Abstract
Background
In luminal breast cancer cell lines, TFAP2C regulates expression of key genes in the estrogen receptor–associated cluster and represses basal-associated genes including CD44. We examined the effect of TFAP2C overexpression in a basal cell line and characterized the expression of TFAP2C and CD44 in breast cancer specimens to determine if expression was associated with clinical response.
Methods
MDA-MB-231 breast cancer cells were treated with a TFAP2C-containing plasmid and evaluated for effects on CD44 expression. Pretreatment biopsy cores from patients receiving neoadjuvant chemotherapy for breast cancer were evaluated for TFAP2A, p53, TFAP2C, and CD44 expression by immunohistochemistry.
Results
Overexpression of TFAP2C in MDA-MB-231 cells resulted in decreased expression of CD44 mRNA and protein, P< 0.05. A pathologic complete response (pCR) following neoadjuvant chemotherapy was achieved in 17% of patients (4/23). Average expression for TFAP2C by immunohistochemistry in patients with a pCR was 93%, compared with 46% in patients with residual disease, P= 0.016; and in tumors that stained at ≥80% for TFAP2C, 4 of 9 (44%) achieved pCR, compared with 0 of 14 below 80%, P= 0.01. Additionally, in tumors that stained ≤80% for CD44, 4 of 10 (40%) achieved pCR, compared with 0 of 13 >80%, P = 0.02. In tumors that stained high for TFAP2C (≥80%) and low for CD44 (≥80%), 4 of 7 (57%) achieved pCR, compared with 0 of 16 in all other groups (P= 0.004).
Conclusions
TFAP2C repressed CD44 expression in basal-derived breast cancer. In primary breast cancer specimens, high TFAP2C and low CD44 expression were associated with pCR after neoadjuvant chemotherapy and could be predictive of tumors that have improved response to neoadjuvant chemotherapy.
Keywords: TFAP2C, Breast cancer, Pathologic complete response, Neoadjuvant chemotherapy, CD44
1. Introduction
Breast cancer is the most common cancer in women, with an estimated 227,000 new cases in the United States in 2012. Despite high rates of long-term survival, breast cancer remains second only to lung cancer as the most common cause of cancer death in women [1]. Because of the continued high level of mortality from this disease, new markers of tumor behavior and treatment strategies are being investigated. Currently, much debate exists over the role of neoadjuvant chemotherapy in breast cancer [2]. Although traditionally used only for patients with inoperable disease, primary chemotherapy has been investigated in resectable tumors for the past 20 y. Multiple studies have shown that neoadjuvant chemotherapy is effective to reduce local tumor burden and increase patient eligibility for breast-conserving treatment [3–9]. Additionally, the triple-negative molecular subtype has been identified as having improved response to neoadjuvant chemotherapy in reduction of locoregional recurrence, higher rates of pathologic complete response (pCR), and improved survival after pCR [10–12]. Based on these findings that molecular expression patterns can be indicative of biologically distinct tumors with differential response to neoadjuvant chemotherapy, we sought to investigate markers of response in our patient population.
Transcription factor activating protein 2-gamma (TFAP2C) is a DNA-binding transcription factor that we have previously shown to be involved in regulation of estrogen receptor (ER) and ER-associated genes [13–15]. TFAP2C provides direct transcriptional regulation of the RET proto-oncogene [16], expression of which has been linked to reduced survival and hormone sensitivity in breast cancer [17]. TFAP2C also regulates glutathione peroxidase, which in vitro influences breast cancer sensitivity to doxorubicin [18]. Additionally, in breast cancer TFAP2C expression is predictive of response to hormone therapy and high expression of TFAP2C correlates to shortened patient survival [19]. TFAP2A is a TFAP family member with a similar DNA binding pattern to TFAP2C, but with distinct transcriptional regulatory activity in both ER-positive and ER-negative breast cancer cell lines [15,20].
