Abstract
Neurons are highly dependent on oxidative metabolism for their energy supply, and cytochrome c oxidase (COX) is a key energy-generating enzyme in the mitochondria. A unique feature of COX is that it is one of only four proteins in mammalian cells that are bigenomically-regulated. Of its thirteen subunits, three are encoded in the mitochondrial genome and ten are nuclear-encoded on nine different chromosomes. The mechanism of regulating this multisubunit, bigenomic enzyme poses a distinct challenge. In recent years, we found that nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) mediate such bigenomic coordination. The latest candidate is the specificity factor (Sp) family of proteins. In N2a cells, we found that Sp1 regulates all 13 COX subunits. However, we discovered recently that in primary neurons, it is Sp4 and not Sp1, that regulates some of the key glutamatergic receptor subunit genes. The question naturally arises as to the role of Sp4 in regulating COX in primary neurons. The present study utilized multiple approaches, including chromatin immunoprecipitation, promoter mutational analysis, knockdown and over-expression of Sp4, as well as functional assays to document that Sp4 indeed functionally regulate all 13 subunits of COX as well as mitochondrial transcription factors A and B.
Keywords: Cytochrome c oxidase, depolarization, mitochondria, transcription factor, TTX, Sp4
Introduction
The brain is a highly energy-demanding organ, and much of the energy utilized by neurons is derived from oxidative metabolism in the mitochondria. Cytochrome c oxidase (COX) plays an important role in this process, as it is one of three proton-pumping, energy-generating enzymes of the inner mitochondrial membrane. It is also the terminal enzyme of the electron transport chain, without which energy metabolism cannot be carried to completion (Wikström et al., 1981; Wong-Riley, 1989).
In neurons, COX activity is controlled by the demands of neuronal activity. A major component of neuronal activity is the repolarization of membrane potential after excitatory depolarization, and it is highly energy-dependent (Wong-Riley, 1989). Energy metabolism and neuronal activity are, therefore, tightly coupled processes (Lowry, 1975, Wong-Riley, 1989). When neuronal activity is increased, so does the demand for ATP from the oxidative pathway, and the level of COX is up-regulated; likewise, reducing neuronal activity leads to the down-regulation of COX (Wong-Riley, 1989; Liang et a., 2006). Thus, COX serves as a sensitive and reliable metabolic marker for neuronal activity.
COX is one of only four known proteins in mammalian cells that are bigenomic, i.e., derived from both the nuclear and the mitochondrial genomes. Three of its 13 subunits are mitochondrial-encoded and they form the catalytic core of the enzyme, whereas the other 10 are nucleus-specified (Kadenbach et al., 1983) on nine different chromosomes. The coordinated regulation of this multisubunit, multi-chromosomal, bigenomic enzyme is indeed challenging. In searching for transcription factor candidates, we found that both nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) directly regulate all 10 nucleus-encoded COX subunit genes (Ongwijitwat et al., 2006; Dhar et al., 2008). The fact that each of them regulates mitochondrial transcription factors A, B1, and B2 (TFAM, TFB1M and TFB2M, respectively), all of which, in turn, regulate the transcription and replication of mitochondrial DNA (mtDNA) (Virbasius and Scarpulla, 1994; Gleyzer et al., 2005), indicates that NRF-1 and NRF-2 indirectly regulate the 3 mitochondrial-encoded COX subunit genes as well. More recently, we found that each of these two transcription factors also regulates key subunit genes of the glutamatergic NMDA receptors (Dhar and Wong-Riley, 2009; Priya et al., 2013a). Thus, NRF-1 and NRF-2 mediate the tight coupling between neuronal synaptic transmission and energy metabolism at the molecular level.
Besides NRF-1 and NRF-2, promoters of a number of nucleus-encoded COX subunit genes reportedly also contain putative binding sites for specificity protein 1 (Sp1) (Grossman and Lomax, 1997; Ongwijitwat and Wong-Riley, 2004). Indeed, we recently documented that Sp1 regulates all COX subunit genes in neuroblastoma N2a cells (Dhar et al., 2013). However, when we started analyzing Sp factors’ role in regulating mediators of synaptic transmission in primary neurons, we found that it is Sp4, and not Sp1, that regulates the expression of NMDA receptor subunit genes (Priya et al., 2013b). We also found that the nuclear level of Sp4 is much higher in primary neurons than in N2a cells, whereas the converse is true for Sp1 (Priya et al., 2013b). These findings compelled us to probe for the role of Sp4 in regulating any or all of COX subunit genes in primary neurons as well as in N2a cells, a cell type commonly used by investigators of neurons. The goal of the present study is to use multiple approaches to elucidate this issue.
Materials and methods
All experiments and animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publications No. 80-23, revised 1996), and all protocols were approved by the Medical College of Wisconsin Animal Care and Use Committee (approval can be provided upon request). All efforts were made to minimize the number of animals used and their suffering. The ARRIVE guidelines have been followed.
Cell culture
Mouse N2a neuroblastoma cells were obtained from ATCC (Manassas, VA) and grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Sigma, St Luis, MO), penicillin (50 units/ml), and streptomycin (100 μg/ml) (Invitrogen, Carlsbad, CA) at 37°C in a humidified incubator supplied with 5% CO2.
Primary visual cortical neurons from 1-day-old mice (C57Bl/6, both sexes, Jackson Laboratory, Bar Harbor, ME) were cultured according to methods described previously for rats (Ongwijitwat and Wong-Riley, 2004). The density of plating in 35 mm poly-L-lysine-coated dishes was ~200,000 cells/dish. The culture media contained Neurobasal-A together with B27 supplement (Invitrogen). Glial cell proliferation was minimized by the addition of Ara-C (Sigma) to the media.
In Silico analysis of Sp4 binding
The NCBI mouse genome database was used to determine DNA sequences 1 kb upstream and 1 kb downstream of the transcription start point (TSP) for all 10 nucleus-encoded COX subunit genes as well as for TFAM, TFB1M, and TFB2M genes. The Sp binding motif of ‘GGGCGG’ and its complement ‘CCGCCC’, or the atypical motif of ‘CACCC’ and its complement ‘GGGTG’, was searched for the promoter of each gene.
NCBI’s Ensembl interface was used to align promoter sequences for the conservation of Sp binding motif in mice, rats, and humans.
In vivo analysis of Sp4 binding by chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation assays were done according to methods described previously (Priya et al., 2013a). Approximately 0.1 g of visual cortical tissue was obtained from fresh murine brains and fixed with 2% formaldehyde at 24°C for 20 min. They were then placed in a swelling buffer (85 mM KCl, 5 mM PIPES, pH 8.0, protease inhibitors, and 1% NP-40), homogenized, centrifuged to obtain the nuclei, and sonicated. The lysate was then immunoprecipitated with either the control anti-NGFR (nerve growth factor receptor) p75 goat polyclonal antibodies (2 μg) (C20, sc-6188, Santa Cruz Biotechnology, Santa Cruz, CA) or Sp4 polyclonal rabbit antibodies (2 μg) (V20, sc-645, Santa Cruz Biotechnology). No antibody served as another immunoprecipitation control. Precipitated chromatin was used in a PCR with primers that encompassed putative SP binding sites identified in silico (Table 1). β-actin exon 5 was the negative control and murine NT3 promoter with known Sp4 binding site (Ramos et al. 2009) was the positive control (see Table 1). DreamTaq polymerase (Thermo-Fisher Scientific, Waltham, MA, USA) was used with cycling parameters as follows: hot-start at 94°C for 120 s followed by denaturation at 94°C for 30 s, annealing at 59.5°C for 30 s, and extension at 72°C for 20 s (with 32–36 cycles per reaction). The addition of betaine improved the quality of PCR. Products were run on 2% agarose gels and visualized with ethidium bromide.
