Abstract
Binding of the glucagon peptide to the glucagon receptor (GCGR) triggers the release of glucose from the liver during fasting, thus GCGR plays an important role in glucose homeostasis. Here we report the crystal structure of the seven transmembrane (7TM) helical domain of human GCGR at 3.4 Å resolution, complemented by extensive site-specific mutagenesis, and a hybrid model of glucagon bound to GCGR to understand the molecular recognition of the receptor for its natural ligand. Beyond the shared 7TM fold, the GCGR transmembrane domain deviates from class A G protein-coupled receptors with a large ligand binding pocket and the first transmembrane helix having a “stalk” region that extends three alpha-helical turns above the plane of the membrane. The stalk orients the extracellular domain (~12 kDa) relative to the membrane to form the glucagon binding site that captures the peptide and facilitates the insertion of glucagon’s N-terminus into the 7TM domain.
The glucagon receptor (GCGR) is one of the 15 members of the secretin-like (class B) family of G protein-coupled receptors (GPCRs)1 in humans. GCGR is activated by the 29 amino acid hormonal peptide, glucagon (Supplementary Fig. 1a), and is a potential drug target for type 2 diabetes2. During fasting, the pancreas dispatches glucagon to activate GCGR in the liver causing the release of glucose into the blood2. Despite less than 15% sequence homology between class A (rhodopsin-like) and class B GPCRs, many of these receptors presumably share a 7 transmembrane (7TM) helical domain and similar signal transduction mechanisms1. Although a structure-function understanding of the class A family of GPCRs has been greatly advanced during the last few years3, a detailed understanding of class B GPCRs has lagged due to the lack of a 7TM domain structure for these receptors.
All class B GPCRs contain a globular N-terminal extracellular domain (ECD) defined by three conserved disulfide bonds4,5 and a 7TM domain. They are activated by hormonal peptides that bind to both the ECD and the 7TM domain4. Structural details of soluble ECDs, including the ECD of GCGR6, and their role in the selective recognition of peptide hormones’ C-termini have been revealed for several class B receptors5,7,8. Although models of class B 7TM domains and ligand binding have been proposed based on site-directed mutagenesis9–11, photo-crosslinking12–14, and structure-based virtual screening studies15, the accuracy of such modeling has been hampered by the low sequence homology between class A and class B GPCRs.
Crystal structure of GCGR 7TM domain
The 7TM domain of human GCGR was fused to the thermally stabilized E. coli apocytochrome b562RIL16 (referred to as BRIL) at residue 123, and the C-terminus of GCGR truncated at residue 432 (Supplementary Fig. 2). This crystallized GCGR construct (BRIL-GCGR-ΔECD-ΔC, Supplementary Fig. 3) has the same binding affinity for NNC0640 (Supplementary Fig. 1b), as the full-length wild-type GCGR (Supplementary Table 1), indicating that the conformation of the 7TM domain of BRIL-GCGR-ΔECD-ΔC is similar to wild-type GCGR. The structure of the BRIL-GCGR-ΔECD-ΔC was determined at 3.4 Å resolution (Methods and Supplementary Table 2). Although GCGR was crystallized in the presence of the antagonist ligand NNC0640, convincing electron density for NNC0640 was not observed. As expected, GCGR adopts a 7TM fold (Fig. 1), with the BRIL fusion protein folded on top of the receptor and mediating most of the crystal contacts (Supplementary Fig. 4).
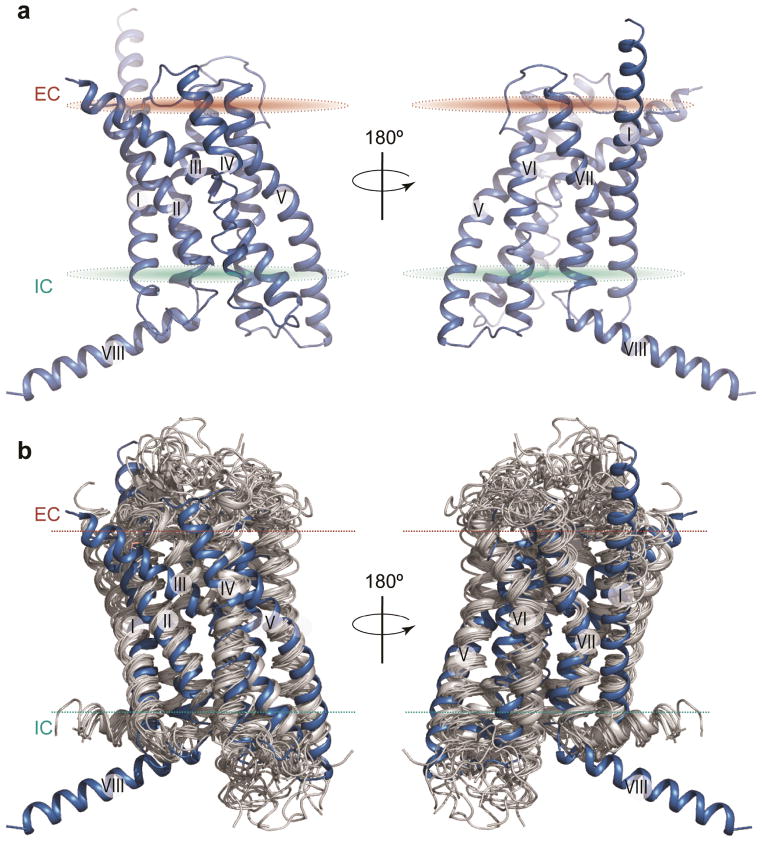
Figure 1. Structure of the 7TM domain of human GCGR and comparison to class A GPCR structures.
a, Cartoon depiction of the 7TM domain structure of GCGR. The two views are rotated 180° relative to each other. The disulfide bond between helix III and extracellular loop 2 (ECL2) is shown as yellow sticks. b, Side view of structural superimposition of 7TM domains of GCGR (blue) and class A GPCRs (grey). Structures of class A GPCRs used (PDB): 1U19, 2RH1, 2YCW, 3RZE, 3PBL, 3UON, 4DAJ, 3EML, 3V2W, 3ODU, 4DJH, 4EA3, 4DKL, 4EJ4, and 3VW7. Extracellular (EC) and intracellular (IC) membrane boundaries are shown as brown and cyan ovals (a) or dotted lines (b), respectively.
Despite the lack of protein sequence conservation, comparison of the GCGR 7TM structure with 15 known class A GPCR structures solved in inactive form shows that orientations and positions of helices in the 7TM bundles are conserved between the two classes (Fig. 1b, Supplementary Fig. 5). The 7TM helices of GCGR superimpose with those of the class A receptors with root-mean-square deviation of Cα backbone atoms (RMSDCα) in the 2.7 – 3.3 Å range, above the 2.2 – 3.0 Å range observed between major branches (α, β, γ and δ) of class A GPCRs. The structural alignment of GCGR with rhodopsin shows an approximate spatial correspondence between residues in the 7TM helices of the two GPCR classes, but also reveals a number of gaps in transmembrane regions reflecting substantial structural deviations in TM helices (Supplementary Fig. 6). The spatial correspondence between 7TM residues makes it possible to project the widely used class A Ballesteros-Weinstein numbering scheme17 (used hereafter for class A as BW number in parenthesis) for comparisons between GPCR classes (Supplementary Table 3). Analysis of sequence and structural features within the class B GPCRs, however, is defined by the Wootten numbering scheme based on class B residue conservation18 (used hereafter for class B receptors as superscript, Supplementary Table 3).