Studies examining binding sites by chromatin immuno-precipitation with direct sequencing (ChIP-seq) and expression changes suggested a role for TFAP2C in repression of CD44 by TFAP2C in the luminal breast cancer cell line MCF-7 (A. Cyr et al., unpublished data, 2013) [15]. Transient or stable knockdown of TFAP2C in MCF-7 cells resulted in increased CD44 expression and an increase in the population of CD44+/CD24– cells (A. Cyr et al., unpublished data, 2013). Cells with the expression pattern of CD44+/CD24– defines the breast cancer stem cell population, which are thought to drive tumor initiation and represent a population more likely to develop metastatic disease and be resistant to chemotherapy [21–24]. Because TFAP2C is involved in regulation of proliferation and oxidative associated genes, and CD44 is a possible TFAP2C target believed to be a marker of chemotherapy resistance, we sought to characterize TFAP2C and CD44 levels to determine if expression correlated to response to neoadjuvant chemotherapy.
2. Methods
2.1. Cell lines
The MDA-MB-231, MCF-7, and BT-474 cell lines were obtained from the American Type Culture Collection (Manassas, VA). MCF-7 and MDA-MB-231 cells were cultured using Dulbecco’s modified Eagle medium and BT-474 were cultured using Dulbecco’s modified Eagle medium/F12. All media were supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin and all cultures grown at 37°C and 5% CO2.
2.2. Chromatin immunoprecipitation sequencing
ChIP-seq was performed as previously described [15]. TFAP2C binding to the CD44 promoter was assessed by searching the University of California Santa Cruz genome browser (http://genome.ucsc.edu/cgi-bin/hgTracks?org=human) with our ChIP-seq tracks overlaid.
2.3. Gel shift assay
Gel shift was performed using the Gel Shift Assay Core System E3050 (Promega, Madison, WI). A 150-bp oligonucleotide probe was created from the sequence underlying each of the two highlighted ChIP-seq peaks using MDA-MB-231 template DNA, and probes were labeled with 32P according to manufacturer protocol. TFAP2C protein was synthesized from pcDNA3.1-AP2C using TNT T7 Quick Coupled Transcription/Translation System 1170 (Promega) [20]. Probe and competitor design are graphically represented in Figure 1 and sequences are provided in Supplemental data. Supershift was performed with TFAP2C antibody SC-12762 (Santa Cruz Biotechnology, Santa Cruz, CA).
Fig. 1.
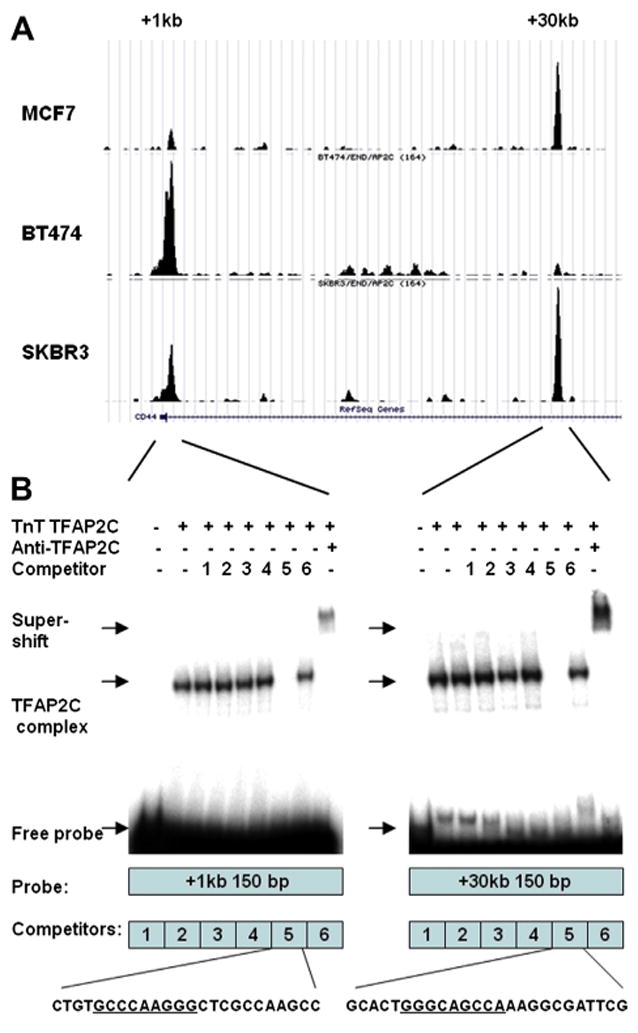
(A) Graphical representation of ChIP-seq data demonstrating locations of TFAP2C binding in the CD44 promoter. Two peaks are identified in the ER-negative SKBR-3 cell line within the first intron at CD44 +kb and CD44 +30kb relative to the transcriptional start site. In ER-positive MCF-7 only the +30kb site is present, and in ER-positive BT-474 only the +1kb site is present. (B) Gel shift assay confirms localization of TFAP2C biding sites at both ChIP-seq peaks. Both probes shift with synthesized TFAP2C protein, demonstrating TFAP2C binding, and supershift with TFAP2C antibody, confirming the specificity of the complex. Beneath each gel shift image is a graphical representation of the probe and competition oligonucleotides. For oligonucleotides found to compete for TFAP2C binding, the sequence is provided with the consensus AP-2 family binding sequence underlined.