Table 1.
Primers for ChIP assays.
| Gene | Sequence | Amplicon length (bp) |
|---|---|---|
| COX4i1 | F: 5′-GAAAACGTCTGCCGGAAAG-3′ R: 5′-GTCACCTGCCACCGCTG-3′ |
284 |
| COX5a | F: 5′-CCACGCAGGAATGTTCACTA-3′ R: 5′-ACGAGAAGCCGGTGTGAG-3′ |
237 |
| COX5b | F: 5′-GCGTTGTTAGACTCCCACCA-3′ R: 5′-AGCTGGTCACGTACCTCCAG-3′ |
200 |
| COX6a1 | F: 5′-GCTGACAAGCAGGGAGATG-3′ R: 5′-GACGCCATCATGGAACTACA-3′ |
215 |
| COX6b | F: 5′-GCCCAGCAACAATAATAAGCA-3′ R: 5′-TAGCAAAGACGCCAATGTCA-3′ |
196 |
| COX6c | F: 5′-GGTGAGAATGGTGGAGAGAGA-3′ R: 5′-GCAAAATACAAGGGGAAACG-3′ |
299 |
| COX7a2 | F: 5′-CGTTTGCTTTCCATTGTGATT-3′ R: 5′-AAACGGAACTCCCTCCTAGC-3′ |
219 |
| COX7b | F: 5′-GATGTTGCCCTTAGCCAAAA-3′ R: 5′-CTAGCTTCCCTTCCCAGTGA-3′ |
194 |
| COX7c | F: 5′-ACATTTCCCACAATCCATCG-3′ R: 5′-ATGGCCGTACCACCTAACTC-3′ |
189 |
| COX8a | F: 5′-TGGGAGCAAAGGTGTCTCAT-3′ R: 5′-GTCCAAGGTCAGGGAGTCAA-3′ |
213 |
| TFAM | F: 5′-GTGACACAAGCCGCAGCAC-3′ R: 5′-CACTACCAGCGTGGGAACTCC-3′ |
331 |
| TFB1M | F: 5′-CTGAAAGAAGTAATGGACGCG-3′ R: 5′-GTGGGACCTTGGAGAAGAC-3′ |
340 |
| TFB2M | F: 5′-GAGGATCGGACACCTCTAGC-3′ R: 5′-CACATTTCACCACCACACTAGG-3′ |
290 |
| NT3 | F: 5′-GAGCAAACTCCAAAATGCCAGG-3′ R: 5′-AAAGTTGCGCCGGGCTATCTC-3′ |
198 |
| β-Actin exon 5 | F: 5′-GCTCTTTTCCAGCCTTCCTT-3′ R: 5′-CGGATGTCAACGTCACACTT-3′ |
187 |
Effect of Sp4 silencing and depolarizing stimulation with KCl
Two Sp4 siRNA sequences (5′-CTGGACAACAGCAGATTATTA-3′ and 5′-CCAGTAACAATCACTAGTGTT-3′) were chosen from the RNAi Consortium’s Public TRC Cloning Database at the Broad Institute and cloned into the pLKO.1 TRC cloning vector (Plasmid 10878, addgene, Cambridge, MA, USA). The pLKO.1 non-mammalian shRNA control vector, which contains a scrambled shRNA sequence that targets no known mammalian genes, was used as the negative control (SHC002, Sigma). An enhanced green fluorescent protein-containing vector (eGFP) was used to identify transfected primary neurons or N2a cells. Primary cultures of murine visual cortex neurons were plated at a density of 2 × 105 cells/dish, and N2a cells at 5 – 8 × 106 cells/dish. Primary neurons were co-transfected 5 days post-plating with 0.5 μg of eGFP vector and either 2 μg of shRNA plasmids (two Sp4 sequences at equal amounts) or with 2 μg of control scrambled vectors using Neurofect (Dhar and Wong-Riley, 2011). In another set of experiments, primary neurons were transfected as above with 0.5 μg of eGFP and 2 μg of Sp1 shRNA plasmids (Santa Cruz Biotechnology) or with 2 μg of control scrambled vectors using Neurofect. N2a cells were co-transfected the day after plating with 1 μg of eGFP and either 3 μg of the shRNA plasmids or 3 μg of the control scrambled vector using Lipofectamine 2000 (Ongwijitwat and Wong-Riley, 2006). On the day after transfection, puromycin was added to the culture medium (at a final concentration of 0.5 μg/ml for primary neurons or 5 μg/ml for N2a cells) for transfection selection. Transfection efficiency monitored by eGFP was 40% to 60% for primary neurons and 40% to 75% for N2a cells. However, puromycin selection effectively yielded 100% of transfected cells. Transfected N2a cells were further stimulated for 5 h with 20 mM KCl as described previously (Liang and Wong-Riley 2006; Yang et al., 2006). Cells were then harvested for either western blot or RNA isolation.
Effect of Sp4 over-expression and impulse blockade with TTX
The Gateway Multisite Cloning kit (Invitrogen) was used to clone the human Sp4 cDNA (Open Biosystems, Lafayette, CO) into the pcDNA-Dest40 vector according to the manufacturer’s instructions and as described previously (Johar et al. 2012). The vector contains neomycin resistance that is selectable with Geneticin in mammalian cells. Primary neurons from mouse visual cortex or N2a cells were plated in 35-mm dishes at the densities described above. Primary neurons were co-transfected 5 days post-plating with 0.5 μg of the eGFP vector and either 1.5 μg of Sp4 expression plasmids or 1 μg of pLKO.1 empty vector controls by means of Neurofect. N2a cells were co-transfected the day after plating with the above vectors by means of Lipofectamine. On the day after transfection, puromycin at a final concentration described above was added to cells transfected with the control empty vectors, whereas Geneticin (Invitrogen) was added to cells transfected with Sp4 expression vectors (at a final concentration of 100 μg/ml for primary neurons or 250 μg/ml for N2a cells) for transfection selection. Transfection efficiency monitored with eGFP was from 40% to 60% for primary neurons and 40% to 75% for N2a cells. However, puromycin or Geneticin selection resulted in 100% transfected cells. Sp4-transfected N2a cells were then subjected to impulse blockade with tetrodotoxin (TTX; 0.4 μM) for 3 days, harvested, and processed for protein and RNA isolation.
RNA isolation, cDNA synthesis, and real-time quantitative PCR
RNeasy kits (Qiagen, Valencia, CA) were used to isolate total RNA, which then underwent DNase I treatment and phenol-chloroform purification. The iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) was used to synthesize cDNA, and the Cepheid Smart Cycler Detection system (Cepheid, Sunnyvale, CA) was used to perform real-time quantitative PCRs, together with IQ Sybr Green SuperMix (170-8880, BioRad). Table 2 shows the primer sequences. PCR cycling parameters were: 2 min hot start at 95°C followed by 15–30 cycles of 10 s denaturation at 95°C, 15 s annealing at the Tm of each primer, and 10 s extension at 72°C. The purity of single PCR product was verified by the melt curve analysis. β-actin and GAPDH were used as internal controls, and the relative amount of mRNAs was determined by the 2−ΔΔCT method (Livak and Schmittgen 2001).
Table 2.
