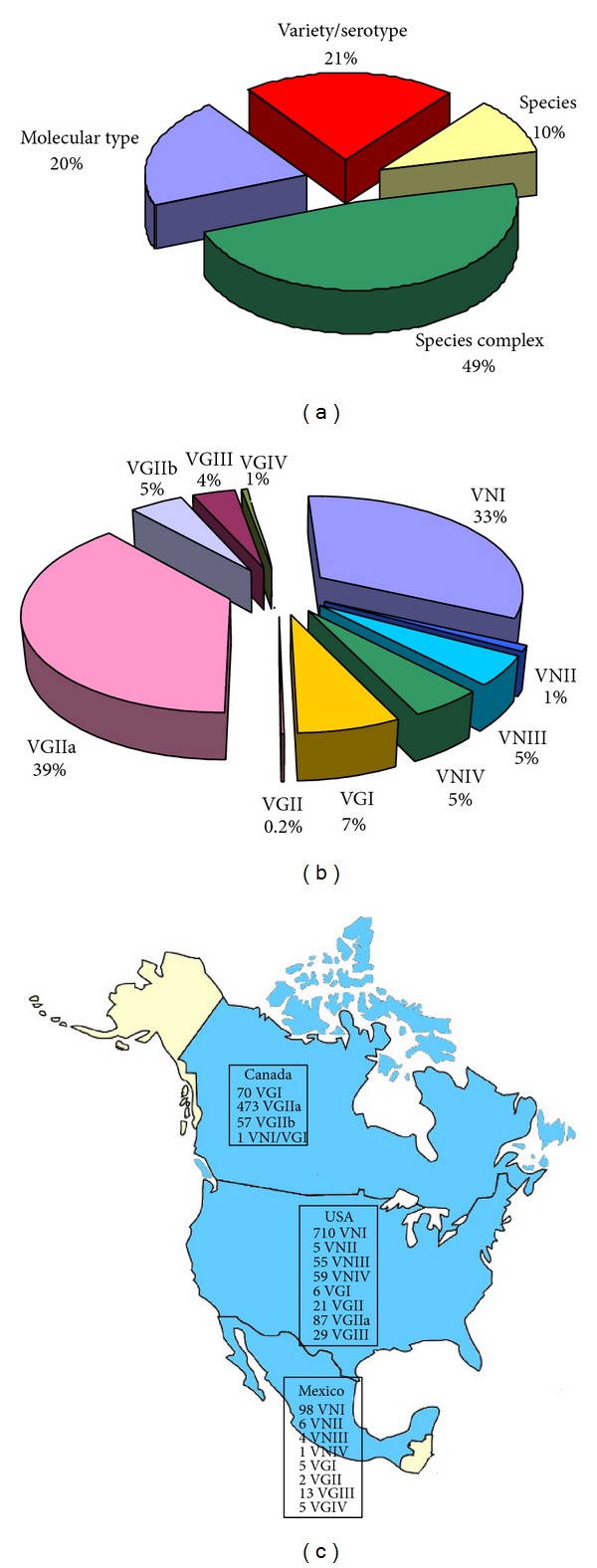
Percentage of Cryptococcus neoformans and Cryptococcus gattii isolates (n = 7,922) identified at species complex, species, variety/serotype, or molecular type level (a). Prevalence of the different VN and VG molecular types among the isolates identified at molecular type level (n = 1,707) (b). Geographic distribution of the molecular types identified in North America (c). Molecular typing data have been combined from the following references: Mexico [64, 120], Canada [121, 122], of the United States [53, 58, 123–130].
4. Concluding Remarks
The present combined analysis shows that about 68,811 C. neoformans/C. gattii isolates were reported in the world till now. The majority of the isolates were reported from Asia and Africa (19,651 and 19,647 isolates, resp.), followed by Central and South America (n = 10,548), Europe (n = 8,736), North America (n = 7,922), and Oceania (n = 2,518). The countries where the isolates were prevalently isolated are South Africa (n = 15,361), China (n = 9,736), USA (n = 6,198), and Brazil (n = 5,709). On the contrary, data are completely lacking from many countries of Africa, Asia, and Eastern Europe. United States is the country where the environment was more extensively surveyed (1089 isolates), followed by Brazil (n = 893), Australia (n = 758), Colombia (n = 742), and India (n = 569). Although 723 environmental isolates were also reported from Canada, they are not representative of the whole country since they were recovered during the monitoring of a restricted territory such as Vancouver Island area. In 38.5% of the isolates (n = 26,473) reported in the literature, the species was not determined, whereas among the isolates identified at least at species level (61.5%, n = 42,338), C. neoformans was about eightfold more frequently isolated than C. gattii (88.6% versus 11.4%). The C. neoformans/C. gattii ratio is variable for each continent being 68 : 1 in Europe, 33 : 1 in Africa, 7.6 : 1 in Asia, 4.5 : 1 in Central and South America, 3.5 : 1 in North America, and 1 : 1.5 in Oceania, where C. gattii is the prevalent species isolated.
Molecular type was determined for 8,077 isolates (12%) representing only a part of the world countries. Molecular data are absent from large parts of Africa, Asia, Eastern Europe, as well as from United Kingdom, Ireland, Norway, and Finland. VNI is the prevalent molecular type worldwide except in Australia and Papua New Guinea, where it is VGI. This latter molecular type was also found in 13.2% of the Asian, in 7% of the North American, in 4% of the Central and South American, and in 3.4% of the European isolates, while only four VGI strains have been reported from Africa. VNII is a rare molecular type which is reported from all the continents, except from Europe, in low percentages. However, a recent MLST study carried out in Italy has showed the presence of one VNB and three VNII strains also among a group of Italian clinical isolates, suggesting that these populations are underestimated in the European continent [215]. In addition, two VNB isolates from Brazil and Colombia, previously reported as VNII, were recognized by MLST analysis, confirming that VNB molecular type is not endemic of Southern Africa [22]. The distribution and prevalence of the VGII molecular type is relevant to elucidate the origin of the Vancouver Island and Northwest Pacific Coast C. gattii outbreak. The present analysis has identified four main reservoirs of VGII molecular type: Brazil (266 isolates), Colombia (n = 127), Australia (n = 121), and Puerto Rico (n = 16). These data confirm the hypotheses suggested by other authors that the Vancouver outbreak could be originated from Australia [14] or from South America [22]. The VGIII molecular type has been prevalently detected in Latin American countries, including Mexico and Sothern California (134 isolates). In the other continents, VGIII is very rare, counting one isolate in Republic of Korea, one in Greece, and six in Oceania. The abundance of VNIII AD hybrids seems to be strictly related to the presence of VNIV molecular type. In Europe and in the USA, where the frequency of isolation of VNIV strains is higher than in other geographical areas (18% and 6%, resp.), a similar percentage of VNIII isolates has been observed, suggesting that in these regions hybridization between VNI and VNIV populations is occurring. VGIV molecular type was reported from Southern Africa (n = 24), India (n = 6), Colombia (n = 12), and Mexico (n = 5), but a recent MLST study, comparing these isolates, has revealed that Indian and Southern African isolates are strictly correlated and different from those from South America [34]. Finally, the interspecies C. neoformans/C. gattii hybrids have been rarely reported from different geographical areas, namely, one from India, one from Colombia, one from Brazil, one from Canada, and three from The Netherlands. However, due to the difficulty to identify these hybrids, it is likely that their prevalence is underestimated.
In conclusion, the present study describes the state of the art of C. neoformans and C. gattii genotyping by a detailed representation of the geographical distribution of the major molecular types, which could be a useful tool to start new epidemiological surveys on the basis of the present knowledge.
Conflict of Interests
The author has no conflict of interests.