2.4. TFAP2C overexpression transfections
TFAP2C was overexpressed using a pcDNA 3.1 plasmid containing a full-length TFAP2C cDNA construct (TFAP2C) and compared with transfection with an empty pcDNA 3.1 plasmid (empty vector). Transfection was performed according to the manufacturer’s protocol using Lipofectamine LTX reagent (Invitrogen, Carlsbad, CA).
2.5. Real-time polymerase chain reaction
Total RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) 48 h after plasmid transfection. Total RNA was converted to cDNA using the Superscript III kit (Invitrogen) utilizing random hexamer primers. Quantitative polymerase chain reaction (PCR) was performed according to standard TaqMan Fast protocol (Applied Biosystems, Carlsbad, CA). TaqMan primer/probe combinations used were TFAP2C (Hs00231476) and CD44 (Hs01075861). GAPDH (Hs02758991) and 18s (Hs99999901) were used as endogenous controls. real-time polymerase chain reaction (RT-PCR) was performed in technical triplicate and averages, standard deviations, and statistical calculations were made using three biologic replicates.
2.6. Western blot
Total protein was isolated 48 h after plasmid transfection using RIPA buffer with Halt protease inhibitor cocktail (Thermo Scientific, Rockford, IL). Antibodies used were TFAP2C (Santa Cruz Biotechnology), CD44 (R&D Systems, Minneapolis, MN), and GAPDH (Santa Cruz Biotechnology). Protein levels were quantified using ImageJ (http://rsb.info.nih.gov/ij/) and statistical calculations were made using three biologic replicates.
2.7. Patients
Twenty-three consecutive patients who underwent neoadjuvant chemotherapy for primary breast cancer and had archived formalin-fixed paraffin-embedded tissue from diagnostic core biopsy specimens were retrospectively enrolled. The institutional review board at the University of Iowa approved this study. Patients with stage IV disease and inflammatory cancer were excluded because they could not be evaluated for pCR and only patients with stage II or stage III disease according to the American Joint Committee on Cancer manual were enrolled. Chemotherapy regimen was determined by the treating medical oncologist in collaboration with the institutional tumor board and was based on clinical trials and national standards at the time of treatment. All data in this study were obtained retrospectively and the decision to undergo neoadjuvant chemotherapy, the treatment regimen, and other clinical and operative decisions were not influenced by this study. The majority of patients were given combination therapy with adriamycin, cyclophosphamide, and paclitaxel. Patients received endocrine therapy and trastuzumab according to institutional standards and receptor status. Chemotherapy regimen, demographic information, and response to neoadjuvant chemotherapy were determined by retrospective chart review. Patients were categorized as having a pCR if on resection they had no evidence of malignant disease, including ductal carcinoma in situ, at the primary tumor site or lymph nodes.
2.8. Immunohistochemistry
Archived formalin-fixed paraffin-embedded pretreatment biopsy specimens were obtained for the enrolled patients and analyzed by our institutional pathology research laboratory. Tissue blocks were sectioned and stained for p53 (Santa Cruz Biotechnology), TFAP2A (Santa Cruz Biotechnology), TFAP2C (Santa Cruz Biotechnology), and CD44 (R&D Systems) with appropriate positive and negative controls. Samples were interpreted by an attending pathologist (R.W.A.) and scored according to percentage of positive cells. Staining intensity was also recorded but was not used in the final analysis.
2.9. Statistical analysis
Univariate statistical analysis was performed on continuous data using the two-sided Student t-test, and for categorical variables frequency association was performed using the two-sided Fisher exact test. All statistical calculations were made using R (www.r-project.org). Significance was defined as P < 0.05.