Primers for real-time quantitative PCR.
| Gene | Sequence | Amplicon (bp) | Tm (°C) |
|---|---|---|---|
| COX1 | F: 5′-GCCTTTGCTTCAAAACGAGA-3′ | 105 | 58 |
| R: 5′-GGTTGGTTCCTCGAATGTGT-3′ | |||
| COX2 | F: 5′-TCTCCCCTCTCTACGCATTC-3′ | 161 | 59 |
| R: 5′-CAGGTTTTAGGTCGTTTGTTG-3′ | |||
| COX3 | F: 5′-ACTTCACCATCCTCCAAGC-3′ | 151 | 59 |
| R: 5′-TGTCGTAGTAGGCAAACAATAAGG-3′ | |||
| COX4i1 | F: 5′-TCTACTTCGGTGTGCCTTCG-3′ | 253 | 59.5 |
| R: 5′-ACTCATTGGTGCCCTTGTTC-3′ | |||
| COX5a | F: 5′-GGAGTTGCGTAAAGGGATGA-3′ | 247 | 60 |
| R: 5′-CACTTTGTCAAGGCCCAGTT-3′ | |||
| COX5b | F: 5′-GGAGGTGGTGTCCCTACTGA-3′ | 241 | 59.5 |
| R: 5′-CAGCCAGAACCAGATGACAG-3′ | |||
| COX6a1 | F: 5′-TCAACGTGTTCCTCAAGTCGC-3′ | 115 | 60 |
| R: 5′-AGGGTATGGTTACCGTCTCCC-3′ | |||
| COX6b | F: 5′-ATGGCCGAAGACATCAAGAC-3′ | 250 | 60 |
| R: 5′-CAGGAAATGTGCCTTCTGCT-3′ | |||
| COX6c | F: 5′-AGCGTCTGCGGGTTCATA-3′ | 154 | 60 |
| R: 5′-GCCTGCCTCATCTCTTCAAA-3′ | |||
| COX7a2 | F: 5′-GAGGACCATCAGCACCACTT-3′ | 234 | 59.5 |
| R: 5′-TGGAGACTGGGATGACGAC-3′ | |||
| COX7b | F: 5′-CGAAGCATTCAGCAAGTGGT-3′ | 209 | 59.5 |
| R: 5′-TGGCATGACTACTGATCTCTCC-3′ | |||
| COX7c | F: 5′-TCTGCCTTCCGTCTCTGC-3′ | 145 | 60 |
| R: 5′-AGAAAGGAGCAGCAAATCCA-3′ | |||
| COX8a | F: 5′-TCCTGCTTCGTGTGTTGTCT-3′ | 70 | 59 |
| R: 5′-TCCCGCTTCTTGTAGCTTTC-3′ | |||
| TFAM | F: 5′-CAGGGCTGCAATTTTCCTAA-3′ | 141 | 58 |
| R: 5′-CCGAAGTGTTTTTCCAGCAT-3′ | |||
| TFB1M | F: 5′-AAGATGGCCCTTTCGTTTATGG-3′ | 102 | 59 |
| R: 5′-GACTGTGCTGTTTGCTTCCTG-3′ | |||
| TFB2M | F: 5′-CCAAAACCCATCCCGTCAAAT-3′ | 135 | 59 |
| R: 5′-AAGGGCTCCAAATGTGGAATAAA-3′ | |||
| SURF1 | F: 5′-GGTTCCTGCTTTTAATCCCTGC-3′ | 119 | 61 |
| R: 5′-GATGGGCTCAGCCATGACT-3′ | |||
| Sp4 | F: 5′-TTGCAGCAAGGCCAGCAGACC-3′ | 130 | 63 |
| R: 5′-GCTTCTTCTTTCCTGGTTCACTGCT-3′ | |||
| NT3 | F: 5′-TCACCACGGAGGAAACGCTAT-3′ | 104 | 61 |
| R: 5′-TCAATGGCTGAGGACTTGTCG-3′ | |||
| Actb | F: 5′-GGCTGTATTCCCTCCATCG-3′ | 154 | 59.5 |
| R: 5′-CCAGTTGGTAACAATGCCATGT-3′ |
Western blot assay
Proteins from whole extracts of mouse brains, cultured mouse primary neurons, N2a cells, Sp4 shRNA, and over-expression samples plus the appropriate controls were loaded onto 10% SDS-PAGE gel and transferred electrophoretically onto polyvinylidene difluoride membranes (Bio-Rad). After blocking, primary antibodies against Sp1 (1:1000; H-225, sc-14027, Santa Cruz Biotechnology), Sp4 (1:1000; V-20, sc-645, Santa Cruz Biotechnology), COX1 (1:1000; Molecular Probes, Life Technologies) and primarily COX4i1 (1: 1000) (Hevner and Wong-Riley 1989) were applied to the blots. β-actin (1:3000; Sigma) was the loading control. Sp1 polyclonal antibodies were raised in rabbits against amino acids 121–345 near the N-terminus of human Sp1, whereas Sp4 polyclonal antibodies raised in rabbits were against a proprietary region within the last 50 amino acids (734–784) at the C-terminus of human Sp4. When these two antigenic sites were compared, there were only 2 consecutive amino acids that were similar between them. When the last 50 amino acids in Sp4 were compared to the entire Sp1 protein, no homology was found. Likewise, no homology was found between N-terminus 121–345 amino acids and the entire Sp4 protein. Human Sp1 and Sp4 proteins share 51% homology, whereas mouse Sp1 and Sp4 share 54% homology, almost all of which are located at scattered intervals between amino acids 430 and 730. On the other hand, there were 95% homology between human and mouse Sp1 and 97% homology between human and mouse Sp4. Sp4 antibodies used in the present study (V-20, sc-645) showed no band in western blots of brain nuclear extracts from Sp4 knockout mice (Göllner et al., 2001), indicating no cross-reactivity with other proteins.
After incubating in goat-anti-rabbit or goat-anti-mouse secondary antibodies (Vector Laboratories, Burlingame, CA, USA), blots were treated with the ECL reagent (Pierce, Rockford, IL, USA) and exposed to autoradiographic film (RPI, Mount Prospect, IL, USA). Relative changes were quantified with an Alpha Imager (Alpha Innotech, San Leandro, CA, USA).
Cytochrome c oxidase histochemistry
Relative activity of cytochrome c oxidase was assayed with histochemistry followed by single neuron optical densitometry (Wong-Riley, 1979) on N2a cells of control, KCl- or TTX-treated, and KCl-plus-Sp4 shRNA-transfected or TTX-plus-Sp4 over-expression-transfected samples. The protocols for KCl, TTX, and transfection were the same as described above.
Statistical analysis
Analysis of variance (ANOVA) was used to determine significance among group means, and Student’s t-tests determined significance between two groups. Significance was set at a P-value of ≤ 0.05.
Results
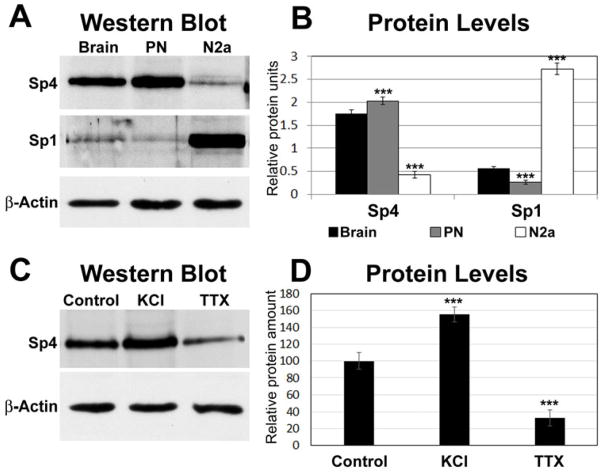
Sp4 is expressed at a higher level in primary neurons and brain than in N2a cells
Western blots of proteins obtained from visual cortices of murine brains, cultured mouse primary neurons (from the visual cortex), and N2a cells are shown in Figure 1A. Sp4 was present in greater amount in brain and primary neurons than in N2a cells, whereas Sp1 was present in greater amount in N2a cells than in brain or primary neurons (P < 0.001 for all) (Fig. 1B). When cultured primary neurons were stimulated with depolarizing KCl or with an impulse blocker TTX, the level of Sp4 was significantly up- or down-regulated, respectively (Fig. 1C, D).