Class A versus B GPCRs
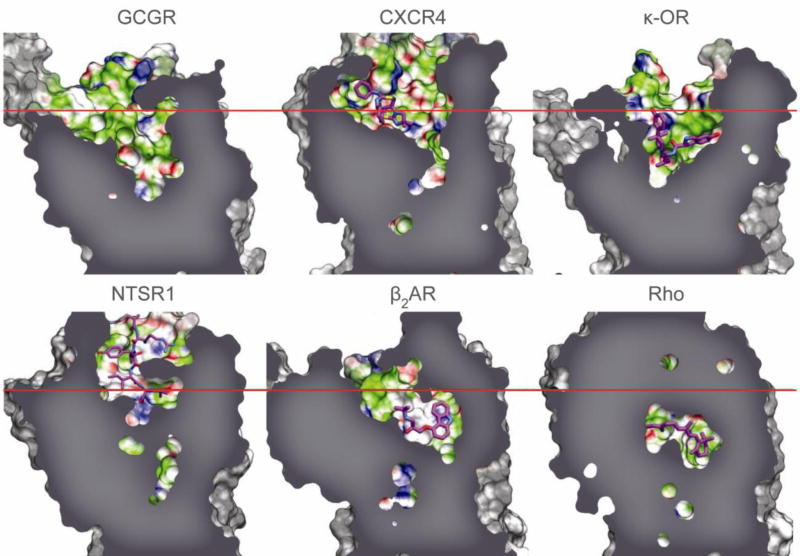
The GCGR structure reveals a number of features in the 7TM domain that are distinct from known class A GPCRs. The N-terminal end of helix I in GCGR is longer than any GPCR structures and extends three additional helical turns (approximately 16 Å) above the extracellular (EC) membrane boundary from Lys136 to Glu126 (Fig. 1). This region of GCGR, referred to as the stalk, may be involved in glucagon binding and defines the orientation of the ECD with respect to the 7TM domain. Extracellular loop 1 (ECL1) of GCGR is 16 residues long, as compared with 4–6 residues in most class A GPCRs. Although the ECL1 residues 201–215 are not resolved in the crystal structure, mutagenesis studies presented here and elsewhere19,20,21 indicate that these residues are involved in interactions with peptide ligands. The distance between the EC tips of 7TM helices II and VI is the largest among GPCR structures, and the distance between the EC tips of helices III and VII is among the largest, except for kappa (κ-OR) and mu opioid receptors (μ-OR)22,23 (Supplementary Fig. 5). The positioning of the EC tips of these 7TM helices creates a wider and deeper cavity in the ligand binding pocket of GCGR, which is larger than any class A receptor structures (Fig. 2, Supplementary Table 4).
Figure 2. Comparison of ligand binding pocket of GCGR with class A GPCRs.
The binding cavity of GCGR compared with the binding cavities of human chemokine receptor CXCR4 (PDB 3ODU), human κ-opioid receptor (κ-OR) (PDB 4DJH), rat neurotensin receptor (NTSR1) (PDB 4GRV), human β2 adrenergic receptor (β2AR) (PDB 2RH1), and bovine rhodopsin (Rho) (PDB 1U19) for comparison purposes (Supplementary Table 4). The approximate position of the EC membrane boundary is shown as a red line, and bound ligands as magenta carbon atoms.
At the intracellular (IC) side, the distances between the helical tips of GCGR are within the same range as those in class A structures, except for an extensive inward shift of the IC tip of helix VII (Supplementary Fig. 5). While the inward shift in the IC part of helix VII is a hallmark of class A receptor activation3, it is not yet clear what role the IC region of helix VII plays in GCGR. The receptor lacks a proline kink in helix VII, which is a part of the conserved NP (BW 7.50) xxY motif in class A GPCRs24; instead, helix VII of GCGR has a glycine residue (Gly3937.50) that allows for a helical bend in this region. This glycine in helix VII of GCGR is part of the FQG7.50xxVxxxY7.57CF motif that is fully conserved in class B receptors (Fig. 3a). This Gly3937.50 induced bend is stabilized by hydrophobic interactions with Phe1842.57 of helix II in GCGR (Fig. 3a).
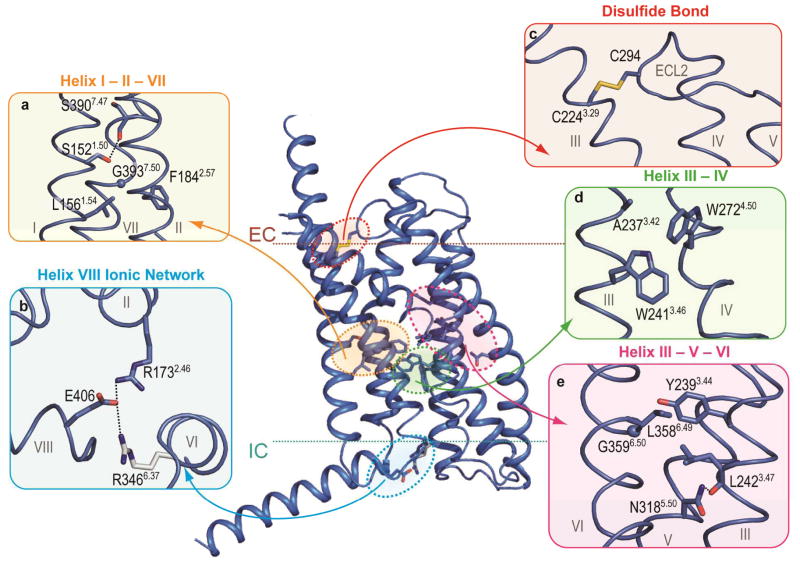
Figure 3. Structural features of class B GPCRs.
Comparison of GCGR and class A GPCRs crystal structures indicates distinct and conserved features. a, d, and e, The homologous GCGR residues involved in helix I – II, III – IV, and III –VI interface interactions as discussed for class A receptors by Venkatakrishnan et al25, and class B GPCR specific residues that mediate helix I – VII, II – VII, and III – V interface interactions. b, GCGR residues Glu406 of helix VIII, Arg1732.46, and Arg3466.37 form a class B receptor specific ionic network. Arg3466.37 (grey) has a weak electron density. c, The disulfide bond between Cys2243.29 and Cys294ECL2 in the GCGR structure, a conserved feature between classes A and B receptors. Hydrogen bond interactions are indicated by black dashed lines. Electron density maps for residues in this figure are shown in Supplementary Fig. 10.
The GCGR structure also includes an IC helix VIII comprised of 20 residues at the C-terminal end of the receptor that tilts approximately 25° away from the membrane as compared with its consensus position in class A (Fig. 1). This tilt is likely a result of crystal packing interactions between molecules (Supplementary Fig. 4). With the packing artifact noted, Glu406 in helix VIII is fully conserved in class B and interacts with conserved residues Arg1732.46 and Arg3466.37, forming two interhelical salt bridges (Fig. 3b). Though the tilt of helix VIII may alter interactions in the region, modeling of this region with helix VIII parallel to the membrane suggests that the Glu406 salt bridges are preserved in this conformation. The salt bridges formed by these residues are likely to be a distinct feature of class B receptors because there is no strong conservation among these residues in class A.