References
- 1.Del Valle L, Piña-Oviedo S. HIV disorders of the brain; pathology and pathogenesis. Frontiers in Bioscience. 2006;11(1):718–732. doi: 10.2741/1830. [DOI] [PubMed] [Google Scholar]
- 2.Abadi J, Nachman S, Kressel AB, Pirofski LA. Cryptococcosis in children with AIDS. Clinical Infectious Diseases. 1999;28(2):309–313. doi: 10.1086/515130. [DOI] [PubMed] [Google Scholar]
- 3.Mamidi A, DeSimone JA, Pomerantz RJ. Central nervous system infections in individuals with HIV-1 infection. Journal of NeuroVirology. 2002;8(3):158–167. doi: 10.1080/13550280290049723. [DOI] [PubMed] [Google Scholar]
- 4.Wright D, Schneider A, Berger JR. Central nervous system opportunistic infections. Neuroimaging Clinics of North America. 1997;7(3):513–525. [PubMed] [Google Scholar]
- 5.Korfel A, Menssen HD, Schwartz S, Thiel E. Cryptococcosis in Hodgkin’s disease: description of two cases and review of the literature. Annals of Hematology. 1998;76(6):283–286. doi: 10.1007/s002770050403. [DOI] [PubMed] [Google Scholar]
- 6.Urbini B, Castellini C, Rondelli R, Prete A, Pierinelli S, Pession A. Cryptococcal meningitis during front-line chemotherapy for acute lymphoblastic leukemia. Haematologica. 2000;85(10):1103–1104. [PubMed] [Google Scholar]
- 7.Reid G. Cryptococcus infection in immunocompetent individuals. International Journal of Infectious Diseases. 2012;16 [Google Scholar]
- 8.Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerging Infectious Diseases. 2008;14(5):755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakim JG, Gangaidzo IT, Heyderman RS, et al. Impact of HIV infection on meningitis in Harare, Zimbabwe: a prospective study of 406 predominantly adult patients. AIDS. 2000;14(10):1401–1407. doi: 10.1097/00002030-200007070-00013. [DOI] [PubMed] [Google Scholar]
- 10.Holmes CB, Losina E, Walensky RP, Yazdanpanah Y, Freedberg KA. Review of human immunodeficiency virus type 1-related opportunistic infections in sub-Saharan Africa. Clinical Infectious Diseases. 2003;36(5):652–662. doi: 10.1086/367655. [DOI] [PubMed] [Google Scholar]
- 11.Boekhout T, Theelen B, Diaz M, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans . Microbiology. 2001;147(4):891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 12.Chen SCA, Brownlee AG, Sorrell TC, et al. Identification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions. Journal of Infectious Diseases. 1996;173(3):754–758. doi: 10.1093/infdis/173.3.754. [DOI] [PubMed] [Google Scholar]
- 13.Latouche GN, Huynh M, Sorrell TC, Meyer W. PCR-restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Applied and Environmental Microbiology. 2003;69(4):2080–2086. doi: 10.1128/AEM.69.4.2080-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437(7063):1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 15.Meyer W, Marszewska K, Amirmostofian M, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20(8):1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, α mating type populations. Eukaryotic Cell. 2008;7(10):1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carriconde F, Gilgado F, Arthur I, et al. Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population—an emerging outbreak in Australia. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0016936.e16936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorrell TC, Brownlee AG, Ruma P, Malik R, Pfeiffer TJ, Ellis DH. Natural environmental sources of Cryptococcus neoformans var. gattii. Journal of Clinical Microbiology. 1996;34(5):1261–1263. doi: 10.1128/jcm.34.5.1261-1263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGill S, Malik R, Saul N, et al. Cryptococcosis in domestic animals in Western Australia: a retrospective study from 1995–2006. Medical Mycology. 2009;47(6):625–639. doi: 10.1080/13693780802512519. [DOI] [PubMed] [Google Scholar]
- 20.Sorrell TC, Chen SCA, Ruma P, et al. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. Journal of Clinical Microbiology. 1996;34(5):1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd SE, Sorrell TC, Meyer W. Isolation of two molecular types of Cryptococcus neoformans var. gattii from insect frass. Medical Mycology. 2003;41(2):171–176. doi: 10.1080/mmy.41.2.171.176. [DOI] [PubMed] [Google Scholar]
- 22.Ngamskulrungroj P, Gilgado F, Faganello J, et al. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS ONE. 2009;4(6) doi: 10.1371/journal.pone.0005862.e5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong HS, Dagg R, Malik R, Chen S, Carter D. In vitro susceptibility of the yeast pathogen cryptococcus to fluconazole and other azoles varies with molecular genotype. Journal of Clinical Microbiology. 2010;48(11):4115–4120. doi: 10.1128/JCM.01271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kluger EK, Karaoglu HK, Krockenberger MB, Della Torre PK, Meyer W, Malik R. Recrudescent cryptococcosis, caused by Cryptococcus gattii (molecular type VGII) over a 13-year period in a Birman cat. Medical Mycology. 2006;44(6):561–566. doi: 10.1080/13693780600582847. [DOI] [PubMed] [Google Scholar]
- 25.Campbell LT, Currie BJ, Krockenberger M, et al. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii . Eukaryotic Cell. 2005;4(8):1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tisdall PLC, Martin P, Malik R. Cryptic disease in a cat with painful and swollen hocks: an exercise in diagnostic reasoning and clinical decision-making. Journal of Feline Medicine and Surgery. 2007;9(5):418–423. doi: 10.1016/j.jfms.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SCA, Currie BJ, Campbell HM, et al. Cryptococcus neoformans var. gattii infection in northern Australia: existence of an environmental source other than known host eucalypts. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1997;91(5):547–550. doi: 10.1016/s0035-9203(97)90021-3. [DOI] [PubMed] [Google Scholar]
- 28.Randhawa HS, Kowshik T, Chowdhary A, et al. The expanding host tree species spectrum of Cryptococcus gattii and Cryptococcus neoformans and their isolations from surrounding soil in India. Medical Mycology. 2008;46(8):823–833. doi: 10.1080/13693780802124026. [DOI] [PubMed] [Google Scholar]
- 29.Aminnejad M, Diaz M, Arabatzis M, et al. Identification of Novel Hybrids Between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia. 2012;173(5-6):337–346. doi: 10.1007/s11046-011-9491-x. [DOI] [PubMed] [Google Scholar]
- 30.Mandal P, Banerjee U, Casadevall A, Nosanchuk JD. Dual infections with pigmented and albino strains of Cryptococcus neoformans in patients with or without human immunodeficiency virus infection in India. Journal of Clinical Microbiology. 2005;43(9):4766–4772. doi: 10.1128/JCM.43.9.4766-4772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, Xu J. Long-distance dispersal and recombination in environmental populations of Cryptococcus neofarmans var. grubii from India. Microbiology. 2008;154(5):1513–1524. doi: 10.1099/mic.0.2007/015594-0. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, Xu J. Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Environmental Microbiology. 2011;13(7):1875–1888. doi: 10.1111/j.1462-2920.2011.02510.x. [DOI] [PubMed] [Google Scholar]