3. Results
3.1. Identification of TFAP2C binding sites in the CD44 regulatory region
Chip-seq data for the genomic binding locations of TFAP2C in the CD44 regulatory region in MCF-7 and BT-474 ER-positive breast cancer cell lines and SKBR-3 ER-negative breast cancer cell line were compared. Figure 1 demonstrates two main binding peaks within the first intron at CD44+1kb and at CD44+30kb in the CD44 gene relative to the transcriptional start site in ER-negative SKBR-3 breast cancer cells. In ER-positive cells, only the +1kb peak was observed in BT-474, and only the +30kb peak was found in MCF-7. To confirm that TFAP2C bound directly to the DNA at those locations, gel shift assay was performed with probes underlying each peak. Gel shift confirmed that cloned TFAP2C protein bound to both sites, with cold competitors localizing TFAP2C binding to consensus AP-2 family binding sequence. These data confirm that TFAP2C binds directly to two locations in the first intron of the CD44 gene, consistent with known mechanisms of TFAP2C direct transcriptional regulation.
3.2. Overexpression of TFAP2C represses CD44 expression
Based on our observations that knockout of TFAP2C in a luminal cell line (high TFAP2C, low CD44–expressing cell line) resulted in increased expression of CD44 and established mechanisms of TFAP2C action through direct DNA binding, we hypothesized that TFAP2C directly represses CD44 expression. To investigate the role of TFAP2C in a basal cell line, we performed a transient overexpression of TFAP2C in a TFAP2C-negative, high-CD44-expressing basal cell line, MDA-MB-231. Transfection of a TFAP2C expression plasmid resulted in a 700-fold increase in TFAP2C mRNA (Fig. 2) and a 20-fold increase in TFAP2C protein expression (Fig. 3) compared to empty vector transfection, showing successful TFAP2C over-expression. At 48 h post transfection, overexpression of TFAP2C resulted in an 80% reduction in CD44 mRNA (Fig. 2) and a 60% reduction in CD44 protein (Fig. 3) compared to empty vector transfection. These results demonstrate that transient TFAP2C expression caused repression of CD44 expression in MDA-MB-231 breast cancer cells.
Fig. 2.
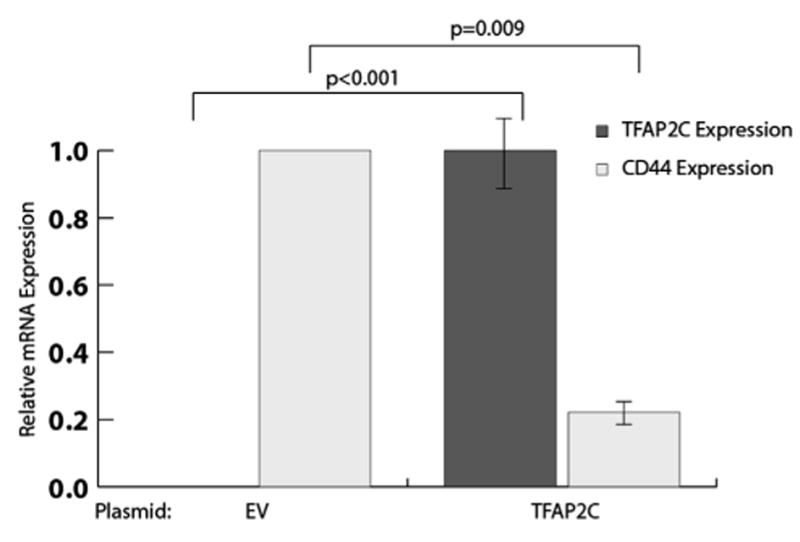
Overexpression of TFAP2C transcriptionally represses CD44 expression. RT-PCR demonstrated the effects of TFAP2C plasmid transfection on TFAP2C and CD44 mRNA expression in the triple-negative breast cancer cell line MDA-MB-231. mRNA levels were normalized to GAPDH and 18s rRNA levels and to empty vector (EV) transfection. RT-PCR was performed in triplicate and averages and standard deviations were calculated using three biologic replicates. Statistical P values are shown using brackets.
Fig. 3.