The GCGR 7TM structure also reveals several structural features that are conserved between class A and B receptors. One such feature is a disulfide bond between Cys294 in ECL2 and Cys2243.29 (BW 3.25, Supplementary Fig. 6), which apparently stabilizes the receptor’s 7TM fold (Fig. 3c). Another conserved feature of a common GPCR fold25 involves similar regions of contacts between helices I-II, I-VII, III-IV, and III-VI in class A and B GPCRs. The two GPCR classes, however, contain different patterns of conserved residues in these positions (Fig. 3, Supplementary Fig. 6). While in class B GPCRs the helix I-II interaction is stabilized by conserved hydrophobic residues Leu1561.54 and Phe1842.57, class A GPCRs contain conserved polar residues Asn (BW 1.50) and Asp (BW 2.50) in this region25. At the helix I-VII interface, Ser1521.50 forms a hydrogen bond with the backbone of Ser3907.47 (Fig. 3a). Mutation of homologous glucagon-like peptide-1 receptor (GLP1R) residues Ser1551.50 and Ser3927.47 reduces expression and alters receptor signaling18. At the GCGR helix III-IV interface (Fig. 3d), the conserved residue Trp2724.50 interacts with Trp2413.46, while in class A structures the Trp residue in helix IV (BW 4.50) interacts with the residue at BW17 position 3.38 in helix III (homologous GCGR residue Ala2373.42) (ref 24,25). The helix III-VI interface (Fig. 3e) in class B GPCRs contains conserved hydrophobic residues Tyr(/Phe)2393.44 and Leu(/Phe)3586.49 which make similar hydrophobic interactions as structurally aligned Ile/Val/Leu (BW 3.40) and Phe (BW 6.44) residues present in most class A GPCRs25 (Supplementary Fig. 6). This interface is further stabilized in class B GPCRs by close contact between the conserved Tyr2393.44 and Gly3596.50. Another class B GPCR specific interhelical hydrogen bond is formed between the conserved Asn3185.50 and the backbone of Leu2423.47 at the helix III-V interface (Fig. 3e).
Recognition between GCGR and glucagon
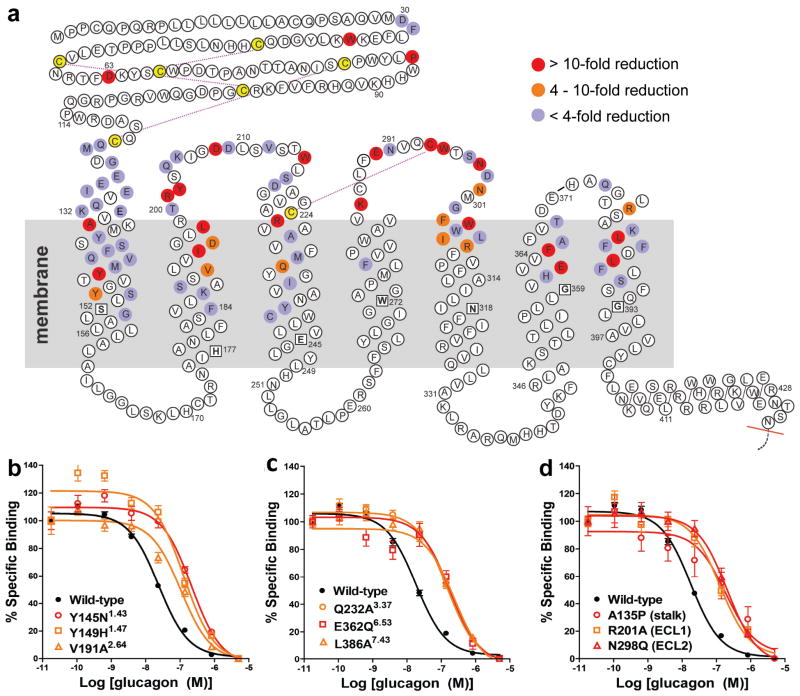
To better understand GCGR-glucagon interactions, we performed a comprehensive mutagenesis and glucagon binding study of GCGR at 90 different residue positions (Fig. 4, Supplementary Table 5). A total of 129 mutants were tested, and of these, 110 covering 85 different positions had expression levels greater than 30% of wild-type GCGR. Of them, 44 mutations covering 31 different positions in the GCGR 7TM domain had more than 4-fold reduction in glucagon binding (IC50 values) relative to wild-type GCGR. The results of these GCGR mutation studies were mapped onto the crystal structure of the GCGR 7TM domain (Fig. 5). Most of the residues that play an important role in glucagon binding face the main cavity in the 7TM core, and form a binding site that covers parts of ECL1, ECL2, ECL3, and helices I, II, III, V, VI, and VII, and extends deep into the 7TM cavity.
Figure 4. Effects of mutation studies in GCGR snake plot.
a, Mutated residues that show < 4 fold (purple), 4–10 fold (orange), and > 10 fold (red) changes of IC50 values for glucagon binding with receptor expression >30% of wild-type (Supplementary Table 5). Mutation studies to investigate peptide ligand binding have been previously reported for several class B GPCRs including GCGR6,19,21,26,30,31,34, GLP1R6,8,18,20,28,36, GIPR9,37, rSCTR11,29,35,38,43, and VPAC1(ref 39–41) (Supplementary Table 6). The most conserved residues in helices I to VII of class B GPCRs18 are boxed and bolded. b, c, and d, Representative binding curves of GCGR mutants with glucagon. Data are expressed as a percentage of specific 125I-glucagon binding in the absence of unlabeled peptide. Each point (± S.E.M.) represents the mean value of three independent experiments done in triplicate (IC50 are shown in Supplementary Table 5).
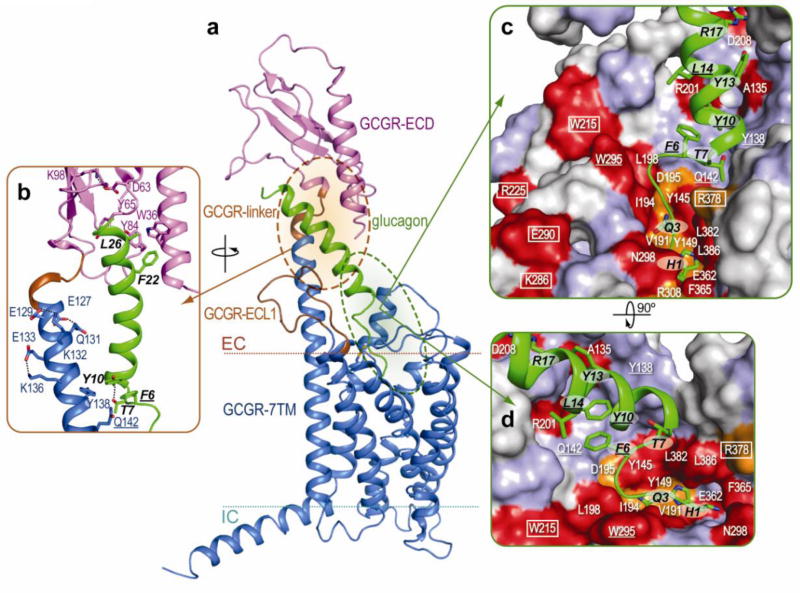
Figure 5. Model of GCGR bound to glucagon.
a, and b, GCGR with the ECD (magenta) and 7TM domain (blue) bound to glucagon (green). Residues 122–126 and 199–218 (brown) are not defined in the GCGR ECD (GCGR-linker) (PDB 4ERS) and 7TM domain (ECL1) crystal structures, respectively. The GCGR ECD structure and the interactions between GCGR ECD and glucagon resemble those in the GCGR ECD (PDB 4ERS)6 and GLP1-GLP1R ECD complex (PDB 3IOL)8 structures, respectively. c, and d, The effects of mutation studies of individual GCGR residues on glucagon (green) binding mapped onto the GCGR binding surface using the color coding presented in Figure 4. Important glucagon residues are labeled black. GCGR residues proposed to be important in stabilizing extracellular loops are boxed. GCGR-glucagon residue pairs that are homologous to residue pairs identified in GLP1R-GLP1 cross-linking studies are underlined12.