- 33.Gugnani HC, Mitchell TG, Litvintseva AP, et al. Isolation of Cryptococcus gattii and Cryptococcus neoformans var. grubii from the flowers and bark of Eucalyptus trees in India. Medical Mycology. 2005;43(6):565–569. doi: 10.1080/13693780500160785. [DOI] [PubMed] [Google Scholar]
- 34.Cogliati M, Chandrashekar N, Esposto MC, Chandramuki A, Petrini B, Viviani MA. Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India. Mycoses. 2012;55:262–268. doi: 10.1111/j.1439-0507.2011.02082.x. [DOI] [PubMed] [Google Scholar]
- 35.Tewari A, Behera B, Mathur P, Xess I. Comparative analysis of the vitek 2 antifungal susceptibility system and E-test with the CLSI M27-A3 broth microdilution method for susceptibility testing of Indian clinical isolates of Cryptococcus neoformans . Mycopathologia. 2012;173(5-6):427–433. doi: 10.1007/s11046-012-9528-9. [DOI] [PubMed] [Google Scholar]
- 36.Jain N, Wickes BL, Keller SM, et al. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. Journal of Clinical Microbiology. 2005;43(11):5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng X, Yao Z, Ren D, Liao W. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. Journal of Medical Microbiology. 2008;57(12):1481–1490. doi: 10.1099/jmm.0.2008/003665-0. [DOI] [PubMed] [Google Scholar]
- 38.Katsu M, Kidd S, Ando A, et al. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Research. 2004;4(4-5):377–388. doi: 10.1016/S1567-1356(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 39.Feng X, Yao Z, Ren D, Liao W, Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Research. 2008;8(6):930–938. doi: 10.1111/j.1567-1364.2008.00422.x. [DOI] [PubMed] [Google Scholar]
- 40.Feng X, Wu J, Ling B, Ren D, Yao Z. Molecular and phenotypic characterization of a VGII genotype Cryptococcus gattii XH91 isolated in China. Acta Microbiologica Sinica. 2010;50(11):1460–1465. [PubMed] [Google Scholar]
- 41.Lui G, Lee N, Ip M, et al. Cryptococcosis in apparently immunocompetent patients. QJM. 2006;99(3):143–151. doi: 10.1093/qjmed/hcl014. [DOI] [PubMed] [Google Scholar]
- 42.Li A-S, Pan W-H, Wu S-X, et al. Ecological surveys of the Cryptococcus species complex in China. Chinese Medical Journal. 2012;125(3):511–516. [PubMed] [Google Scholar]
- 43.Cai X, Liu K, Liang Y, Yu H, Lv F, Liang X. Isolated biliary cryptococcosis manifesting as obstructive jaundice in an immunocompetent adult. International Journal of Medical Sciences. 2012;9(3):200–206. doi: 10.7150/ijms.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sriburee P, Khayhan S, Khamwan C, Panjaisee S, Tharavichitkul P. Serotype and PCR-fingerprints of clinical and environmental isolates of Cryptococcus neoformans in Chiang Mai, Thailand. Mycopathologia. 2004;158(1):25–31. doi: 10.1023/b:myco.0000038435.14281.f4. [DOI] [PubMed] [Google Scholar]
- 45.Simwami SP, Khayhan K, Henk DA, et al. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathogens. 2011;7(4) doi: 10.1371/journal.ppat.1001343.e1001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngamskulrungroj P, Serena C, Gilgado F, Malik R, Meyer W. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clinical Microbiology and Infection. 2011;17(2):251–258. doi: 10.1111/j.1469-0691.2010.03222.x. [DOI] [PubMed] [Google Scholar]
- 47.Ngamwongsatit P, Sukroongreung S, Nilakul C, Prachayasittikul V, Tantimavanich S. Electrophoretic karyotypes of C. neoformans serotype A recovered from Thai Patients with AIDS. Mycopathologia. 2005;159(2):189–197. doi: 10.1007/s11046-004-6671-y. [DOI] [PubMed] [Google Scholar]
- 48.Annual Report of the Forest Insect and Disease Survey. Canadian Forestry Service, 1973; 1974. [Google Scholar]
- 49.Tay ST, Lim HC, Tajuddin TH, Rohani MY, Hamimah H, Thong KL. Determination of molecular types and genetic heterogeneity of Cryptococcus neoformans and C. gattii in Malaysia. Medical Mycology. 2006;44(7):617–622. doi: 10.1080/13693780600857330. [DOI] [PubMed] [Google Scholar]
- 50.Tay ST, Rohani MY, Soo Hoo TS, Hamimah H. Epidemiology of cryptococcosis in Malaysia. Mycoses. 2010;53(6):509–514. doi: 10.1111/j.1439-0507.2009.01750.x. [DOI] [PubMed] [Google Scholar]
- 51.Day JN, Hoang TN, Duong AV, et al. Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VN1. Journal of Clinical Microbiology. 2011;49(2):658–664. doi: 10.1128/JCM.01985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liaw SJ, Wu HC, Hsueh PR. Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clinical Microbiology and Infection. 2010;16(6):696–703. doi: 10.1111/j.1469-0691.2009.02930.x. [DOI] [PubMed] [Google Scholar]
- 53.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172(4):2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamoto K, Hatakeyama S, Itoyama S, et al. Cryptococcus gattii genotype VGIIa infection in man, Japan, 2007. Emerging Infectious Diseases. 2010;16(7):1155–1157. doi: 10.3201/eid1607.100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi YH, Ngamskulrungroj P, Varma A, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Research. 2010;10(6):769–778. doi: 10.1111/j.1567-1364.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viviani MA, Cogliati M, Esposto MC, et al. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Research. 2006;6(4):614–619. doi: 10.1111/j.1567-1364.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- 57.Wiesner DL, Corcoran JM, Mcdonald T, et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio. 2012;3(5) doi: 10.1128/mBio.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. Journal of Infectious Diseases. 2005;192(5):888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 59.Bell M, Archibald LK, Nwanyanwu O, et al. Seasonal variation in the etiology of bloodstream infections in a febrile inpatient population in a developing country. International Journal of Infectious Diseases. 2001;5(2):63–69. doi: 10.1016/s1201-9712(01)90027-x. [DOI] [PubMed] [Google Scholar]
- 60.Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, Mitchell TG. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019688.e19688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Litvintseva AP, Mitchell TG. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathogens. 2012;8(2) doi: 10.1371/journal.ppat.1002495.e1002495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Botes A, Boekhout T, Hagen F, Vismer H, Swart J, Botha A. Growth and mating of Cryptococcus neoformans var. grubii on woody debris. Microbial Ecology. 2009;57(4):757–765. doi: 10.1007/s00248-008-9452-1. [DOI] [PubMed] [Google Scholar]
- 63.Miglia KJ, Govender NP, Rossouw J, et al. Analyses of pediatric isolates of Cryptococcus neoformans from South Africa. Journal of Clinical Microbiology. 2011;49(1):307–314. doi: 10.1128/JCM.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer W, Castañeda A, Jackson S, et al. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerging Infectious Diseases. 2003;9(2):189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frasés S, Ferrer C, Sánchez M, Colom-Valiente MF. Molecular epidemiology of isolates of the Cryptococcus neoformans species complex from Spain. Revista Iberoamericana de Micologia. 2009;26(2):112–117. doi: 10.1016/S1130-1406(09)70021-X. [DOI] [PubMed] [Google Scholar]