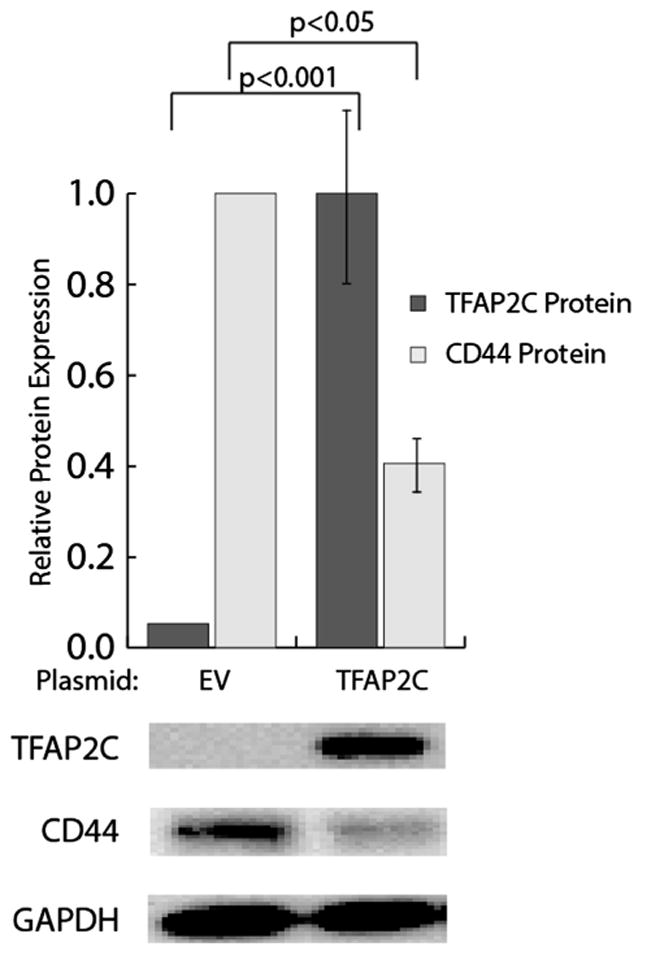
Overexpression of TFAP2C in MDA-MB-231 cells reduces CD44 protein levels. Western blot test demonstrated the effects of TFAP2C plasmid transfection on TFAP2C and CD44 protein levels in MDA-MB-231 cells. Overexpression of TFAP2C was associated with a significant reduction in CD44 levels. Western blots were performed in biologic triplicate and protein levels quantified using ImageJ.
3.3. Patient demographics
In order to investigate the potential role of expression of TFAP2C and target genes on response to neoadjuvant chemotherapy, 23 patients treated with neoadjuvant chemotherapy followed by surgical resection for stage II and III breast cancer and for which archived pretreatment core biopsy tissue was available were analyzed. Four of 23 patients (17%) achieved a pCR following neoadjuvant chemotherapy. Patient demographics, tumor receptor status data, and staging are listed in the Table and were significant only for a negative correlation between ER-positive tumors and achieving a pCR.
Table.
Patient demographics, receptor status, and stage as a function of response to neoadjuvant chemotherapy.
| Partial response | pCR | P value | |
|---|---|---|---|
| Patients | 19 | 4 | |
| Age | 50.7 ± 11.5 | 40.0 ± 10.7 | 0.14 |
| ER positive | 16 (84%) | 1 (25%) | 0.04 |
| Her2 positive | 3 (16%) | 2 (50%) | 0.19 |
| Triple negative | 2 (11%) | 1 (25%) | 0.45 |
| Stage II | 9 (47%) | 2 (50%) | 1.0 |
| Stage III | 10 (53%) | 2 (50%) | 1.0 |
| Adriamycin-based chemotherapy | 16 (84%) | 3 (75%) | 1.0 |
3.4. TFAP2C and CD44 expression were associated with pCR
Scored immunohistochemistry data were analyzed according to tumor receptor status and response to chemotherapy. Expression of p53, TFAP2A, TFAP2C, and CD44 was not statistically different in ER-positive (luminal), HER2-overexpressing, or triple-negative (basal) tumors. Expression data for p53, TFAP2A, TFAP2C, and CD44 separated by response to chemotherapy is shown in Figure 4. Expression of p53 and TFAP2A did not differ between the partial response and pCR groups. However, mean TFAP2C expression was significantly higher in the pCR group (92.5%) compared with the partial response (46.3%, P =0.02), and pCR tumors were significantly more likely to have ≥80% cells positive for TFAP2C (4/4, 100%) compared with partial response tumors (5/19, 26%, P = 0.01). There was additionally a trend in the patients that converted from a positive to a negative axilla following neoadjuvant chemotherapy for higher mean TFAP2C expression (65%) compared with patients with residual axillary disease (38%, P = 0.06), further indicating an association between increased TFAP2C expression and improved response to neoadjuvant chemotherapy.