To investigate the recognition between glucagon and GCGR, we built a glucagon-bound GCGR structure model, based on the GCGR 7TM domain crystal structure, the GCGR ECD structure (PDB accession: 4ERS)6, and the ECD structure of the GCGR homolog GLP1R bound to the GLP1 peptide (PDB 3IOL)8 (Fig. 5a). The model further included several experimentally supported distance restraints between GCGR and glucagon based on photo-crosslinking studies between GLP1R and GLP1 (ref 12).
The predicted binding mode of glucagon to the ECD of GCGR (Fig. 5b) is in line with the current (Fig. 4, Supplementary Table 5) and previously reported mutation studies on GCGR6,26 and GLP1R8. Figure 5b shows how GCGR residues Asp63, Tyr65, and Lys98 function in stabilizing the ECD as observed in the GCGR ECD crystal structure6 and supported by mutagenesis studies6,26 (Fig. 4, Supplementary Table 5). The Trp36 side chain is an important hydrophobic interaction site for the C-terminal region of glucagon, similar to Trp39 in the GLP1R-GLP1 ECD crystal structure8. The stalk, observed in helix I of the GCGR 7TM crystal structure, links the ECD and 7TM domain in the model (Figs. 5a, b). The α-helical conformation of the stalk is supported by intrahelical interactions in the crystal structure (Glu133–Lys136) and model (Glu127–Gln131 and Glu129–Lys132), and is likely to be further stabilized by interactions with the extended ECL1 and the α-helical portion of glucagon. The potential function of the α-helical stalk in glucagon binding is supported by a reduced glucagon affinity to the Ala135Pro mutant (Fig. 4d, Supplementary Table 5), which likely distorts the α-helical conformation of the stalk region. The stalk may also function as part of the binding site for the previously proposed middle hinge region of glucagon27. Interestingly, an α-helical conformation in this region of GLP1R in complex with the GLP1 peptide was recently proposed based on crosslinking data between receptor and peptide residues12. In the GCGR-glucagon model, the corresponding pairs of residues, F6–Gln1421.40 and Y10–Tyr1381.36 (one letter amino acid abbreviation is used to designate glucagon residues), are located in close proximity and point towards each other, supporting a similar interaction mode as proposed in the GLP1R-GLP1 complex12 (Fig. 5b). The 12Å distance between L14–Trp295ECL2 in the GCGR-glucagon model exceeds the range of crosslinking distances in previous GLP1R-GLP1 models (8–9Å)12, though this may reflect differences between GCGR and GLP1R ligand binding modes.
There is no clear consensus on the binding site location of peptide ligands in the 7TM domain of class B GPCRs, which has been associated either with the extracellular loop regions4,7, or with a pocket in the 7TM domain11,12,28. The GCGR-glucagon model illustrates a way to account for the extensive interactions of the peptide with extracellular loops, as well as residues deep in the 7TM domain (Fig. 5c,d). First of all, the GCGR crystal structure reveals that some of the binding site residues previously positioned at the top of 7TM helices or in extracellular loops29–31 are in fact located deeper in the 7TM domain. Second, an extended flexible conformation of the first five residues allows glucagon to reach deep into the pocket. Our model of glucagon incorporates a hypothetical N-capping conformation32 of the peptide helix in residues F6-T7-Y10, similar to the one observed in the receptor bound pituitary adenylate cyclase activating polypeptide (PACAP)33, though other conformations of this region are possible.
Most interactions predicted by the GCGR-glucagon binding model are supported by mutagenesis (Figs. 4, 5c,d, Supplementary Table 5) and photo-cross-linking studies on GCGR6,19,21,26,30,31 and other class B GPCRs8–20,26,28,29,31,34–36. In the 7TM domain, many residues predicted to interact with glucagon show dramatic effects on glucagon binding without reducing receptor expression (Fig. 4, Supplementary Table 5). Figure 5 shows how these mutations line the 7TM binding site in the GCGR-glucagon model and include residues that are located deep in the pocket (Tyr1491.47, Val1912.64, Gln2323.37, Glu3626.53 and Leu3867.43). These results strongly support extension of the N-terminus of glucagon deep into the GCGR pocket, a region that could be equally important for ligand binding as in class A GPCRs.
In the loop region, residues Arg201, Tyr202, Asp208, and Trp215 of GCGR either stabilize the ECL1 conformation and/or directly interact with glucagon (Fig. 5c,d). The GCGR-glucagon binding model further suggests that residues Trp295 and Asn298 directly interact with glucagon, as mutation of these ECL2 residues strongly affects ligand binding. Although mutations of Asp218 (ref 21), Cys224 (ref 21, 26), Arg225, Lys286, Glu290, and Cys294 (Fig. 4, Supplementary Table 5) also affect ligand binding, these residues do not directly interact with glucagon in the model, but can play a role in stabilizing the loop conformation compatible with glucagon binding. For example, ECL2 and ECL1 are stabilized by a disulfide bridge between Cys294 and Cys2243.29, and potential salt bridges between Lys286 and Glu290, and between Asp218 and Arg225, respectively. Similarly, Arg378 is proposed to play a role in glucagon binding indirectly by stabilizing the ECL3 conformation, while Trp3045.36 stabilizes ECL2 at the interface between helices V and VI.
The GCGR-glucagon model presented in Figure 5 is based on crystallographic evidence and is consistent with the results of extensive mutation binding studies (Fig. 4, Supplementary Table 5)6,19,21,26,30,31,34; and thus provides insight into recognition between ligand and receptor. The hypothetical model of the complex can provide a useful platform for the design of biochemical and biophysical experiments detailing the complex structure, as well as the design of stabilized constructs that may lead to solution of the full-length receptor-ligand complex.