- 66.Gonzalez A, Linares C, Peñataro JS, Boekhout T, Sánchez M. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Medical Mycology. 2012;50(1):67–73. doi: 10.3109/13693786.2011.574239. [DOI] [PubMed] [Google Scholar]
- 67.Ropstad EO, Leiva M, Peña T, Morera N, Martorell J. Cryptococcus gattii chorioretinitis in a ferret. Veterinary Ophthalmology. 2011;14(4):262–266. doi: 10.1111/j.1463-5224.2011.00885.x. [DOI] [PubMed] [Google Scholar]
- 68.Morera N, Juan-Sallés C, Torres JM, Sánchez M, Zamora MÁ, Francisca Colom M. Cryptococcus gattii infection in a Spanish pet ferret (Mustela putorius furo) and asymptomatic carriage in ferrets and humans from its environment. Medical Mycology. 2011;49(7):779–784. doi: 10.3109/13693786.2011.564216. [DOI] [PubMed] [Google Scholar]
- 69.Guinea J, Hagen F, Peláez T, et al. Antifungal susceptibility, serotyping, and genotyping of clinical Cryptococcus neoformans isolates collected during 18 years in a single institution in Madrid, Spain. Medical Mycology. 2010;48(7):942–948. doi: 10.3109/13693781003690067. [DOI] [PubMed] [Google Scholar]
- 70.Cejudo MATG, Gallego AG, Lacasa EC, et al. Evaluation of the VITEK 2 system to test the susceptibility of Candida spp., Trichosporon asahii and Cryptococcus neoformans to amphotericin B, flucytosine, fluconazole and voriconazole: a comparison with the M27-A3 reference method. Medical Mycology. 2010;48(5):710–719. doi: 10.3109/13693780903473343. [DOI] [PubMed] [Google Scholar]
- 71.Colom MF, Frasés S, Ferrer C, et al. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. Journal of Clinical Microbiology. 2005;43(7):3548–3550. doi: 10.1128/JCM.43.7.3548-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desnos-Ollivier M, Patel S, Spaulding AR, et al. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans . mBio. 2010;1(1) doi: 10.1128/mBio.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genetics and Biology. 2008;45(4):400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Chowdhary A, Meis JF. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerging Infectious Diseases. 2012;18(1):172–174. doi: 10.3201/eid1801.111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagen F, Colom MF, Swinne D. Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerging Infectious Diseases. 2012;18:1918–1624. doi: 10.3201/eid1810.120068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Georgi A, Schneemann M, Tintelnot K, et al. Cryptococcus gattii meningoencephalitis in an immunocompetent person 13 months after exposure. Infection. 2009;37(4):370–373. doi: 10.1007/s15010-008-8211-z. [DOI] [PubMed] [Google Scholar]
- 77.Tintelnot K, Lemmer K, Losert H, Schär G, Polak A. Follow-up of epidemiological data of cryptococcosis in Austria, Germany and Switzerland with special focus on the characterization of clinical isolates. Mycoses. 2004;47(11-12):455–464. doi: 10.1111/j.1439-0507.2004.01072.x. [DOI] [PubMed] [Google Scholar]
- 78.Cogliati M, Esposto MC, Clarke DL, Wickes BL, Viviani MA. Origin of Cryptococcus neoformans var. neoformans Diploid strains. Journal of Clinical Microbiology. 2001;39(11):3889–3894. doi: 10.1128/JCM.39.11.3889-3894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Posteraro B, Vella A, Cogliati M, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii . Journal of Clinical Microbiology. 2012;50(7):2472–2476. doi: 10.1128/JCM.00737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viviani MA, Esposto MC, Cogliati M, Montagna MT, Wickes BL. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Medical Mycology. 2001;39(5):383–386. doi: 10.1080/mmy.39.5.383.386. [DOI] [PubMed] [Google Scholar]
- 81.Romeo O, Scordino F, Chillemi V, Criseo G. Cryptococcus neoformans/Cryptococcus gattii species complex in Southern Italy: an overview on the environmental diffusion of serotypes, genotypes and mating-types. Mycopathologia. 2012;174(4):283–291. doi: 10.1007/s11046-012-9547-6. [DOI] [PubMed] [Google Scholar]
- 82.Romeo O, Scordino F, Criseo G. Environmental Isolation of Cryptococcus gattii Serotype B, VGI/MATα Strains in Southern Italy. Mycopathologia. 2011;171(6):423–430. doi: 10.1007/s11046-010-9389-z. [DOI] [PubMed] [Google Scholar]
- 83.Iatta R, Hagen F, Montagna MT. Cryptococcus gattii infection in an immunocompetent patient from Southern Italy. Mycopathologia. 2012;174(1):87–92. doi: 10.1007/s11046-011-9493-8. [DOI] [PubMed] [Google Scholar]
- 84.Barchiesi F, Cogliati M, Esposto MC, et al. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. Journal of Infection. 2005;51(1):10–16. doi: 10.1016/j.jinf.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 85.Pernice I, Lo Passo C, Criseo G, Pernice A, Todaro-Luck F. Molecular subtyping of clinical and environmental strains of Cryptococcus neoformans variety neoformans serotype A isolated from southern Italy. Mycoses. 1998;41(3-4):117–124. doi: 10.1111/j.1439-0507.1998.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 86.Lindberg J, Hagen F, Laursen A, Stenderup J, Boekhout T. Cryptococcus gattii risk for tourists visiting Vancouver Island, Canada. Emerging Infectious Diseases. 2007;13(1):178–179. doi: 10.3201/eid1301.060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Velegraki A, Kiosses VG, Pitsouni H, Toukas D, Daniilidis VD, Legakis NJ. First report of Cryptococcus neoformans var. gattii serotype B from Greece. Medical Mycology. 2001;39(5):419–422. doi: 10.1080/mmy.39.5.419.422. [DOI] [PubMed] [Google Scholar]
- 88.Mlinarić-Missoni E, Hagen F, Chew WHM, Važić-Babić V, Begovac J. In vitro antifungal susceptibilities and molecular typing of sequentially isolated clinical Cryptococcus neoformans strains from Croatia. Journal of Medical Microbiology. 2011;60(10):1487–1495. doi: 10.1099/jmm.0.031344-0. [DOI] [PubMed] [Google Scholar]
- 89.Illnait-Zaragozi MT, Martínez-Machín GF, Fernández-Andreu CM, Boekhout T, Meis JF, Klaassen CHW. Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009124.e9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martínez-Machín GF, Perurena-Lancha MR, Theelen B, Boekhout T, Meis JF, Klaassen CH. Environmental isolation and characterisation of Cryptococcus species from living trees in Havana city, Cuba. Mycoses. 2012;55(3):e138–e144. doi: 10.1111/j.1439-0507.2012.02168.x. [DOI] [PubMed] [Google Scholar]
- 91.Illnait-Zaragozí MT, Hagen F, Martínez-Machín GF, et al. Reactivation of a Cryptococcus gattii infection in a cheetah (Acinonyx jubatus) held in the National Zoo, Havana, Cuba. Mycoses. 2011;54(6):e889–e892. doi: 10.1111/j.1439-0507.2011.02046.x. [DOI] [PubMed] [Google Scholar]
- 92.Loperena-Alvarez Y, Ren P, Li X, et al. Genotypic characterization of environmental isolates of Cryptococcus gattii from Puerto Rico. Mycopathologia. 2010;170(4):279–285. doi: 10.1007/s11046-010-9296-3. [DOI] [PubMed] [Google Scholar]
- 93.Escandón P, Sánchez A, Martínez M, Meyer W, Castañeda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Research. 2006;6(4):625–635. doi: 10.1111/j.1567-1364.2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 94.Firacative C, Torres G, Claudia Rodriguez M, Escandon P. First environmental isolation of Cryptococcus gattii serotype B, from Cucuta, Colombia. Biomedica. 2011;31:118–123. doi: 10.1590/S0120-41572011000100014. [DOI] [PubMed] [Google Scholar]