Fig. 1.
Mouse brain (visual cortex) and cultured primary neurons (from mouse visual cortex) express more Sp4 than N2a cells. A, Western blots of proteins obtained from mouse brain, cultured mouse primary neurons (PN), and N2a cells labeled with Sp1 or Sp4 antibodies. β actin served as the loading control. N = 3 for each group. B, Quantification of bands indicated that both brain and primary neurons have much more Sp4 than N2a cells. On the other hand, N2a cells have significantly more Sp1 than either brain or primary neurons. C, Western blots of Sp4 proteins from cultured primary neurons subjected to KCl or TTX. β actin was the loading control. N = 3 for each group. D, Quantification of bands indicated that Sp4 protein was up-regulated by KCl depolarizing stimulation and down-regulated by TTX impulse blockade as compared to controls. ***= P < 0.001.
In vivo binding of Sp4 to promoters of all 10 nuclear COX subunits and 3 mitochondrial transcription factors
Promoters of TFAM, TFB1M, TFB2M, and all ten COX subunit genes were analyzed for possible in vivo Sp4 binding by means of chromatin immunoprecipitation (ChIP) assays. The positive control was NT3 with a known Sp4 binding site and the negative control was β-actin exon 5 (Actb). Anti-nerve growth factor receptor (NGFR) p75 antibodies served as the negative control for immunoprecipitation with the same stock of cell lysate derived from mouse visual cortical tissue. An additional no antibody control was also used. The efficiency of PCR was indicated by a 0.5% dilution of input chromatin obtained before immunoprecipitation.
Immunoprecipitation with Sp4 antibodies and amplifications with semi-quantitative PCRs revealed positive bands for the three mitochondrial transcription factors, TFAM, TFB1M, and TFB2M, as well as for the ten nucleus-encoded COX subunit genes (4i1, 5a, 5b, 6a1, 6b, 6c, 7a2, 7b, 7c, and 8a) (Fig. 2). The positive control NT3 also showed a distinct band, whereas the negative control β-actin (Actb) did not. The immunoprecipitation control NGFR (NGFR lanes) also yielded no detectable band, neither did the no-antibody control (Blank lanes) (Fig. 2).
Fig. 2.
Sp4 binds to the promoters of all 10 nucleus-encoded COX subunits as well as TFAM, TFB1M, and TFB2M in vivo. ChIP assays were done on mouse visual cortical tissue immunoprecipitated with anti-Sp4 antibodies (Sp4 lanes), anti-nerve growth factor receptor p75 antibodies (negative control, NGFR lanes), or no antibody (negative control, Blank). Input lanes indicate 0.5% of input chromatin without immunoprecipitation. NT3 promoter served as the positive control and β-actin (Actb) served as the negative control for Sp factors. Promoters of all 10 nucleus-encoded COX subunit, TFAM, TFB1M, and TFB2M were immunoprecipitated with anti-Sp4 antibodies and not with anti-NGRF or without exogenous antibodies.
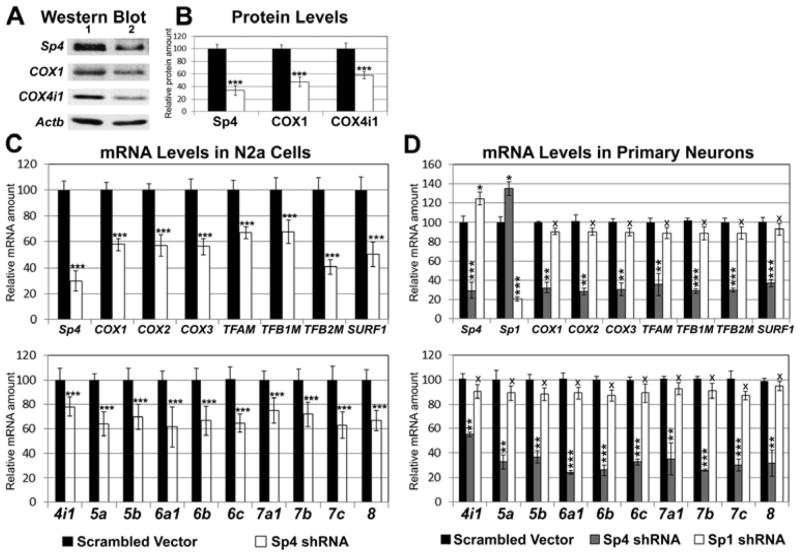
Knockdown of Sp4 reduces the levels of all thirteen COX subunits
Transfection of N2a cells with Sp4 shRNA led to a 67% decrease in their Sp4 protein (P < 0.001, Fig. 3A, B) and a 70% decrease in their Sp4 message (P < 0.001, Fig. 3C). The levels of TFAM, TFB1M, and TFB2M as well as of the 3 mitochondrial-encoded COX1, COX2, and COX3 subunits were significantly reduced (P < 0.001 for all, Fig. 3C upper panel). Relative to empty vector controls, the message of a COX assembly factor, surfeit 1 (SURF1), with a reported Sp factor binding site (Cole and Gaston, 1997) was also down-regulated (P < 0.001, Fig. 3C upper panel), as were those of 10 nucleus-encoded COX subunit genes (COX4i1 to COX8a; P < 0.001 for all, Fig. 3C, lower panel). Sp4 knockdown also caused a 53% and 40% reduction in COX1 and COX4i1 proteins, respectively (P < 0.001 for both, Fig. 3A, B).
Fig. 3.
Silencing of Sp4 in N2a cells (A–C) or in primary neurons (D) suppressed expression of all thirteen COX subunits. A, Down-regulation of Sp4, COX1, and COX4i1 protein levels in Sp4-silenced cells as compared to those transfected with scrambled vector controls. Lane 1, scrambled vectors; lane 2, Sp4 shRNA. β-actin (Actb) served as a loading control. N = 6 for each group. B, Quantification of protein levels shown in A. Solid bars, controls; open bars, Sp4 shRNA. C, Sp4 shRNA-transfected N2a cells had a significant down-regulation of transcript levels for Sp4, the mitochondrial-encoded COX subunits (COX1, COX2, and COX3), mitochondrial transcription factors (TFAM, TFB1M, and TFB2M), SURF1, and nucleus-encoded COX subunits (COX4i1 - COX8a) as compared to those transfected with scrambled vectors. N = 5 for each data point; ***= P < 0.001 as compared to scrambled vectors. D, Sp4 shRNA-transfected primary neurons also had a significant down-regulation of Sp4, mitochondrial encoded COX subunits (COX1, COX2, and COX3), mitochondrial transcription factors (TFAM, TFB1M, and TFB2M), SURF1, and nucleus-encoded subunits (COX4i1 - COX8a) compared with scrambled vector controls. Silencing of Sp4 induced a slight but significant increase in the expressions of Sp1. Silencing of Sp1 also induced a slight but significant increase in the transcripts of Sp1. However, Sp1 shRNA did not lead to any change in the expressions of all 13 COX subunit genes, nor those of TFAM, TFB1M, TFB2M, and SURF1 in primary neurons. N = 3 for each data point; *= P < 0.05; **= P < 0.01; ***= P < 0.001. X= Non-significant as compared to scrambled vector controls.