Related class B GPCRs
The GCGR-glucagon model can be informative for understanding common features that determine ligand recognition of other class B receptors. The GCGR mutation data (Fig. 4, Supplementary Table 5, and previous studies6,19,21,26,30,31,34 are paralleled by mutagenesis of homologous residues in other class B GPCRs. Supplementary Table 6 shows an overview of 274 previously reported mutants of GCGR6,19,21,26,30,31,34, GLP1R6,8,18,20,28,36, glucose-dependent insulinotropic polypeptide receptor (GIPR)9,37, rat secretin receptor (rSCTR)11,29,35,38, and vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide receptor 1 (VPAC1) (ref 39–41). For example, mutations of other class B GPCRs in residues that align to GCGR residues Tyr65ECD (8), Tyr84ECD (8), Leu85ECD (8), Tyr1451.43 (35), Tyr1491.47 (28,35), Lys1872.60 (9,18,20,28,29,39), Ile1942.67 (9,18,20,28,29,39), Asp1952.68 (11,20,28,29), Leu1982.71 (11,21), Arg2253.30 (20), Gln2323.37 (9,28), Lys2864.64 (36), Glu290ECL2 (36), Trp295ECL2 (11,36), Asn298ECL2 (11,36), Phe3656.56 (9,11), and Leu3867.43 (40) have been shown to affect peptide ligand binding and/or potency, supporting the GCGR-glucagon model in Figure 5. Interestingly, the GCGR-glucagon model demonstrates that residues which have been identified to interact with the homologous residues Q3 of glucagon, D3 of secretin, and D3 of vasoactive intestinal polypeptide are located within the same vicinity in the 7TM domain of GCGR (Lys1872.60 and Ile1942.67) (ref 30, 31), rSCTR (Tyr1281.47, Arg1662.60, Lys1732.67, and Asp1742.68) (ref 29,35), and VPAC1 (Arg1882.60 and Lys1952.67) (ref 39), respectively (Supplementary Table 6).
The distinct structural features and larger binding pocket of the GCGR 7TM domain provide new insights into the molecular details of peptide ligand binding and a more reliable structural template for the design of specific and potent small molecules for the treatment of type 2 diabetes. Moreover, the apparent overlap of class B GPCR binding sites suggests that, despite possible structural differences between class B GPCRs, the GCGR crystal structure might offer new opportunities to construct structural models to describe interactions between peptide ligands and other class B GPCRs. This is particularly exciting for those receptors involved in glucose regulation, including GLP1R and GIPR.
Materials and Methods
BRIL-GCGR-ΔECD-ΔC construct design and Sf9 expression
The human wild-type GCGR DNA was synthesized by DNA 2.0 and codon optimized for expression in Spodoptera frugiperda (Sf9) insect cells. The BRIL-GCGR-ΔECD-ΔC fusion construct was made by deleting N-terminal residues 1–122, fusing the thermally stabilized apocytochrome b562RIL (M7W, H102I, R106L) (referred to as BRIL) from E. coli at residue 123 (ref 16), and truncating the C-terminus at residue 432 to create the final construct for crystallization (Supplementary Fig. 2). This chimeric construct was obtained after screening 60 constructs of different BRIL junction and C-terminal truncation sites to generate crystals with diffraction data of the highest quality and resolution. The construct was cloned into a modified pFastBac1 vector (Invitrogen) containing an expression cassette with haemagglutinin signal sequence at the N-terminus, and a PreScission protease site, 10× His, and Flag tag at the C-terminus. The BRIL-GCGR-ΔECD-ΔC fusion construct was expressed in Sf9 cells using the Bac-to-Bac Baculovirus Expression System as described previously44. Sf9 cells at density of 2–3 × 106 cells ml−1 were infected with P1 or P2 virus at a multiplicity of infection (m.o.i.) of 7.5. Cells were harvested less than 48 h post-infection and cell pellets were stored at −80 °C until use.
BRIL-GCGR-ΔECD-ΔC fusion construct purification
Sf9 membranes were prepared with 1 wash cycle of hypotonic buffer (25 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl) in the presence of EDTA-free protease inhibitor cocktail tablets (Roche) and 4 wash cycles of high-salt buffer (25 mM HEPES pH 7.5, 1 M NaCl, 10 mM MgCl2, 20 mM KCl). Membrane pellets were homogenized in storage buffer (25 mM HEPES pH 7.5, 500 mM NaCl, 40% glycerol), flash frozen in liquid nitrogen, and stored at −80 °C until use.
The GCGR antagonist ligand NNC0640 (Supplementary Fig. 1b) was essential for purification and crystallization of the BRIL-GCGR-ΔECD-ΔC fusion construct. Two grams of washed membranes containing the BRIL-GCGR-ΔECD-ΔC fusion construct were incubated with 270 μM of compound NNC0640 for 30 min at room temperature. The receptor was solubilized with 1/0.1% (w/v) of n-dodecyl-β-D-maltopyranoside (Anatrace)/cholesteryl hemisuccinate (Sigma) (DDM/CHS) for 2 h at 4 °C. The insoluble material was pelleted by ultracentrifugation in a Ti70 rotor at 70,000 rpm for 30 min at 4 °C. The NaCl and DDM/CHS concentrations of the supernatant were adjusted to 800 mM and 0.5/0.05%, respectively, by adding equal volume of talon binding buffer (25mM HEPES pH 7.0, 1.475 M NaCl, 10% glycerol). Protein was bound to 2 ml of talon superflow resin slurry (Clontech) overnight at 4 °C on a rotator in the presence of 15 mM imidazole pH 7.5 and 100 μM NNC0640.
The talon resin was washed with 10× bed volume of wash buffer 1 (25 mM HEPES pH 7.0, 800 mM NaCl, 10% glycerol, 0.04/0.008% DDM/CHS, 30 μM NNC0640, 40 mM imidazole pH 7.5). Detergent concentration was lowered by washing the resin with 20× bed volume of wash buffer 2 (25 mM HEPES pH 7.0, 500 mM NaCl, 10% glycerol, 0.02/0.004% DDM/CHS, 30 μM NNC0640). The BRIL-GCGR-ΔECD-ΔC fusion construct was eluted with 2.5 ml of elution buffer (25 mM HEPES pH 7.0, 150 mM NaCl, 10% glycerol, 0.02/0.004% DDM/CHS, 30 μM NNC0640, 300 mM imidazole pH 7.5). The eluted BRIL-GCGR-ΔECD-ΔC fusion construct was desalted with PD-10 desalting column (GE Healthcare Life Sciences) to remove the imidazole. His-tagged PreScission protease was added to the samples and incubated overnight at 4 °C to remove the C-terminal 10× His and Flag tags. Reverse talon purification was performed to isolate the cleaved BRIL-GCGR-ΔECD-ΔC fusion construct by flowing the sample through 200 μL talon superflow resin twice. The flow-through material, containing the cleaved BRIL-GCGR-ΔECD-ΔC fusion construct, was concentrated to 80 mg/ml using a Vivaspin centrifuge concentrator (GE Healthcare) with a 100,000 kDa molecular weight cut-off.
BRIL-GCGR-ΔECD-ΔC fusion construct lipidic cubic phase (LCP) crystallization
For LCP crystallization, the BRIL-GCGR-ΔECD-ΔC construct was mixed with molten lipid at a ratio of 40/60 % (v/v) using a mechanical syringe mixer42. Due to the high detergent concentration, 10–15% (volume of LCP) of 5 M NaCl was added after the lipid and protein were mixed to convert a destabilized lipidic mesophase into LCP45. The host lipid for the LCP reconstitution was monoolein (Sigma) with 10% (w/w) of cholesterol (AvantiPolar Lipids). Crystallization trials were setup as previously described46. LCP-FRAP was used to identify initial crystallization conditions that led to GCGR crystals47. Crystals were obtained at 20 °C in 100 mM MES pH 6.0, 140–200 mM Na/K tartrate tetrahydrate, 9–17% (v/v) PEG 400, 0.35–0.55% (v/v) Jeffamine M-600 pH 7.0, 200 μM NNC0640; grown to a final size of 50–100 μm in the longest dimension in about 5 d (Supplementary Fig. 3); and harvested from the LCP matrix using 50 μm MiTeGen micromounts and immediately flash frozen in liquid nitrogen.