- 95.Escandón P, Sánchez A, Firacative C, Castañeda E. Isolation of Cryptococcus gattii molecular type VGIII, from Corymbia ficifolia detritus in Colombia. Medical Mycology. 2010;48(4):675–678. doi: 10.3109/13693780903420633. [DOI] [PubMed] [Google Scholar]
- 96.Cortés JA, Reyes P, Gómez C, Buitrago G, Leal AL. Fungal bloodstream infections in tertiary care hospitals in Colombia. Revista Iberoamericana de Micologia. 2011;28(2):74–78. doi: 10.1016/j.riam.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 97.Casali AK, Goulart L, Rosa E Silva LK, et al. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Research. 2003;3(4):405–415. doi: 10.1016/S1567-1356(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 98.Lugarini C, Goebel CS, Condas LAZ, et al. Cryptococcus neoformans isolated from Passerine and Psittacine bird excreta in the state of Paraná, Brazil. Mycopathologia. 2008;166(2):61–69. doi: 10.1007/s11046-008-9122-3. [DOI] [PubMed] [Google Scholar]
- 99.de Jesus MS, Rodrigues WC, Barbosa G, et al. Cryptococcus neoformans carried by Odontomachus bauri ants. Memorias do Instituto Oswaldo Cruz. 2012;107(4):466–469. doi: 10.1590/s0074-02762012000400004. [DOI] [PubMed] [Google Scholar]
- 100.Trilles L, Lazéra M, Wanke B, Theelen B, Boekhout T. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Medical Mycology. 2003;41(5):383–390. doi: 10.1080/1369378031000137206. [DOI] [PubMed] [Google Scholar]
- 101.Trilles L, Lazéra MDS, Wanke B, et al. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Memorias do Instituto Oswaldo Cruz. 2008;103(5):455–462. doi: 10.1590/s0074-02762008000500008. [DOI] [PubMed] [Google Scholar]
- 102.do PSE Costa S, dos S Lazéra M, Santos WRA, et al. First isolation of Cryptococcus gattii molecular type VGII and Cryptococcus neoformans molecular type VNI from environmental sources in the city of Belém, Pará, Brazil. Memorias do Instituto Oswaldo Cruz. 2009;104(4):662–664. doi: 10.1590/s0074-02762009000400023. [DOI] [PubMed] [Google Scholar]
- 103.Abegg MA, Cella FL, Faganello J, Valente P, Schrank A, Vainstein MH. Cryptococcus neoformans and Cryptococcus gattii isolated from the excreta of psittaciformes in a Southern Brazilian Zoological Garden. Mycopathologia. 2006;161(2):83–91. doi: 10.1007/s11046-005-0186-z. [DOI] [PubMed] [Google Scholar]
- 104.Andrade-Silva L, Ferreira-Paim K, Mora DJ, et al. RAPD analysis with the primer L15996 of Brazilian clinical and environmental Cryptococcus neoformans isolates. Mycopathologia. 2012;174(1):53–59. doi: 10.1007/s11046-011-9515-6. [DOI] [PubMed] [Google Scholar]
- 105.Freire AKL, dos Santos Bentes A, Salem JI, Wanke B, de Souza JVB. Molecular characterisation of the causative agents of Cryptococcosis in patients of a tertiary healthcare facility in the state of Amazonas-Brazil. Mycoses. 2012;55(3):e145–e150. doi: 10.1111/j.1439-0507.2012.02173.x. [DOI] [PubMed] [Google Scholar]
- 106.Matos CS, De Souza Andrade A, Oliveira NS, Barros TF. Microbiological characteristics of clinical isolates of Cryptococcus spp. in Bahia, Brazil: molecular types and antifungal susceptibilities. European Journal of Clinical Microbiology and Infectious Diseases. 2012;31(7):1647–1652. doi: 10.1007/s10096-011-1488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khell Da Silva B, Freire AK, De Lima Sampaio I, Silva Dos Santos M, De Souza JV. Characterization of clinical isolates of the Cryptococcus neoformans-Cryptococcus gattii species complex from the Amazonas State in Brazil. Revista Iberoamericana de Micologia. 2012;29(1):40–43. doi: 10.1016/j.riam.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 108.Pinto Junior VL, Pone MVS, Pone SM, et al. Cryptococcus gattii molecular type VGII as agent of meningitis in a healthy child in Rio de Janeiro, Brazil: report of an autochthonous case. Revista da Sociedade Brasileira de Medicina Tropical. 2010;43(6):746–748. doi: 10.1590/s0037-86822010000600032. [DOI] [PubMed] [Google Scholar]
- 109.Dos Santos WRA, Meyer W, Wanke B, et al. Primary endemic Cryptococcosis gattii by molecular type VGII in the state of Pará, Brazil. Memorias do Instituto Oswaldo Cruz. 2008;103(8):813–818. doi: 10.1590/s0074-02762008000800012. [DOI] [PubMed] [Google Scholar]
- 110.Martins MA, Matos D. Susceptibility to antifungal agents and genotypes of Brazilian clinical and environmental Cryptococcus gattii strains. Diagnostic Microbiology and Infectious Disease. 2012;72(4):332–339. doi: 10.1016/j.diagmicrobio.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 111.Andrade-Silva L, Ferreira-Paim K, Silva-Vergara ML, Pedrosa AL. Molecular characterization and evaluation of virulence factors of Cryptococcus laurentii and Cryptococcus neoformans strains isolated from external hospital areas. Fungal Biology. 2010;114(5-6):438–445. doi: 10.1016/j.funbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 112.Ferreira-Paim K, Pedrosa AL, Silva-Vergara ML. Genotyping of Cryptococcus neoformans isolated from captive birds in Uberaba, Minas Gerais, Brazil. Mycoses. 2011;54(5):e294–e300. doi: 10.1111/j.1439-0507.2010.01901.x. [DOI] [PubMed] [Google Scholar]
- 113.Souza LKH, Souza Junior AH, Costa CR, et al. Molecular typing and antifungal susceptibility of clinical and environmental Cryptococcus neoformans species complex isolates in Goiania, Brazil. Mycoses. 2010;53(1):62–67. doi: 10.1111/j.1439-0507.2008.01662.x. [DOI] [PubMed] [Google Scholar]
- 114.Ribeiro MA, Ngamskulrungroj P. Molecular characterization of environmental Cryptococcus neoformans isolated in Vitoria, ES, Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2008;50(6):315–320. doi: 10.1590/s0036-46652008000600001. [DOI] [PubMed] [Google Scholar]
- 115.Ribeiro AM, Silva LKRE, Schrank IS, Schrank A, Meyer W, Vainstein MH. Isolation of Cryptococcus neoformans var. neoformans serotype D from Eucalypts in South Brazil. Medical Mycology. 2006;44(8):707–713. doi: 10.1080/13693780600917209. [DOI] [PubMed] [Google Scholar]
- 116.Martins LMS, Wanke B, Lazéra MS, et al. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piauí (northeastern Brazil) Memorias do Instituto Oswaldo Cruz. 2011;106(6):725–730. doi: 10.1590/s0074-02762011000600012. [DOI] [PubMed] [Google Scholar]
- 117.Mora DJ, Pedrosa AL, Rodrigues V, et al. Genotype and mating type distribution within clinical Cryptococcus neoformans and Cryptococcus gattii isolates from patients with cryptococcal meningitis in Uberaba, Minas Gerais, Brazil. Medical Mycology. 2010;48(4):561–569. doi: 10.3109/13693780903358317. [DOI] [PubMed] [Google Scholar]
- 118.Igreja RP, Dos Santos Lazéra M, Wanke B, Gutierrez Galhardo MC, Kidd SE, Meyer W. Molecular epidemiology of Cryptococcus neoformans isolates from AIDS patients of the Brazilian city, Rio de Janeiro. Medical Mycology. 2004;42(3):229–238. doi: 10.1080/13693780310001644743. [DOI] [PubMed] [Google Scholar]
- 119.Refojo N, Perrotta D, Brudny M, Abrantes R, Hevia AI, Davel G. Isolation of Cryptococcus neoformans and Cryptococcus gattii from trunk hollows of living trees in Buenos Aires City, Argentina. Medical Mycology. 2009;47(2):177–184. doi: 10.1080/13693780802227290. [DOI] [PubMed] [Google Scholar]