Fig. 4.
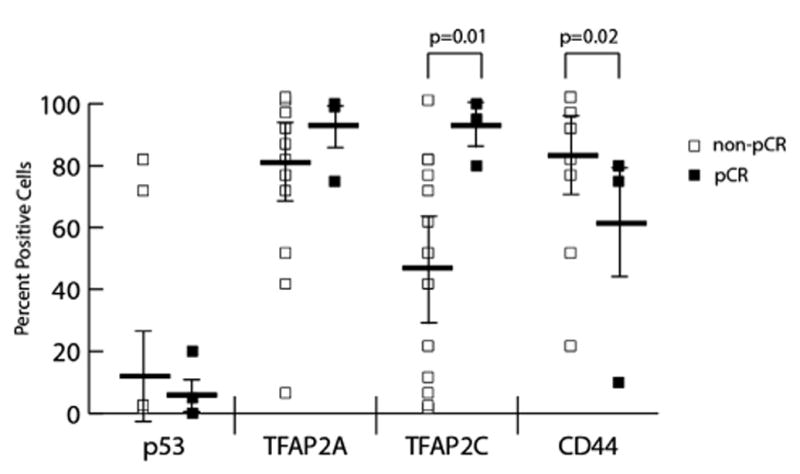
Immunohistochemistry expression patterns of patients treated with neoadjuvant chemotherapy. Samples were scored by percentage of cells staining positive for p53, TFAP2A, TFAP2C, and CD44 and compared in patients having a pCR (n = 4) versus a partial response (n = 19) to chemotherapy. Increased TFAP2C expression and decreased CD44 positivity were significantly associated with a pCR. TFAP2A and p53 expression were not associated with response to neoadjuvant chemotherapy. Horizontal bars represent the mean and error bars standard deviation. Data points with identical values are overlaid and each point might not be visible for all tests.
Mean CD44 expression did not differ statistically between the two groups; however, in the pCR group, tumors were more likely to have ≤80% CD44 positivity (4/4, 100%) compared with partial response tumors (6/19, 31%, P = 0.02). When combined, seven tumors stained at or above 80% positive for TFAP2C and at or below 80% for CD44. Of those seven, 4 (57%) achieved a pCR, compared with none of the 13 tumors (0%) with other expression patterns, P = 0.004, Figure 5.
Fig. 5.
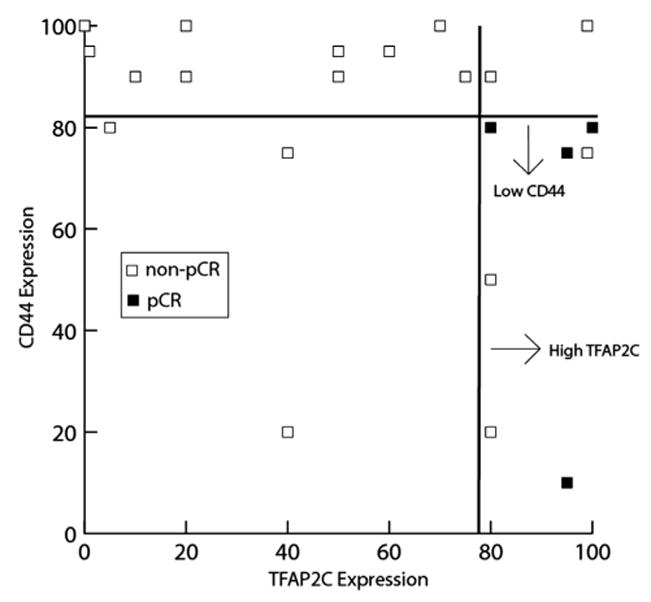
pCR following neoadjuvant chemotherapy is associated with high TFAP2C and low CD44 expression. Data points represent patients graphed according to TFAP2C and CD44 positivity on immunohistochemistry. Patients with high TFAP2C (≥80%) and low CD44 (≤80%) were significantly more likely to achieve a pCR (57%) compared with all other expression patterns (0%, P = 0.004).