Data collection and processing
X-ray data were collected at the 23ID-D beamline (GM/CA CAT) at the Advanced Photon Source (Argonne, IL) using a 20 μm minibeam at a wavelength of 1.0330 Å and a MarMosaic 300 CCD detector. Crystals were aligned and data collected using a strategy similar to other GPCR structures48. Typically, 10–15 frames at 1° oscillation and 1–2 s exposure with non-attenuated beam were collected per crystal due to the fast onset of radiation damage. A 93.9% complete at 3.3 Å data set was obtained by indexing, integrating, scaling, and merging data sets from 14 crystals using HKL2000 (ref 49). Analysis of the final data set by the UCLA diffraction anisotropy server, (http://www.doe.mbi.ucla.edu/~sawaya/anisoscale/), indicated that diffraction along the b* axis was weaker than in the other two directions, therefore, reflections were subjected to a mild anisotropic truncation with resolution limits of 3.3, 3.4, and 3.3 Å along a*, b* and c*, respectively, before using them in the refinement.
Experimental phasing
Initial attempts to find a molecular replacement solution using previous class A GPCR structures as search models in Phaser50 did not generate any reliable solutions. Therefore, experimental phasing for the BRIL-GCGR-ΔECD-ΔC fusion construct was attempted by soaking the crystals with different heavy atoms. After screening many crystals, a (single wavelength anomalous dispersion) SAD data set was obtained from 1 crystal that was soaked overnight with 100 mM tantalum bromide (Ta6Br12) cluster (Jena Bioscience). The data were collected at 23ID-D beamline (GM/CA CAT) at the Advanced Photon Source using the peak wavelength from the tantalum L3 edge (9.880 keV). A beam size of 10 μm with 5× attenuation with 1° oscillation and 1 s exposure per frame was used. A complete 360° data set was acquired from a single crystal by collecting wedges of 30° with direct and inverse beam and translating 6 times along the crystal length to expose a fresh portion of the crystal for each wedge. The SAD data set was integrated and scaled at 4 Å resolution using HKL2000. PHENIX.AutoSol51 was used to search for the heavy atom sites with anomalous signals at 6 Å resolution (Supplementary Fig. 7), yielding initial electron density maps.
Structure determination and refinement
While the experimental phasing work was underway, “mixed model” molecular replacement (MR) search templates were generated based on all known GPCR structures using PHENIX.ROSETTA52. The “mixed model” templates were then superimposed together and manually trimmed to remove structurally poorly conserved regions. Parallel MR searches with Phaser were then carried out using these GPCR models along with the high resolution BRIL structure (PDB 1M6T) as search models on a linux cluster53. The search template based on rhodopsin (PDB 2Z73) produced a potential solution (TFZ=8.5). This MR solution was validated by the experimental phasing maps (Supplementary Fig. 8), and by the appearance of density not present in the search model. The experimental SIRAS phases calculated from the heavy atom were good up to 6–7 Å. However, SIRAS phases did not improve the MR maps, and thus were not used in the final refinement.
All refinement was performed using the MR solution with rounds of REFMAC5 (ref 54), autoBUSTER55, and PHENIX.AutoBuild56, followed by manual examination and rebuilding of refined coordinates in COOT57 using both 2|FO| - |FC| and |FO| - |FC| maps, as well as omit maps calculated using the Bhat procedure58 (Supplementary Fig. 9). We state 3.4 Å as the overall effective resolution of this structure; however, data to 3.3 Å were included in refinement, which improved the R/Rfree statistics (Supplementary Table 2).
At 3.4 Å resolution, the electron densities for the majority of residues in the GCGR 7TM structure are visible, except for residues Arg201–Trp215 (corresponding to ECL1), and therefore these residues were not built into the GCGR 7TM structure. Residues Gly269–Met276, Thr296–Asp299, Ile315–Ile317, and Phe365–Glu371 were built into the GCGR structure, but they contained breaks in the electron densities of the Cα backbone. Hence, other conformations are possible for these residues.
Although we do not observe density for NNC0640 in the canonical ligand binding pocket, this ligand is required to obtain diffraction quality crystals of the BRIL-GCGR-ΔECD-ΔC construct. There are two electron density blobs outside of canonical ligand binding pocket, one at the bottom of helix VI and VII near Lys3496.40, and the other at helix I near Trp1451.43. However, both of them are too small to accommodate NNC0640.
Energy-based conformational modeling of GCGR-glucagon complex
Glucagon was docked into the crystal structure of the GCGR ECD (PDB 4ERS, residues 28–123) (ref 6) using the crystal structure of the closely related GLP1R-GLP1 complex (PDB 3IOL)8 as a template. All molecular modeling and docking was performed using ICM molecular modeling software (Molsoft LLC)59. The initial α-helical conformation of glucagon peptide residues 11–29 was modeled based on GLP1 residues 17–35, and soft tethers between corresponding backbone Cα atoms of glucagon and GLP1 were applied. Conformation of the glucagon peptide and the interacting side chains in the ECD binding pocket were optimized (3 independent simulations of 106 steps) using ICM global optimization procedure in internal coordinates59,60 with improved conformational energy terms for protein and peptides4 and “tether weight” = 0.1.
The model of the ECD-glucagon complex was then docked and optimized with the crystal structure of the 7TM domain, completed with all side chains and ECL1. This flexible energy-based docking/optimization procedure involved all torsion coordinates in the regions, which are not defined by crystal structures, including protein backbone in the residues 1 to 10 of glucagon, and GCGR residues in ECL1 (199–218), and linker (122–126). In addition, side chain torsion variables were set free in all glucagon residues and the following regions of the GCGR model: helix I stalk region (125–136), ECL2 region (289–310), ECL3 (368–377), as well as 31 other residues lining the 7TM binding pocket.
The following three soft harmonic restraints derived from experimental crosslinking data in GLP1R and GLP1 (ref 12) were applied between glucagon and GCGR side chains to guide docking: F6(cb)–Gln1421.40(cd), Y10(cb)–Tyr1381.36(oh), L14(cb)–Trp295ECL2(ch2). Two intramolecular harmonic restraints were also applied to glucagon residues, T7(og1)–Y10(n) and F6(cz)-Y10(cz), to facilitate N-capped formation in glucagon, as suggested by previous comparative studies of class B peptide ligands32. Finally, a restraint was applied between the positively charged N-terminal amino group of glucagon and the carboxyl group of Glu3626.53. The importance of the carboxyl group of Glu3626.53, which is the only negatively charged residue in the 7TM binding pocket, is supported by GCGR (Supplementary Table 5) and GLP1R28 mutation studies (Supplementary Table 6). Since the N-terminus is the only basic moiety in the first 10 residues of glucagon, a potential Glu3626.53 salt bridge with the glucagon N-terminus is the most likely explanation for the mutation effects in Glu3626.53. A total of 164 torsion variables were systematically sampled with ICM Monte-Carlo global optimization, and 455 were locally minimized in the course of this procedure. The special “local” sampling option was applied to the ECL1 region backbone to allow efficient optimization. Three independent runs of the global optimization procedure (107 steps each) resulted in similar best energy conformations within 2.5 Å RMSD for the glucagon peptide non-hydrogen atoms.