- 120.Olivares LRC, Martínez KM, Cruz RMB, et al. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Medical Mycology. 2009;47(7):713–721. doi: 10.3109/13693780802559031. [DOI] [PubMed] [Google Scholar]
- 121.Bartlett KH, Hoang L, Kidd S, et al. A decade of experience: Cryptococcus gattii in British Columbia. Mycopathologia. 2012;173(5-6):311–319. doi: 10.1007/s11046-011-9475-x. [DOI] [PubMed] [Google Scholar]
- 122.Bovers M, Hagen F, Kuramae EE, et al. AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerging Infectious Diseases. 2008;14(7):1105–1108. doi: 10.3201/eid1407.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.MacDougall L, Kidd SE, Galanis E, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerging Infectious Diseases. 2007;13(1):42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Datta K, Bartlett KH, Baer R, et al. Spread of Cryptococcus gattii into Pacific Northwest Region of the United States. Emerging Infectious Diseases. 2009;15(8):1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Byrnes Edmond J. EJ, Li W, Ren P, et al. A diverse population of Cryptococcus gattii molecular type VGIII in Southern Californian HIV/AIDS patients. PLoS Pathogens. 2011;7(9) doi: 10.1371/journal.ppat.1002205.e1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Litvintseva AP, Mitchell TG. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infection and Immunity. 2009;77(8):3188–3195. doi: 10.1128/IAI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sellers B, Hall P, Cine-Gowdie S. Cryptococcus gattii: an emerging fungal pathogen in the Southeastern United States. American Journal of the Medical Sciences. 2012;343:1547–1559. doi: 10.1097/MAJ.0b013e3182464bc7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Walraven CJ, Gerstein W, Hardison SE, et al. Fatal disseminated cryptococcus gattii infection in New Mexico. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028625.e28625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Byrnes EJ, Li W, Lewit Y, et al. First reported case of Cryptococcus gattii in the southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS ONE. 2009;4(6) doi: 10.1371/journal.pone.0005851.e5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Upton A, Fraser JA, Kidd SE, et al. First contemporary case of human infection with Cryptococcus gattii in puget sound: evidence for spread of the Vancouver Island outbreak. Journal of Clinical Microbiology. 2007;45(9):3086–3088. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infection and Immunity. 2001;69(1):115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. Journal of Clinical Microbiology. 1999;37(3):838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ansheng L, Nishimura K, Taguchi H, Tanaka R, Shaoxi W, Miyaji M. The isolation of Cryptococcus neoformans from pigeon droppings and serotyping of naturally and clinically sourced isolates in China. Mycopathologia. 1993;124(1):1–5. doi: 10.1007/BF01103049. [DOI] [PubMed] [Google Scholar]
- 134.Agha Kuchak Afshari S, Shokohi T, Aghili R, Badali H. Epidemiology and molecular characterization of Cryptococcus neoformans isolated from pigeon excreta in Mazandaran province, northern Iran. Journal de Mycologie Medicale. 2012;22(2):160–166. doi: 10.1016/j.mycmed.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 135.Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryotic Cell. 2007;6(6):949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu Y, Du P-C, Li W-G, Lu J-X. Identification and molecular analysis of pathogenic yeasts in droppings of domestic pigeons in Beijing, China. Mycopathologia. 2012;174(3):203–214. doi: 10.1007/s11046-012-9536-9. [DOI] [PubMed] [Google Scholar]
- 137.Randhawa HS, Kowshik T, Preeti Sinha K, et al. Distribution of Cryptococcus gattii and Cryptococcus neoformans in decayed trunk wood of Syzygium cumini trees in north-western India. Medical Mycology. 2006;44(7):623–630. doi: 10.1080/13693780600860946. [DOI] [PubMed] [Google Scholar]
- 138.Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Current Infectious Disease Reports. 2008;10(1):58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- 139.Bovers M, Hagen F, Kuramae EE, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii . FEMS Yeast Research. 2006;6(4):599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 140.Hagen F, Meis JF, Curfs-Breuker I, et al. Extensive genetic diversity within the Dutch clinical Cryptococcus neoformans population. Journal of Clinical Microbiology. 2012;50(6):1918–1926. doi: 10.1128/JCM.06750-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68(4):943–946. [PubMed] [Google Scholar]
- 142.Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans . Mycologia. 1976;68(4):821–833. [PubMed] [Google Scholar]
- 143.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annual Review of Microbiology. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 144.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans . Nature. 2005;434(7036):1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 145.Gillece JD, Schupp JM, Balajee SA, et al. Whole genome sequence analysis of cryptococcus gattii from the pacific northwest reveals unexpected diversity. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028550.e28550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heitman J, Casadevall A, Lodge JK, Perfect JR. The Cryptococcus neoformans genome sequencing project. Mycopathologia. 1999;148(1):1–7. doi: 10.1023/a:1007136602930. [DOI] [PubMed] [Google Scholar]
- 147.Brandt ME, Hutwagner LC, Klug LA, et al. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Journal of Clinical Microbiology. 1996;34(4):912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Varma A, Kwon-Chung KJ. DNA probe for strain typing of Cryptococcus neoformans . Journal of Clinical Microbiology. 1992;30(11):2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ruma P, Chen SCA, Sorrell TC, Brownlee AG. Characterization of Cryptococcus neoformans by random DNA amplification. Letters in Applied Microbiology. 1996;23(5):312–316. doi: 10.1111/j.1472-765x.1996.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 150.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans . Journal of Clinical Microbiology. 1993;31(9):2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Diaz MR, Boekhout T, Kiesling T, Fell JW. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans . FEMS Yeast Research. 2005;5(12):1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 152.Firacative C, Meyer W. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0037566.37566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of Clinical Microbiology. 2011;49(8):3050–3053. doi: 10.1128/JCM.00651-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Meyer W, Aanensen DM, Boekhout T, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii . Medical Mycology. 2009;47(6):561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Viviani MA, Wen H, Roverselli A, et al. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. Journal of Medical and Veterinary Mycology. 1997;35(5):355–360. [PubMed] [Google Scholar]