4. Discussion
TFAP2C is a key transcription factor involved in the regulation of many genes associated with the luminal phenotype in breast cancer. We identify TFAP2C to also be a direct repressor of the stem cell marker CD44 in both ER-positive and ER-negative breast cancer cell lines, further identifying a key role for TFAP2C in regulating gene expression in breast cancer. Interestingly, in this study, in which three of the four pCR tumors were ER-negative, we have identified an important role for TFAP2C in defining a subset of nonluminal cancers that are more likely to respond to neoadjuvant chemotherapy. Our findings indicate that increased expression of functional TFAP2C is associated with pCR following neoadjuvant chemotherapy in breast cancer. Additionally, high-TFAP2C-expressing tumors become even more strongly associated with pCR when they concurrently express low levels of CD44. This finding is interesting for two reasons, the first being that concurrent high TFAP2C and low CD44 expression is consistent with transcriptional repression of CD44 by TFAP2C, indicating functional TFAP2C with chromatin structure that is accessible to TFAP2C at that location. Next, this finding is consistent with the hypothesis that CD44 expression is associated with cancer stem cells and that decreased populations of cancer stem cells result in tumors that are more responsive to neoadjuvant chemotherapy.
Current indications for neoadjuvant chemotherapy for locally advanced breast cancer are not well defined and represent an evolving field of intense investigation [2]. Improved delivery of neoadjuvant chemotherapy will require stratification to identify tumors that have a worse prognosis and require more aggressive treatment, as well as tumors that have a molecular expression profile that indicates improved responsiveness to chemotherapy. Triple-negative tumors are clinically more aggressive and have better neoadjuvant response outcomes compared to other tumor subtypes. This has led to increased selection of triple-negative tumors for neoadjuvant chemotherapy, and this approach has shown improved survival in patients having achieved a pCR [12].
Based on those findings, research is being directed at finding markers that identify distinct tumor phenotypes within the current receptor classifications that predict tumor behavior and response to chemotherapy. High expression of TFAP2C has been identified in multiple studies as being linked, both directly and indirectly through target genes, to decreased survival [19,25]. Our findings indicate, however, that although these tumors have a distinct, more aggressive behavior, they also could have an improved response to neoadjuvant chemotherapy. Potentially, identifying aggressive high-TFAP2C-expressing breast cancers that also express low levels of CD44 could identify tumors that have lower populations of cancer stem cells and are therefore more responsive to neoadjuvant chemotherapy. Functionally, measurement of these two markers could identify a distinct tumor biology, which is more likely to achieve a pCR with neoadjuvant chemotherapy.
Our study characterized only 23 patients from a single institution. These results need to be validated in a larger study with a more diverse patient population. However, our results indicate that high TFAP2C and low CD44 expression could represent a distinct biology with functionally intact TFAP2C. With further study, identification of high-TFAP2C-expressing/low-CD44-expressing tumors could indicate tumors with more pronounced response to neoadjuvant chemotherapy, which could improve outcomes with this treatment strategy for select patients.
Supplementary Material
Acknowledgments
The authors would like to thank Junlin Liao, PhD, for his help with statistical calculations.
This work was supported by the National Institutes of Health grants R01CA109294 (PI: R.J. Weigel) and T32CA148062 (PI: R. J. Weigel) and by a generous gift from the Kristen Olewine Milke Breast Cancer Research Fund. P.M.S. was supported by National Institutes of Health grant T32CA148062.
Footnotes
Presented as an oral presentation at the 8th Annual Academic Surgical Congress, New Orleans, Louisiana, February 5–7, 2013.
Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jss.2013.04.042.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann M, von Minckwitz G, Mamounas EP, et al. Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol. 2012;19:1508. doi: 10.1245/s10434-011-2108-2. [DOI] [PubMed] [Google Scholar]
- 3.Bear HD, Anderson S, Brown A, et al. The effect on tumor response of adding sequential preoperative docetaxel to preoperative doxorubicin and cyclophosphamide: preliminary results from National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2003;21:4165. doi: 10.1200/JCO.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Powles TJ, Hickish TF, Makris A, et al. Randomized trial of chemoendocrine therapy started before or after surgery for treatment of primary breast cancer. J Clin Oncol. 1995;13:547. doi: 10.1200/JCO.1995.13.3.547. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham JD, Weiss SE, Ahmed S, et al. The efficacy of neoadjuvant chemotherapy compared to postoperative therapy in the treatment of locally advanced breast cancer. Cancer Invest. 1998;16:80. doi: 10.3109/07357909809039761. [DOI] [PubMed] [Google Scholar]
- 6.Fisher B, Bryant J, Wolmark N, et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol. 1998;16:2672. doi: 10.1200/JCO.1998.16.8.2672. [DOI] [PubMed] [Google Scholar]
- 7.Cance WG, Carey LA, Calvo BF, et al. Long-term outcome of neoadjuvant therapy for locally advanced breast carcinoma: effective clinical downstaging allows breast preservation and predicts outstanding local control and survival. Ann Surg. 2002;236:295. doi: 10.1097/01.SLA.0000027526.67560.64. discussion 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonadonna G, Valagussa P, Brambilla C, et al. Primary chemotherapy in operable breast cancer: eight-year experience at the Milan Cancer Institute. J Clin Oncol. 1998;16:93. doi: 10.1200/JCO.1998.16.1.93. [DOI] [PubMed] [Google Scholar]
- 9.Spanheimer PM, Carr JC, Thomas A, et al. The response to neoadjuvant chemotherapy predicts clinical outcome and increases breast conservation in advanced breast cancer. Am J Surg. 2013 doi: 10.1016/j.amjsurg.2012.10.025. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyers MO, Klauber-Demore N, Ollila DW, et al. Impact of breast cancer molecular subtypes on locoregional recurrence in patients treated with neoadjuvant chemotherapy for locally advanced breast cancer. Ann Surg Oncol. 2011;18:2851. doi: 10.1245/s10434-011-1665-8. [DOI] [PubMed] [Google Scholar]
- 11.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 12.Fisher CS, Ma CX, Gillanders WE, et al. Neoadjuvant chemotherapy is associated with improved survival compared with adjuvant chemotherapy in patients with triple-negative breast cancer only after complete pathologic response. Ann Surg Oncol. 2012;19:253. doi: 10.1245/s10434-011-1877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodfield GW, Horan AD, Chen Y, Weigel RJ. TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res. 2007;67:8439. doi: 10.1158/0008-5472.CAN-07-2293. [DOI] [PubMed] [Google Scholar]
- 14.Woodfield GW, Hitchler MJ, Chen Y, Domann FE, Weigel RJ. Interaction of TFAP2C with the estrogen receptor-alpha promoter is controlled by chromatin structure. Clin Cancer Res. 2009;15:3672. doi: 10.1158/1078-0432.CCR-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer. 2010;49:948. doi: 10.1002/gcc.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanheimer PM, Woodfield GW, Cyr AR, et al. Expression of the RET proto-oncogene is regulated by TFAP2C in breast cancer independent of the estrogen receptor. Ann Surg Oncol. 2012 doi: 10.1245/s10434-012-2570-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Mayer JA, Mazumdar A, Brown PH. The rearranged during transfection/papillary thyroid carcinoma tyrosine kinase is an estrogen-dependent gene required for the growth of estrogen receptor positive breast cancer cells. Breast Cancer Res Treat. 2012;133:487. doi: 10.1007/s10549-011-1775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulak MV, Cyr AR, Woodfield GW, et al. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.400. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gee JM, Eloranta JJ, Ibbitt JC, et al. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol. 2009;217:32. doi: 10.1002/path.2430. [DOI] [PubMed] [Google Scholar]
- 20.McPherson LA, Weigel RJ. AP2alpha and AP2gamma:a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrison BJ, Schmidt CW, Lakhani SR, Reynolds BA, Lopez JA. Breast cancer stem cells: implications for therapy of breast cancer. Breast Cancer Res. 2008;10:210. doi: 10.1186/bcr2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei W, Hu H, Tan H, Chow LW, Yip AY, Loo WT. Relationship of CD44+CD24-/low breast cancer stem cells and axillary lymph node metastasis. J Transl Med. 2012;10(Suppl 1):S6. doi: 10.1186/1479-5876-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HE, Kim JH, Kim YJ, et al. An increase in cancer stem cell population after primary systemic therapy is a poor prognostic factor in breast cancer. Br J Cancer. 2011;104:1730. doi: 10.1038/bjc.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulay A, Breuleux M, Stephan C, et al. The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res. 2008;68:3743. doi: 10.1158/0008-5472.CAN-07-5100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