It should be noted that in the absence of glucagon, the ECD is likely to be more flexible, sampling multiple orientations relative to the 7TM domain6. The model also does not attempt to infer a specific functional state of the receptor, partially because such state is not precisely defined for the 7TM crystal structure itself. For instance, NNC0640 used to stabilize the 7TM receptor fragment is a competitive antagonist to glucagon, which may have an effect on the crystallized conformation, even though NNC0640 is absent in the final structure. The accuracy of the GCGR-glucagon model may also be limited by the weak electron density of ECL2 and the top of helix V (residues 289-310), and the assumption that glucagon binds GCGR in an N-capped conformation7,32.
Glucagon binding to GCGR mutants
Construction of GCGR mutants and cell culture transfection
The cDNA encoding the human GCGR was originally obtained from GeneCopoeia Inc. (Rockville, MD) and cloned into the expression vector pcDNA3.1/V5-His-TOPO (Invitrogen) at the Hind III and EcoRI sites. The single and double mutants were constructed by PCR-based site directed mutagenesis. CHO-K1 cells were seeded onto 96-well poly-D-lysine treated cell culture plates (PerkinElmer, Waltham, MA) at a density of 2.7 × 104 per well. After overnight culture, the cells were transiently transfected with wild-type or mutant GCGR DNA using Lipofectamine 2000 transfection reagent (Invitrogen).
Whole-cell glucagon binding assay
Cells were harvested 24 h after transfections, washed twice, and incubated with blocking buffer (F12 supplemented with 33 mM HEPES pH 7.4 and 0.1% bovine serum albumin (BSA)) for 2 h at 37 °C. For homogeneous binding, the cells were incubated in binding buffer with constant concentration of 125I-glucagon (40 pM) and different concentrations of unlabeled glucagon (0.02 nM ~ 5 μM) at room temperature for 3 h. Cells were washed three times with ice-cold PBS and lysed by 50 μl lysis buffer (PBS supplemented with 20 mM Tris-HCl, 1% Triton X-100, pH 7.4). The plates were subsequently counted for radioactivity (counts per minute, CPM) in a scintillation counter (MicroBeta2 Plate Counter, PerkinElmer) using a scintillation cocktail (OptiPhase SuperMix, PerkinElmer). Specific binding was determined by subtracting non-specific binding observed in the presence of 5 μM unlabeled glucagon.
Expression level quantitation of constructed GCGR in cells by flow cytometry
Approximately 1 × 105 transfected CHO-K1 cells were blocked with PBS containing 5% BSA at room temperature for 15 min and then incubated with 1:100 diluted primary antibody (anti-GCGR, Epitomics, Burlingame, CA, USA) at room temperature for 1 h. The cells were then washed three times with PBS containing 1% BSA followed by a 1 h incubation with anti-rabbit Alexa-488-conjugated secondary antibody (1:300, Invitrogen) at 4 °C in the dark. After washes, the cells were resuspended in 200 μl of PBS containing 1% BSA for detection in a flow cytometer (Accuri™ C6, BD Biosciences, San Jose, CA) utilizing laser excitation and emission wavelengths of 488 and 519 nm, respectively. For each measurement, approximately 20,000 cellular events were collected and fluorescence intensity of positive expression cell population calculated.
NNC0640 binding assay (cell membrane based binding)
NNC0640 binding was analyzed using plasma membranes prepared from HEK293T cells transiently expressing GCGR constructs. Approximately 1.2 × 108 transfected HEK293T cells were harvested, suspended in 10 ml ice-cold membrane binding buffer (25 mM Tris-HCl, 0.1% BSA and 1 mM EDTA, pH 7.4) and centrifuged for 5 min at 200 g. The resulting pellet was resuspended in cold membrane binding buffer, pulled through a 25G×1″ needle four times and centrifuged for 5 min at 20,000 g. The precipitate containing the plasma membranes was suspended in membrane binding buffer containing protease inhibitor (Sigma-Aldrich, St. Louis, MO) and stored at −80 °C. Protein concentration was determined using a protein BCA assay kit (Pierce Biotechnology, Pittsburgh, PA).
For homogeneous binding, cell membrane homogenates (20 μg protein per well) were incubated in membrane binding buffer with constant concentration of 3H-NNC0640 (50 nM, labeled by PerkinElmer) and serial dilutions of unlabeled NNC0640 (1.26 nM~100 μM) at room temperature for 5 h. Nonspecific binding was determined in the presence of 100 μM NNC0640. Following incubation, the samples were filtered rapidly in vacuum through glass fiber filter plates (Millipore, Billerica, MA). After soaking and rinsing 4 times with ice-cold binding buffer, the filters were dried and counted for radioactivity in a scintillation counter (PerkinElmer).
Western blot
Protein samples were prepared as above, separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. After a 2 h incubation with blocking buffer, the membranes were incubated with 1:1000 primary antibody (anti-V5, Sigma) overnight. The membranes were then washed three times with TBS-T buffer (0.05M Tris, 0.15M NaCl, 0.1% (v/v) Tween) followed by a 2 h incubation with anti-mouse horseradish peroxidase-conjugated secondary antibody (1:1000, Cell Signaling Technology, Danvers, MA). The membranes were washed again and then detected with SuperSignal West Dura Substrate (ThermoScientific, Rockford, IL) according to the manufacturer’s instructions. Each membrane was exposed to X-ray film for detecting the blots. Bands were quantified with Quantity One Software (Bio-Rad Laboratories, Hercules, CA).
Statistical analysis
Results are presented as Means ± SEM. Changes in specific radiolabeled ligands binding and cell surface expression of GCGR constructs were normalized to those measured with wild-type GCGR control (100%). IC50 values in binding assay were determined by nonlinear regression analysis using the Prism 5 software (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
This work was supported by NIH Roadmap grant P50 GM073197 for technology development (V.C. and R.C.S.) and PSI: Biology grant U54 GM094618 for biological studies and structure production (target GPCR-49) (V.K., V.C. and R.C.S.); The Ministry of Health grants 2012ZX09304-011 and 2013ZX09507002 (M.-W.W.), Shanghai Science and Technology Development Fund 11DZ2292200 (M.-W.W.); Novo Nordisk-Chinese Academy of Sciences Research Fund NNCAS-2011-7 (M.-W.W.); Thousand Talent Program in China (R.C.S. and M.-W.W.); NIH Postdoctoral Training Grant (NRSA) F32 DK088392 (F.Y.S.); The Netherlands Organization for Scientific Research (NWO) through a VENI grant (Grant 700.59.408 to C.d.G.). We also thank V. Hruby and M. Cai for advice with the glucagon binding assay and general discussions; J. Velasquez for help on molecular biology; T. Trinh and M. Chu for help on baculovirus expression; K. Kadyshevskaya for assistance with figure preparation; X.Q. Cai, J. Wang, Y. Feng, A.T. Dai, Y. Zhou, J.J. Deng, Y.B. Dai, and J.W. Zhao for technical assistance in mutation studies; A. Walker for assistance with manuscript preparation; and J. Smith, R. Fischetti for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104.
Footnotes
Supplementary Information is linked to the online version of the paper at (web).