In cultured primary neurons transfected with Sp4 shRNA, Sp4 mRNA levels were reduced by 71% (P < 0.001, Figure 3D). Likewise, the mRNA levels of all 13 COX subunits, the three mitochondrial transcription factors, and SURF1 were significantly reduced by 65 – 75% (except for a 47% reduction in COX 4i1; P < 0.001 for all, Fig. 3D) as compared to neurons transfected with scrambled vectors. Knocking down Sp4 induced a slight but significant increase in the expressions of Sp1 (P < 0.05); likewise, knocking down Sp1 caused a slight but significant increase in Sp4 transcripts (P < 0.05) (Fig. 3D). However, silencing of Sp1 had no significant effect on the expressions of any of the 13 COX subunit genes, nor those of TFAM, TFB1M, TFB2M, or SURF1 genes in primary neurons (Fig. 3D).
The greater reduction of transcript levels in primary neurons than in N2a cells could be related to the fact that N2a cells have a greater abundance of Sp1 than primary neurons and, therefore, could support a higher target gene expression with Sp4 knockdown.
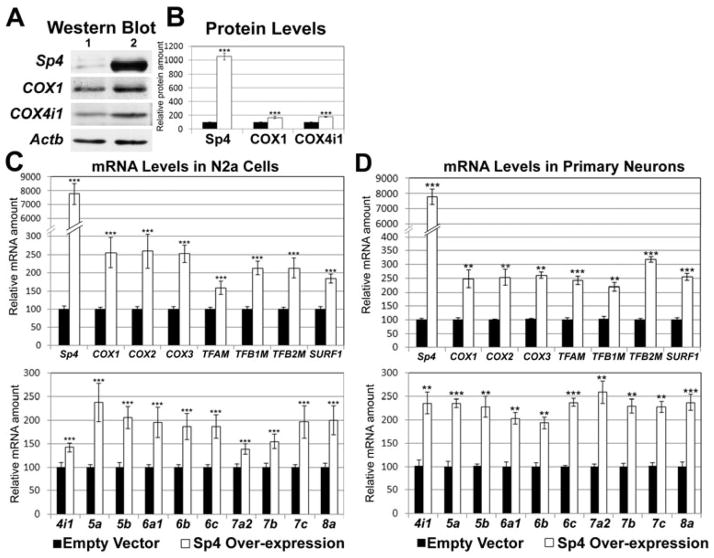
Over-expression of Sp4 up-regulates all thirteen COX subunits
In N2a cells transfected with Sp4 expression vectors, there was a 10-fold increase in Sp4 protein (P < 0.001, Fig. 4A, B) and a 77-fold increase in Sp4 message (P < 0.001, Fig. 4C). The message levels of COX1, COX2, COX3, TFAM, TFB1M, TFB2M and SURF1 were all significantly up-regulated (P < 0.001 for all, Fig. 4C upper panel), as were those of the 10 nucleus-encoded COX subunits (P < 0.001 for all, Fig. 4C lower panel). Over-expression of Sp4 also led to a 68% increase in the protein level of COX1 (P < 0.001, Fig. 4A) and a 78% increase in that of COX4i1 (P < 0.01, Fig. 4A, B).
Fig. 4.
Over-expression of Sp4 in N2a cells (A – C) or in primary neurons (D) increased expression of all thirteen COX subunits. A, Up-regulation of Sp4, COX1, and COX4i1 protein levels in cells transfected with Sp4 expression vectors as compared to empty vector controls. Lane 1, empty vectors; Lane 2, Sp4 over-expression. N = 6 for each group. B, Quantification of protein levels shown in A. Solid bars, controls; open bars, Sp4 over-expression. C, Sp4 over-expression led to a significant up-regulation of Sp4, mitochondrial encoded COX subunits (COX1, COX2, and COX-3), mitochondrial transcription factors (TFAM, TFB1M, and TFB2M), SURF1, and nucleus-encoded COX subunits (COX4i1 - COX8a) as compared to empty vector controls. N = 6 for each group. All * P values were compared to empty vectors. ** P < 0.01 and *** P < 0.001 when compared to empty vectors. D, Sp4 over-expression in primary neurons also led to a significant up-regulation of Sp4, mitochondrial encoded COX subunits (COX1, COX2, and COX-3), mitochondrial transcription factors (TFAM, TFB1M, and TFB2M), SURF1, and all 10 nucleus-encoded subunits (COX4i1 - COX8a) compared to empty vector controls. N = 3 for each group. All * P values were compared to empty vectors. ** P < 0.01 and *** P < 0.001.
In cultured primary neurons transfected with Sp4 expression vectors, a 28-fold increase in their Sp4 message was noted (P < 0.001, Fig. 4D). There was also a significant increase in their transcript levels of the 3 mitochondrial COX subunits, 3 mitochondrial transcription factors, SURF1, and 10 nucleus-encoded COX subunits relative to empty vector controls (P < 0.01 – 0.001 for all, Fig. 4D).
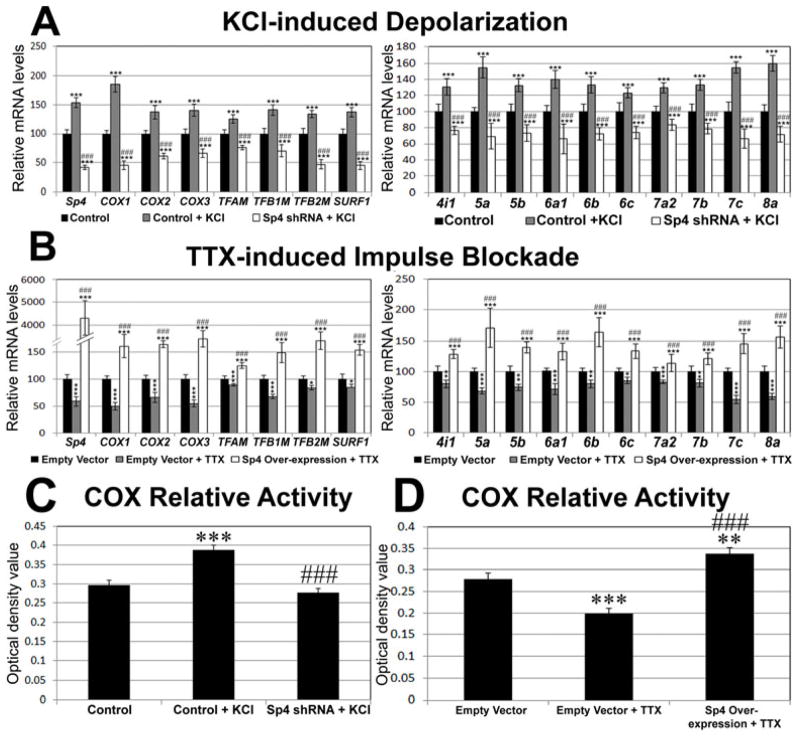
Regulation of COX expression by Sp4 is related to neuronal activity
To elucidate the role of Sp4 in activity-dependent expressions of COX subunit genes, N2a cells were transfected with either Sp4 shRNA or scrambled vectors before being stimulated by 20 mM KCl-induced depolarization for 5 hours. Such KCl treatment was found to be effective in up-regulating gene expression of COX in primary neurons (Liang and Wong-Riley, 2006; Yang et al. 2006). Indeed, depolarizing stimulation led to a significant increase in the mRNA levels of Sp4, 3 mitochondrial COX subunits, 3 mitochondrial transcription factors, SURF1, and 10 nucleus-encoded COX subunits as compared to scrambled vector controls (P < 0.001 for all, Fig. 5A). However, in the presence of Sp4 shRNA, all of these target genes failed to respond to KCl stimulation (P < 0.001 for all as compared to scrambled vector controls and to control + KCl) (Fig. 5A).
Fig. 5.