- 156.Lemmer K, Naumann D, Raddatz B, Tintelnot K. Molecular typing of Cryptococcus neoformans by PCR fingerprinting, in comparison with serotyping and Fourier transform infrared-spectroscopy-based phenotyping. Medical Mycology. 2004;42(2):135–147. doi: 10.1080/13693780310001624565. [DOI] [PubMed] [Google Scholar]
- 157.Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cogliati M, Allaria M, Tortorano AM, Viviani MA. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Medical Mycology. 2000;38(2):97–103. doi: 10.1080/mmy.38.2.97.103. [DOI] [PubMed] [Google Scholar]
- 159.Cogliati M, Esposto MC, Tortorano AM, Viviani MA. Cryptococcus neoformans population includes hybrid strains homozygous atmating-type locus. FEMS Yeast Research. 2006;6(4):608–613. doi: 10.1111/j.1567-1364.2006.00085.x. [DOI] [PubMed] [Google Scholar]
- 160.Cogliati M, Esposto MC, Liberi G, Tortorano AM, Viviani MA. Cryptococcus neoformans typing by PCR fingerprinting using (GACA) 4 primers based on C. neoformans genome project data. Journal of Clinical Microbiology. 2007;45(10):3427–3430. doi: 10.1128/JCM.01199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cogliati M, Tortorano AM. Heterozygosis and Pathogenicity of Cryptococcus neoformans AD-Hybrid Isolates. Mycopathologia. 2012;173(5-6):347–357. doi: 10.1007/s11046-011-9467-x. [DOI] [PubMed] [Google Scholar]
- 162.Keller SM, Viviani MA, Esposto MC, Cogliati M, Wickes BL. Molecular and genetic characterization of a serotype A MAta Cryptococcus neoformans isolate. Microbiology. 2003;149(1):131–142. doi: 10.1099/mic.0.25921-0. [DOI] [PubMed] [Google Scholar]
- 163.Viviani MA, Antinori S, Cogliati M, et al. European Confederation of Medical Mycology (ECMM) prospective survey of cryptococcosis: report from Italy. Medical Mycology. 2002;40(5):507–517. [PubMed] [Google Scholar]
- 164.Viviani MA, Nikolova R, Esposto MC, Prinz G, Cogliati M. First European case of serotype a MATa Cryptococcus neoformans infection. Emerging Infectious Diseases. 2003;9(9):1179–1180. doi: 10.3201/eid0909.020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23(21):4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taylor JW, Fisher MC. Fungal multilocus sequence typing—it’s not just for bacteria. Current Opinion in Microbiology. 2003;6(4):351–356. doi: 10.1016/s1369-5274(03)00088-2. [DOI] [PubMed] [Google Scholar]
- 167.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryotic Cell. 2005;4(10):1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sanfelice F. Contributo alla morfologia e biologia dei blastomiceti che si sviluppano nei succhi di alcuni frutti. Annali di Igiene. 1894;4:463–495. [Google Scholar]
- 169.Meyer W, Trilles L, Firacative C, et al. Global molecular population genetic analysis of the Cryptococcus neoformans/C. gattii species complex—steps towards a global MLST database. Mycoses. 2012;55 [Google Scholar]
- 170.Malik R, Alderton B, Finlaison D, et al. Cryptococcosis in ferrets: a diverse spectrum of clinical disease. Australian Veterinary Journal. 2002;80(12):749–755. doi: 10.1111/j.1751-0813.2002.tb11343.x. [DOI] [PubMed] [Google Scholar]
- 171.Vaughan RJ, Vitali SD, Eden PA, et al. Cryptococcosis in Gilbert’s and long-nosed potoroo. Journal of Zoo and Wildlife Medicine. 2007;38(4):567–573. doi: 10.1638/2007-0005R.1. [DOI] [PubMed] [Google Scholar]
- 172.Connolly JH, Krockenberger MB, Malik R, Canfield PJ, Wigney DI, Muir DB. Asymptomatic carriage of Cryptococcus neoformans in the nasal cavity of the koala (Phascolarctos cinereus) Medical Mycology. 1999;37(5):331–338. doi: 10.1046/j.1365-280x.1999.00236.x. [DOI] [PubMed] [Google Scholar]
- 173.Riley CB, Bolton JR, Mills JN, Thomas JB. Cryptococcosis in seven horses. Australian veterinary journal. 1992;69(6):135–139. doi: 10.1111/j.1751-0813.1992.tb07482.x. [DOI] [PubMed] [Google Scholar]
- 174.Malik R, Krockenberger MB, Cross G, et al. Avian cryptococcosis. Medical Mycology. 2003;41(2):115–124. doi: 10.1080/mmy.41.2.115.124. [DOI] [PubMed] [Google Scholar]
- 175.Rotstein DS, West K, Levine G, et al. Cryptococcus gattii VGI in a spinner dolphin (Stenella longirostris) from Hawaii. Journal of Zoo and Wildlife Medicine. 2010;41(1):181–183. doi: 10.1638/2009-0145.1. [DOI] [PubMed] [Google Scholar]
- 176.Vilcins I, Krockenberger M, Agus H, Carter D. Environmental sampling for Cryptococcus neoformans var. gattii from the Blue Mountains National Park, Sydney, Australia. Medical Mycology. 2002;40(1):53–60. doi: 10.1080/mmy.40.1.53.60. [DOI] [PubMed] [Google Scholar]
- 177.Grover N, Nawange SR, Naidu J, Singh SM, Sharma A. Ecological niche of Cryptococcus neoformans var. grubii and Cryptococcus gattii in decaying wood of trunk hollows of living trees in Jabalpur City of Central India. Mycopathologia. 2007;164(4):159–170. doi: 10.1007/s11046-007-9039-2. [DOI] [PubMed] [Google Scholar]
- 178.Makimura K, Karasawa M, Hosoi H, et al. A Queensland koala kept in a Japanese zoological park was carrier of an imported fungal pathogen, Filobasidiella neoformans var. bacillispora (Cryptococcus neoformans var. gattii) Japanese Journal of Infectious Diseases. 2002;55(1):31–32. [PubMed] [Google Scholar]
- 179.Kido N, Shibata E, Yamamoto Y. Long-term surveillance and treatment of subclinical cryptococcosis and nasal colonization by Cryptococcus neoformans and C. gattii species complex in captive koalas (Phascolarctes cinereus) Medical Mycology. 2012;50(3):291–298. doi: 10.3109/13693786.2011.594967. [DOI] [PubMed] [Google Scholar]
- 180.Gokulshankar S, Ranganathan S, Ranjith MS, Ranjithsingh AJA. Prevalence, serotypes and mating patterns of Cryptococcus neoformans in the pellets of different avifauna in Madras, India. Mycoses. 2004;47(7):310–314. doi: 10.1111/j.1439-0507.2004.00995.x. [DOI] [PubMed] [Google Scholar]
- 181.Randhawa HS, Kowshik T, Khan ZU. Efficacy of swabbing versus a conventional technique for isolation of Cryptococcus neoformans from decayed wood in tree trunk hollows. Medical Mycology. 2005;43(1):67–71. doi: 10.1080/13693780410001712025. [DOI] [PubMed] [Google Scholar]
- 182.Girish Kumar CP, Prabu D, Mitani H, Mikami Y, Menon T. Environmental isolation of Cryptococcus neoformans and Cryptococcus gattii from living trees in Guindy National Park, Chennai, South India: letter to the editor. Mycoses. 2010;53(3):262–264. doi: 10.1111/j.1439-0507.2009.01699.x. [DOI] [PubMed] [Google Scholar]
- 183.Misra VC, Randhawa HS. Occurrence and significance of Cryptococcus neoformans in vegetables and fruits. The Indian Journal of Chest Diseases & Allied Sciences. 2000;42(4):317–321. [PubMed] [Google Scholar]
- 184.Pal M, Mehrotra BS. Studies on the isolation of Cryptococcus neoformans from fruits and vegetables. Mykosen. 1985;28(4):200–205. doi: 10.1111/j.1439-0507.1985.tb02115.x. [DOI] [PubMed] [Google Scholar]
- 185.Singh SM, Naidu J, Sharma A, Nawange SR, Singh K. First case of cryptococcosis in a new species of bandicoot (Bandicota indica) caused by Cryptococcus neoformans var. grubii. Medical Mycology. 2007;45(1):89–93. doi: 10.1080/13693780600967253. [DOI] [PubMed] [Google Scholar]
- 186.Okabayashi K, Imaji M, Osumi T, et al. Antifungal activity of itraconazole and voriconazole against clinical isolates obtained from animals with mycoses. Japanese Journal of Medical Mycology. 2009;50(2):91–94. doi: 10.3314/jjmm.50.091. [DOI] [PubMed] [Google Scholar]