Sp4 and its target genes are all regulated by neuronal activity in N2a cells. A. KCl-induced depolarization significantly increased mRNA levels of Sp4, 3 mitochondrial-encoded COX subunits, 3 mitochondrial transcription factors, SURF1, and 10 nucleus-encoded COX subunits. In the presence of Sp4 shRNA, however, KCl was not able to up-regulate the levels of all transcripts. B. TTX-induced impulse blockade led to reduced levels of Sp4, 3 mitochondrial-encoded COX subunits, 3 mitochondrial transcription factors, SURF1, and 10 nucleus-encoded COX subunits. Over-expression of Sp4, however, rescued all 13 COX subunits and 3 mitochondrial transcription factors as well as SURF1 from TTX-induced suppression N = 6 for each data point; *= P < 0.05; **= P < 0.01; ***= P < 0.001 when compared to either scrambled (A) or empty vector (B) controls. ### = P < 0.001 when compared to control + KCl (A) or control + TTX (B). C. Optical densitometric analysis of reaction product of cytochrome c oxidase histochemistry in N2a cells. KCl induced a significant increase in the relative activity of COX as compared to scrambled vector controls. However, no up-regulation was observed in cells transfected with Sp4 shRNA. N for optical densitometric (OD) analysis = 122 cells for control, 125 for control + KCl, and 128 for Sp4 shRNA + KCl. *** = P < 0.001 when compared to controls. ### = P < 0.001 when compared to control + KCl. No significance was found between Sp4 shRNA + KCl and controls. D. Three days of TTX treatment led to significant reduction of COX relative activity. However, in N2a cells transfected with Sp4 expression vectors, the activity was increased rather than down-regulated. N for OD analysis = 115 cells for empty vector controls, 162 for empty vector + TTX, and 207 for Sp4 over-expression + TTX. **= P < 0.01; ***= P < 0.001 when compared to controls. ### = P < 0.001 when compared to control + KCl.
At a concentration of 0.4 μM, the impulse blocker TTX has been found to reduce COX subunit transcript levels and COX enzyme activity both in vivo and in primary neurons (Hevner and Wong-Riley 1993; DeYoe et al. 1995; Liang and Wong-Riley 2006). To determine if over-expressing Sp4 would rescue the down-regulation of COX subunits imposed by impulse blockade, N2a cells were transfected with either Sp4 expression vectors or empty vector controls were treated with 0.4 μM TTX for 3 days. As shown in Fig. 5B, TTX alone reduced the mRNA levels of Sp4 (P < 0.001), 3 mitochondrial COX subunits (P < 0.001 for all), 3 mitochondrial transcription factors (P < 0.001 for TFAM and TFB1M; P < 0.05 for TFB2M), SURF1 (P < 0.05), and 10 nucleus-encoded COX subunits (P < 0.001 for COX5a, 6a1, 7a2, 7c, and 8a; P < 0.01 for COX4i1, 5b, 6b, and 7b; P < 0.05 for COX6c). Thus, there was an overall suppressive effect of TTX on all COX subunit gene expression in neurons. On the other hand, Sp4 over-expression rescued TTX-induced reductions in the mRNA levels of all of the above genes tested (P < 0.001 for all as compared to empty vector controls and to control + TTX) (Fig. 5B).
The effect of knockdown or over-expression of Sp4 on COX enzyme activity was tested in primary neurons. As shown in Figs. 5C and 5D, Sp4 shRNA prevented the up-regulation of activity by KCl depolarization, whereas Sp4 over-expression rescued COX enzyme activity from being down-regulated by TTX blockade.
Conservation of Sp4 binding sites among mice, rats, and humans
The functional Sp4 binding sites on 10 nucleus-encoded COX subunits, TFAM, TFB1M, and TFB2M are conserved among humans, mice, and rats (data not shown). The homology ranged between 60 to 100% (mainly between 80 to 90%). As Sp1 and Sp4 recognize and bind to the same cis- motif, the sites for Sp4 are consistent with our recently published sites for Sp1 (Dhar et al., 2013).
Discussion
In the past, Sp1 has been the focus of most studies on Sp factors. Analysis of COX subunit genes have pointed to the existence of Sp1 binding sites without consideration for Sp4’s potential role in regulation (Grossman and Lomax, 1997; Ongwijitwat and Wong-Riley, 2004). The present study, however, documents by means of multiple approaches that in primary neurons, it is Sp4, and not Sp1, that plays a more prominent role in regulating all 13 subunits of COX. Sp4 is more abundant in both mouse visual cortex and cultured primary neurons than in N2a cells, whereas the reverse is true for Sp1, a more ubiquitously expressed member of the Sp family. Despite its abundance in neurons, Sp4 has received very little attention in the past. The present study revealed that Sp4 directly regulates all ten nucleus-encoded COX subunit genes and indirectly controls the expression of three mitochondria-encoded COX genes by regulating the three transcription factors (TFAM, TFB1M and TFB2M) critical in the transcription and replication of mitochondrial DNA. Sp4 also regulates an important COX assembly factor SURF1 in neurons. Significantly, knocking down Sp4 prevented the up-regulation of COX message, protein, and activity induced by depolarizing stimulation of neuronal activity, and over-expression of Sp4 rescued COX message, protein, and activity from being down-regulated by neuronal impulse blockade with TTX. The effect of Sp4 silencing is much greater in primary neurons than in N2a cells.
Neurons depend almost solely on oxidative metabolism for their energy supply, and COX plays a critical role in this process, as it carries oxidative metabolism to completion and helps in the generation of ATP by actively pumping H+ from the inside to the outside of mitochondria in setting up the proton gradient (Kadenbach et al. 1983; Wong-Riley 1989; 2012). Mitochondria-encoded subunits of COX (COX1, COX2, and COX3) are the largest and form the catalytic core of the enzyme (Kadenbach et al. 2000; Napiwotzki et al. 1997). Functions of the 10 nucleus-encoded COX subunits are beginning to be explored. They reportedly include the regulation of the catalytic activity, assembly, or stability of the enzyme (Kadenbach et al. 2000; Hüttermann et al. 2001; 2003). At high ATP/ADP ratio in the mitochondria, COX4 binds to ATP and allosterically inhibits enzyme activity (Arnold and Kadenbach 1997; Napiwotzki et al. 1997). Likewise, COX5b and COX6a regulate COX activity according to the ATP/ADP ratio (Frank and Kadenbach 1996; Bender and Kadenbach 2000). COX5a can bind to 3,4-diiodothyronine (T2) and relieve the intramitochondrial ATP-induced allosteric inhibition of COX activity (Arnold et al. 1998). COX6b assists in stabilizing the dimeric COX enzyme, and COX6c binds to cytochrome C with low affinity (Tsukihara et al. 1996). COX6a, 7a, and 8 have isoforms that are tissue-specific that differentially regulate COX activity (Kadenbach et al. 1990; 2000). The precise functions of COX subunits 7a, 7b, 7c, and 8a are not fully understood at this time.