- 187.Irokanulo EOA, Makinde AA, Akuesgi CO, Ekwonu M. Cryptococcus neoformans var neoformans isolated from droppings of captive birds in Nigeria. Journal of Wildlife Diseases. 1997;33(2):343–345. doi: 10.7589/0090-3558-33.2.343. [DOI] [PubMed] [Google Scholar]
- 188.Swinne D, Kayembe K, Niyimi M. Isolation of saprophytic Cryptococcus neoformans var. neoformans in Kinshasa, Zaïre. Annales de la Societe Belge de Medecine Tropicale. 1986;66(1):57–61. [PubMed] [Google Scholar]
- 189.Mseddi F, Sellami A, Jarboui MA, Sellami H, Makni F, Ayadi A. First environmental isolations of Cryptococcus neoformans and Cryptococcus gattii in Tunisia and review of published studies on environmental isolations in Africa. Mycopathologia. 2011;171(5):355–360. doi: 10.1007/s11046-010-9381-7. [DOI] [PubMed] [Google Scholar]
- 190.Millward IR, Williams MC. Cryptococcus neoformans granuloma in the lung and spinal cord of a free-ranging cheetah (Acinonyx jubatus). A clinical report and literature review. Journal of the South African Veterinary Association. 2005;76(4):228–232. doi: 10.4102/jsava.v76i4.432. [DOI] [PubMed] [Google Scholar]
- 191.Montagna MT, Santacroce MP, Caggiano G, Tatò D, Ajello L. Cavernicolous habitats harbouring Cryptococcus neoformans: results of a speleological survey in Apulia, Italy, 1999-2000. Medical Mycology. 2003;41(5):451–455. doi: 10.1080/13693780310001602785. [DOI] [PubMed] [Google Scholar]
- 192.Mantovani A, Mazzoni A, Morganti L. Cryptococcus neoformans in habitat of birds and bats in Emilia-Romagna. Rivista Italiana d'igiene. 1970;29(3):205–211. [PubMed] [Google Scholar]
- 193.Belluco S, Thibaud JL, Guillot J, et al. Spinal cryptococcoma in an immunocompetent cat. Journal of Comparative Pathology. 2008;139(4):246–251. doi: 10.1016/j.jcpa.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lagrou K, Van Eldere J, Keuleers S, et al. Zoonotic transmission of Cryptococcus neoformans from a magpie to an immunocompetent patient. Journal of Internal Medicine. 2005;257(4):385–388. doi: 10.1111/j.1365-2796.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 195.Bauwens L, Vercammen F, Wuytack C, Van Looveren K, Swinne D. Isolation of Cryptococcus neoformans in Antwerp Zoo’s nocturnal house. Mycoses. 2004;47(7):292–296. doi: 10.1111/j.1439-0507.2004.00991.x. [DOI] [PubMed] [Google Scholar]
- 196.Acosta B, Alvarez P, Deniz S, Rodriguez L, Real F, Rosario I. Cryptococcal lymphadenitis in a dog. Revista Iberoamericana de Micologia. 1999;16(3):155–157. [PubMed] [Google Scholar]
- 197.Campisi E, Mancianti F, Pini G, Faggi E, Gargani G. Investigation in central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis . European Journal of Epidemiology. 2003;18(4):357–362. doi: 10.1023/a:1023652920595. [DOI] [PubMed] [Google Scholar]
- 198.Ruiz A, Velez D, Fromtling RA. Isolation of saprophytic Cryptococcus neoformans from Puerto Rico: distribution and variety. Mycopathologia. 1989;106(3):167–170. doi: 10.1007/BF00443058. [DOI] [PubMed] [Google Scholar]
- 199.Leite DP, Yamamoto ACA, Leal-Santos FA, Takahara DT, Hahn RC. Cryptococcus spp isolated from dust microhabitat in Brazilian libraries. Journal of Occupational Medicine and Toxicology. 2012;7(1, article 11) doi: 10.1186/1745-6673-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Passoni LFC, Wanke B, Nishikawa MM, Lazéra MS. Cryptococcus neoformans isolated from human dwellings in Rio de Janeiro, Brazil: an analysis of the domestic environment of AIDS patients with and without cryptococcosis. Medical Mycology. 1998;36(5):305–311. [PubMed] [Google Scholar]
- 201.Duarte A, Ordoñez N, Castañeda E. Association of yeasts of the Cryptococcus genus with Eucalyptus species in Santafé de Bogotá. Revista do Instituto de Medicina Tropical de Sao Paulo. 1994;36(2):125–130. [PubMed] [Google Scholar]
- 202.Reimão JQ, Drummond ED, Terceti MDS, Lyon JP, Franco MC, De Siqueira AM. Isolation of Cryptococcus neoformans from hollows of living trees in the city of Alfenas, MG, Brazil. Mycoses. 2007;50(4):261–264. doi: 10.1111/j.1439-0507.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- 203.Riet-Correa F, Krockenberger M, Dantas AFM, Oliveira DM. Bovine cryptococcal meningoencephalitis. Journal of Veterinary Diagnostic Investigation. 2011;23(5):1056–1060. doi: 10.1177/1040638711416624. [DOI] [PubMed] [Google Scholar]
- 204.Lemos LS, de AS, Vieira-da-Motta O, Texeira GN, de Carvalho ECQ. Pulmonary cryptococcosis in slaughtered sheep: anatomopathology and culture. Veterinary Microbiology. 2007;125(3-4):350–354. doi: 10.1016/j.vetmic.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 205.Escandón P, Quintero E, Granados D, Huérfano S, Ruiz A, Castañeda E. Isolation of Cryptococcus gattii serotype B from detritus of Eucalyptus trees in Colombia. Biomédica. 2005;25(3):390–397. [PubMed] [Google Scholar]
- 206.Callejas A, Ordoñez N, Rodriguez MC, Castañeda E. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Medical Mycology. 1998;36(5):341–344. [PubMed] [Google Scholar]
- 207.Fortes ST, Lazéra MS, Nishikawa MM, Macedo RCL, Wanke B. First isolation of Cryptococcus neoformans var. gattii from a native jungle tree in the Brazilian Amazon rainforest. Mycoses. 2001;44(5-6):137–140. doi: 10.1046/j.1439-0507.2001.00651.x. [DOI] [PubMed] [Google Scholar]
- 208.Lazéra MS, Cavalcanti MAS, Trilles L, Nishikawa MM, Wanke B. Cryptococcus neoformans var. gattii—evidence for a natural habitat related to decaying wood in a pottery tree hollow. Medical Mycology. 1998;36(2):119–122. [PubMed] [Google Scholar]
- 209.Raso TF, Werther K, Miranda ET, Mendes-Giannini MJS. Cryptococcosis outbreak in psittacine birds in Brazil. Medical Mycology. 2004;42(4):355–362. doi: 10.1080/13693780410001712061. [DOI] [PubMed] [Google Scholar]
- 210.Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans . Journal of Clinical Microbiology. 2005;43(2):556–564. doi: 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jong SC, Bulmer GS, Ruiz A. Serologic grouping and sexual compatibility of airborne Cryptococcus neoformans . Mycopathologia. 1982;79(3):185–188. doi: 10.1007/BF01837197. [DOI] [PubMed] [Google Scholar]
- 212.Lopez-Martinez R, Castanon-Olivares LR. Isolation of Cryptococcus neoformans var. neoformans from bird droppings, fruits and vegetables in Mexico City. Mycopathologia. 1995;129(1):25–28. doi: 10.1007/BF01139333. [DOI] [PubMed] [Google Scholar]
- 213.Hanley CS, MacWilliams P, Giles S, Paré J. Diagnosis and successful treatment of Cryptococcus neoformans variety grubii in a domestic ferret. Canadian Veterinary Journal. 2006;47(10):1015–1017. [PMC free article] [PubMed] [Google Scholar]
- 214.Licea BA, Garza DG, Urbieta VF, Olivares RAC. Isolation and characterization of Cryptococcus neoformans var. gattii from samples of Eucalyptus camaldulensis in Mexico city. Revista Iberoamericana de Micologia. 1999;16(1):40–42. [PubMed] [Google Scholar]
- 215.Cogliati M, Zamfirova R, Tortorano A, Viviani M. Network FC. Population structure of Italian Cryptococcus neoformans var. grubii clinical isolates. Proceedings of the 8th International Conference on Cryptococcus and Cryptococcosis; 2011; Charleston, SC, USA. p. p. 5. [Google Scholar]