In the Sp family of transcription factors, Sp1, Sp3, and Sp4 are structurally-related zinc finger proteins. These factors recognize the same GC-rich sequences in CpG islands and bind with similar affinity; hence, they compete or cooperate with each other. To date, at least 12,000 Sp-binding sites have been found in the human genome, most of which have been associated with genes involved in many cellular functions (Safe and Abdelrahim 2005; Suske 1999; Li and Davie 2010). Sp1 and Sp3 are ubiquitously expressed, whereas Sp4 is expressed mainly in neurons and testes (Suske 1999; Wierstra 2008). Sp4 has both activating and repressing domains for transcription, and both are operating in neurons (Suske 1999; Wierstra 2008). Sp1 knockout mouse embryos die at about 10 days of gestation (Marin et al., 1997), whereas Sp4 knockout mice survive up to birth. After birth, however, two-thirds of them die within 4 weeks, and surviving ones have abnormalities in their reproductive system and reproductive behavior (Göllner et al., 2001). Thus, physiological functions are not identical between these two Sp factors. Once neurons are differentiated and become postmitotic, Sp4 exerts transcriptional repression on genes involved in mitosis, and the expression of Sp1, which regulates many mitotic proteins, is down-regulated (Mao et al. 2006; 2007; 2009). Recently, we found that in N2a cells, Sp1 regulates the expression of all thirteen COX subunit genes (Dhar et al. 2013). However, as shown in the present study, Sp4 plays a more dominant role in regulating COX in primary neurons. Knocking down Sp1 caused a slight but significant increase in the expression of Sp4 in primary neurons, and vice versa. However, whereas silencing Sp4 is detrimental to the expressions of all COX subunit genes, suggesting that the slight increase in Sp1 expression was not sufficient to rescue these target genes in primary neurons, silencing Sp1 essentially had no effect on the target genes, indicating that they are regulated mainly by Sp4 and not by Sp1 in primary neurons.
Moreover, we recently found that in primary neurons, only Sp4 and not Sp1 regulates the expression of key glutamatergic NMDA receptor Grin1, Grin2a, and Grin2b genes (Priya et al. 2013b). Thus, Sp4 is able to co-regulate mediators of energy metabolism (COX) as well as mediators of synaptic transmission (NMDA receptor subunit genes), thereby enabling the coupling of energy metabolism and neuronal activity at the transcriptional level. This attests to the critical role of Sp4 in normal neuronal functioning.
Our earlier studies have shown that levels of COX transcripts, proteins, and activity are regulated by neuronal activity (Wong-Riley 1989; Hevner and Wong-Riley 1993; DeYoe et al. 1995; Wong-Riley et al. 1998; Liang and Wong-Riley 2006; Yang et al. 2006). The present study indicates that Sp4 itself is also regulated by neuronal activity. Sp4 message and protein are up-regulated by depolarizing stimulation and down-regulated by impulse blockade. Thus, when Sp4’s own expression is knocked down by shRNA, it can no longer support a heightened level of target gene transcription induced by KCl stimulation. Likewise, when Sp4 is over-expressed, it can rescue its target gene expression suppressed by TTX blockade. Thus, Sp4 mediates the activity-dependent regulation of all 13 COX subunit genes, the mitochondrial transcription factor genes, and SURF1 gene in neurons. In mature neurons, when the level of Sp1 is normally reduced (Mao et al. 2007; present study), Sp4 becomes the dominant Sp factor.
COX is a multisubunit, bigenomic, and multichromosomal protein whose transcriptional regulation presents a true operational challenge. To achieve a fine-tuned regulation in neurons, multiple transcription factors are likely involved and a well-orchestrated mechanism must be in place. Besides Sp4, neurons also rely on NRF-1 and NRF-2 to regulate all 10 of its nucleus-encoded subunit genes (Ongwijitwat and Wong-Riley 2005; Ongwijitwat et al. 2006; Dhar et al. 2008). These transcription factors also regulate TFAM, TFB1M, and TFB2M, thereby controlling the expression of the 3 mitochondrial COX subunit genes (Virbasius and Scarpulla 1994; Gleyzer et al. 2005; Dhar et al. 2013). Remarkably, the 10 nucleus-encoded COX subunit genes and genes for TFAM, TFB1M, and TFB2M are all transcribed in the same dynamic transcription factory (Dhar et al. 2009).
The involvement of multiple transcription factors in regulating the same set of genes (13 in the case of COX) necessitates one of three possible mechanisms: complementary (each factor regulating complementary sets of target genes), concurrent and parallel (all regulating the same set of target genes in the same positive or negative direction), or a combination of complementary and concurrent/parallel. NRF-1 and NRF-2 have previously been found to operate in a concurrent and parallel manner in regulating the same set of genes in neurons (Ongwijitwat and Wong-Riley, 2005; Dhar and Wong-Riley, 2008, 2009; Priya et al. 2013a). The present study revealed that Sp4 operates in a concurrent and parallel mechanism with NRF-1 and NRF-2 in regulating all 13 COX subunit genes in neurons. As the binding sites for Sp4, NRF-1, and NRF-2 can be in close proximity to one another for a number of COX subunit genes (our unpublished observations), it is possible that they may interact with each other while regulating their target genes in neurons. The verification or dismissal of such interactions awaits future investigation. Besides transcription factors, coactivators such as peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) are likely to be involved in the regulation of COX in neurons. PGC-1α powerfully coactivates NRF-1 and NRF-2 and is a master regulator of mitochondrial biogenesis (Wu et al. 1999; Scarpulla 2008). It responds to changes in neuronal activity earlier than those of NRF-1 and NRF-2 (Liang and Wong-Riley 2006; Liang et al. 2010). Future studies will determine if PGC-1α interacts directly with Sp4 in neurons.
Acknowledgments
Supported by National Institutes of Health Grants R01 EY018441 and T32 EY14537. We thank Dr. Bindu Nair for her professional assistance. All authors have no conflict of interest to declare.
Abbreviations
- ChIP
chromatin immunoprecipitation
- COX
cytochrome c oxidase
- EMSA
electrophoretic mobility shift assay
- NRF-1
nuclear respiratory factor 1
- NRF-2
nuclear respiratory factor 2
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator 1α
- Sp1
specificity protein 1
- Sp3
specificity protein 3
- Sp4
specificity protein 4
- Surf1
Surfeit locus protein 1
- TFAM
transcription factor A of mitochondria
- TFB1M
transcription factor B1 of mitochondria
- TFB2M
transcription factor B2 of mitochondria
- TSP
transcription start point
References
- Arnold S, Kadenbach B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur J Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- Arnold S, Goglia F, Kadenbach B. 3,5-Diiodothyronine binds to subunit Va of cytochrome-c oxidase and abolishes the allosteric inhibition of respiration by ATP. Eur J Biochem. 1998;252:325–330. doi: 10.1046/j.1432-1327.1998.2520325.x. [DOI] [PubMed] [Google Scholar]
- Basu A, Park K, Atchison ML, Carter RS, Avadhani NG. Identification of a transcriptional initiator element in the cytochrome c oxidase subunit Vb promoter which binds to transcription factors NF-E1 (YY-1, delta) and Sp1. J Biol Chem. 1993;268:4188–4196. [PubMed] [Google Scholar]
- Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase is reversible switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466:730–734. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- Chu C, Zavala K, Fahimi A, Lee J, Xue Q, Eilers H, Schumacher MA. Transcription factors Sp1 and Sp4 regulate TRPV1 gene expression in rat sensory neurons. Mol Pain. 2011;7:44. doi: 10.1186/1744-8069-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole EG, Gaston K. A functional YY1 binding site is necessary and sufficient to activate Surf-1 promoter activity in response to serum growth factors. Nuc Acids Res. 1997;25:3705–3711. doi: 10.1093/nar/25.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoe EA, Trusk TC, Wong-Riley MTT. Activity correlates of cytochrome oxidase-defined compartments in granular and supragranular layers of primary visual cortex of the macaque monkey. Vis Neurosci. 1995;12:629–639. doi: 10.1017/s0952523800008920. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT. The kinesin superfamily protein KIF17 is regulated by the same transcription factor (NRF-1) as its cargo NR2B in neurons. Biochim Biophys Acta. 2011;1813:403–411. doi: 10.1016/j.bbamcr.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: Role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Chromosoms conformation capture of all 13 genomic loci in the transcriptional regulation of the multi-subunit bigenomic cytochrome c oxidase in neurons. J Biol Chem. 2009;284:18644–18650. doi: 10.1074/jbc.M109.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Johar K, Wong-Riley MT. Bigenomic transcriptional regulation of all thirteen cytochrome c oxidase subunit genes by specificity protein 1. Open Biol. 2013;3:120176. doi: 10.1098/rsob.120176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank V, Kadenbach B. Regulation of the H+/e− stoichiometry of cytochrome c oxidase from bovine heart by intramitochondrial ATP/ADP ratios. FEBS Lett. 1996;382:121–124. doi: 10.1016/0014-5793(96)00096-8. [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göllner H, Bouwman P, Mangold M, Karis A, Braun H, Rohner I, Del Rey A, Besedovsky HO, Meinhardt A, van den Broek M, Cutforth T, Grosveld F, Philipsen S, Suske G. Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells. 2001;6:689–697. doi: 10.1046/j.1365-2443.2001.00455.x. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Lomax MI. Nuclear genes for cytochrome c oxidase. Biochim Biophys Acta. 1997;1352:174–192. doi: 10.1016/s0167-4781(97)00025-0. [DOI] [PubMed] [Google Scholar]
- Gunter TE, Gunter KK, Sheu SS, Gavin CE. Mitochondrial calcium transport: physiological and pathological relevance. Am J Physiol. 1994;267:C313–C339. doi: 10.1152/ajpcell.1994.267.2.C313. [DOI] [PubMed] [Google Scholar]
- Hagen G, Müller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Brain cytochrome oxidase: purification, antibody production and immunohistochemical/histochemical correlations in the CNS. J Neurosci. 1989;9:3884–3898. doi: 10.1523/JNEUROSCI.09-11-03884.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Mitochondrial and nuclear gene expression for cytochrome oxidase subunits are disproportionately regulated by functional activity in neurons. J Neurosci. 1993;13:1805–1819. doi: 10.1523/JNEUROSCI.13-05-01805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttermann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–1213. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- Hüttermann M, Schmidt TR, Grossman LI. A third isoform of cytochrome c oxidase subunit VIII is present in mammals. Gene. 2003;312:95–102. doi: 10.1016/s0378-1119(03)00604-8. [DOI] [PubMed] [Google Scholar]
- Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M. Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem. 2007;97:520–531. doi: 10.1111/j.1471-4159.2006.04216.x. [DOI] [PubMed] [Google Scholar]
- Johar K, Priya A, Wong-Riley MTT. Regulation of Na+/K+-ATPase by nuclear respiratory factor 1. Implication in the tight coupling of neuronal activity, energy generation, and energy consumption. J Biol Chem. 2012;287:40381–40390. doi: 10.1074/jbc.M112.414573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B, Jarausch J, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Stroh A, Becker A, Eckerskorn C, Lottspeich F. Tissue- and species-specific expression of cytochrome c oxidase isozymes in vertebrates. Biochim Biophys Acta. 1990;1015:368–372. doi: 10.1016/0005-2728(90)90042-3. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Hüttemann M, Arnold S, Lee I, Bender E. Mitochondrial energy metabolism is regulated via nuclear-coded subunits of cytochrome c oxidase. Free Radic Biol Med. 2000;29:211–221. doi: 10.1016/s0891-5849(00)00305-1. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Barsh GS, Clayton DA. Structure and chromosomal localization of the mouse mitochondrial transcription factor A gene (Tfam) Mamm Genome. 1997;8:139–140. doi: 10.1007/s003359900373. [DOI] [PubMed] [Google Scholar]
- Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat. 2010;192:275–283. doi: 10.1016/j.aanat.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Liang HL, Wong-Riley MTT. Activity-dependent regulation of nuclear respiratory factor-1, nuclear respiratory factor-2 and peroxisome proliferator-activated receptor gamma coactivator-1 in neurons. NeuroReport. 2006;17:401–405. doi: 10.1097/01.wnr.0000204980.98876.11. [DOI] [PubMed] [Google Scholar]
- Liang HL, Ongwijitwat S, Wong-Riley MTT. Bigenomic functional regulation of all 13 cytochrome c oxidase subunit transcripts in rat neurons in vitro and in vivo. Neurosci. 2006;140:177–190. doi: 10.1016/j.neuroscience.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Liang HL, Dhar SS, Wong-Riley MTT. p38 mitogen-activated protein kinase and calcium channels mediate signaling in depolarization-induced activation of peroxisome proliferator-activated receptor gamma coactivator-1alpha in neurons. J Neurosci Res. 2010;88:640–649. doi: 10.1002/jnr.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma L, Song L, Radoi GE, Harrison NL. Transcriptional regulation of the mouse gene encoding the α-4 subunit of the GABAA receptor. J Biol Chem. 2004;279:40451–40461. doi: 10.1074/jbc.M406827200. [DOI] [PubMed] [Google Scholar]
- Mao X, Moerman-Herzog AM, Wang W, Barger SW. Differential transcriptional control of the superoxide dismutase-2 kappa B element in neurons and astrocytes. J Biol Chem. 2006;281:35863–35872. doi: 10.1074/jbc.M604166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Yang SH, Simpkins JW, Barger SW. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. J Neurochem. 2007;100:1300–1314. doi: 10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Moerman-Herzog AM, Chen Y, Barger SW. Unique aspects of transcriptional regulation in neurons – nuances in NFkappaB and Sp1-related factors. J Neuroinflammation. 2009;6:16. doi: 10.1186/1742-2094-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Napiwotzki J, Shinzawa-Itoh K, Yoshikawa S, Kadenbach B. ATP and ADP bind to cytochrome c oxidase and regulate its activity. Biol Chem. 1997;378:1013–1021. doi: 10.1515/bchm.1997.378.9.1013. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MTT. Functional analysis of the rat cytochrome c oxidase subunit 6A1 promoter in primary neurons. Gene. 2004;337:163–171. doi: 10.1016/j.gene.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MTT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MTT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Priya A, Johar K, Wong-Riley MTT. Nuclear respiratory factor 2 regulates the expression of the same NMDA receptor subunit genes as NRF-1: Both factors act by a concurrent and parallel mechanism to couple energy metabolism and synaptic transmission. Biochim Biophys Acta. 2013a;1833:48–58. doi: 10.1016/j.bbamcr.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya A, Johar K, Wong-Riley MTT. Specificity protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B. Biochim Biophys Acta. 2013b doi: 10.1016/j.bbamcr.2013.07.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Valin A, Sun X, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Mol Cell Neurosci. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann NY Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I. Sp1: emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Wikström M, Krab K, Saraste M. A Synthesis. Academic Press; New York: 1981. Cytochrome oxidase. [Google Scholar]
- Wong-Riley M. Changes in the visual system of monocularly sutured or enucleated cats demonstrable with cytochrome oxidase histochemistry. Brain Research. 1979;171:11–28. doi: 10.1016/0006-8993(79)90728-5. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv Exp Med Biol. 2012;748:283–304. doi: 10.1007/978-1-4614-3573-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley M, Anderson B, Liebl W, Huang Z. Neurochemical organization of the macaque striate cortex: Correlation of cytochrome oxidase with Na+K+ATPase, NADPH-diaphorase, nitric oxide synthase, and NMDA receptor subunit 1. Neurosci. 1998;83:1025–1045. doi: 10.1016/s0306-4522(97)00432-6. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yamada M, Amuro N, Goto Y, Okazaki T. Structural organization of the rat cytochrome c oxidase subunit IV gene. J Biol Chem. 1990;265:7687–7692. [PubMed] [Google Scholar]
- Yang SJ, Liang HL, Wong-Riley MTT. Activity-dependent transcriptional regulation of nuclear respiratory factor-1 in cultured rat visual cortical neurons. Neurosci. 2006;141:1181–1192. doi: 10.1016/j.neuroscience.2006.04.063. [DOI] [PubMed] [Google Scholar]