Author Contributions F.Y.S. designed, characterized, and screened constructs and ligands for crystallization. F.Y.S purified and crystallized the receptor in LCP, optimized crystallization conditions, grew crystals, collected diffraction data, and prepared manuscript. G.W.H. and Q.X. solved and refined the structure, and prepared the manuscript. V.C. collected and processed diffraction data, and prepared manuscript. M.H., D.Y., Z.Z., C.Z. and M.-W.W. expressed the receptor, and performed the mutagenesis and ligand-binding assay. V.K. and C.d.G. designed and analyzed the receptor mutagenesis studies, constructed the receptor-ligand model, and prepared the manuscript. D.W. and J.S.J. collected and processed SAD data and determined an initial electron density map from experimental phases. W.L. and V.C. trained and assisted in LCP crystallization. J.L. provided ligands for GCGR and prepared the manuscript. R.C.S., F.Y.S., M.-W.W., V.K., V.C., and C.d.G. were responsible for the overall project strategy and management and wrote the manuscript.
Author Information The coordinates and the structure factors have been deposited in the Protein Data Bank under the accession (4L6R). Reprints and permissions information is available at (web). The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at (web).
References
- 1.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 2.Cho YM, Merchant CE, Kieffer TJ. Targeting the glucagon receptor family for diabetes and obesity therapy. Pharm Ther. 2012;135:247–278. doi: 10.1016/j.pharmthera.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov Today. 2005;10:417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 5.Pal K, Melcher K, Xu HE. Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. Acta pharmacologica Sinica. 2012;33:300–311. doi: 10.1038/aps.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koth CM, et al. Molecular basis for negative regulation of the glucagon receptor. Proc Natl Acad Sci U S A. 2012;109:14393–14398. doi: 10.1073/pnas.1206734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parthier C, Reedtz-Runge S, Rudolph R, Stubbs MT. Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem Sci. 2009;34:303–310. doi: 10.1016/j.tibs.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Underwood CR, et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J Biol Chem. 2010;285:723–730. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yaqub T, et al. Identification of determinants of glucose-dependent insulinotropic polypeptide receptor that interact with N-terminal biologically active region of the natural ligand. Mol Pharmacol. 2010;77:547–558. doi: 10.1124/mol.109.060111. [DOI] [PubMed] [Google Scholar]
- 10.Miller LJ, Dong M, Harikumar KG, Gao F. Structural basis of natural ligand binding and activation of the Class II G-protein-coupled secretin receptor. Biochem Soc Trans. 2007;35:709–712. doi: 10.1042/BST0350709. [DOI] [PubMed] [Google Scholar]
- 11.Dong M, et al. Mapping spatial approximations between the amino terminus of secretin and each of the extracellular loops of its receptor using cysteine trapping. FASEB J. 2012;26:5092–5105. doi: 10.1096/fj.12-212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller LJ, et al. Refinement of glucagon-like peptide 1 docking to its intact receptor using mid-region photolabile probes and molecular modeling. J Biol Chem. 2011;286:15895–15907. doi: 10.1074/jbc.M110.217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong M, et al. Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore. J Biol Chem. 2011;286:23888–23899. doi: 10.1074/jbc.M111.245969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gensure RC, Shimizu N, Tsang J, Gardella TJ. Identification of a contact site for residue 19 of parathyroid hormone (PTH) and PTH-related protein analogs in transmembrane domain two of the type 1 PTH receptor. Mol Endocrinol. 2003;17:2647–2658. doi: 10.1210/me.2003-0275. [DOI] [PubMed] [Google Scholar]
- 15.de Graaf C, Rein C, Piwnica D, Giordanetto F, Rognan D. Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors. ChemMedChem. 2011;6:2159–2169. doi: 10.1002/cmdc.201100317. [DOI] [PubMed] [Google Scholar]
- 16.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of g protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros JA, Weinstein H. In: Methods in Neurosciences. Sealfon Stuart C, editor. Vol. 25. Academic Press; 1995. pp. 366–428. [Google Scholar]
- 18.Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc Natl Acad Sci U S A. 2013;110:5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unson CG, et al. Roles of specific extracellular domains of the glucagon receptor in ligand binding and signaling. Biochemistry. 2002;41:11795–11803. doi: 10.1021/bi025711j. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Q, Jeng W, Wheeler MB. Characterization of glucagon-like peptide-1 receptor-binding determinants. J Mol Endocrinol. 2000;25:321–335. doi: 10.1677/jme.0.0250321. JME00951 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Roberts DJ, Vertongen P, Waelbroeck M. Analysis of the glucagon receptor first extracellular loop by the substituted cysteine accessibility method. Peptides. 2011;32:1593–1599. doi: 10.1016/j.peptides.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 25.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 26.Prevost M, et al. Mutational and cysteine scanning analysis of the glucagon receptor N-terminal domain. J Biol Chem. 2010;285:30951–30958. doi: 10.1074/jbc.M110.102814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn JM, Medeiros M, Trivedi D, Hruby VJ. Development of potent truncated glucagon antagonists. J Med Chem. 2001;44:1372–1379. doi: 10.1021/jm000453e. [DOI] [PubMed] [Google Scholar]
- 28.Coopman K, et al. Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor. Mol Endocrinol. 2011;25:1804–1818. doi: 10.1210/me.2011-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Paolo E, et al. Contribution of the second transmembrane helix of the secretin receptor to the positioning of secretin. FEBS Lett. 1998;424:207–210. doi: 10.1016/s0014-5793(98)00175-6. [DOI] [PubMed] [Google Scholar]
- 30.Perret J, et al. Mutational analysis of the glucagon receptor: similarities with the vasoactive intestinal peptide (VIP)/pituitary adenylate cyclase-activating peptide (PACAP)/secretin receptors for recognition of the ligand’s third residue. Biochem J. 2002;362:389–394. doi: 10.1042/0264-6021:3620389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Runge S, et al. Three distinct epitopes on the extracellular face of the glucagon receptor determine specificity for the glucagon amino terminus. J Biol Chem. 2003;278:28005–28010. doi: 10.1074/jbc.M301085200. [DOI] [PubMed] [Google Scholar]
- 32.Neumann JM, et al. Class-B GPCR activation: is ligand helix-capping the key? Trends Biochem Sci. 2008;33:314–319. doi: 10.1016/j.tibs.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Inooka H, et al. Conformation of a peptide ligand bound to its G-protein coupled receptor. Nat Struct Biol. 2001;8:161–165. doi: 10.1038/84159. [DOI] [PubMed] [Google Scholar]
- 34.Cascieri MA, et al. Characterization of a novel, non-peptidyl antagonist of the human glucagon receptor. J Biol Chem. 1999;274:8694–8697. doi: 10.1074/jbc.274.13.8694. [DOI] [PubMed] [Google Scholar]
- 35.Di Paolo E, et al. Mutations of aromatic residues in the first transmembrane helix impair signalling by the secretin receptor. Receptors Channels. 1999;6:309–315. [PubMed] [Google Scholar]
- 36.Koole C, et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. J Biol Chem. 2012;287:3642–3658. doi: 10.1074/jbc.M111.309328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng CC, Lin L. A point mutation in the glucose-dependent insulinotropic peptide receptor confers constitutive activity. Biochem Biophys Res Commun. 1997;232:96–100. doi: 10.1006/bbrc.1997.6231. [DOI] [PubMed] [Google Scholar]
- 38.Ganguli SC, et al. Protean effects of a natural peptide agonist of the G protein-coupled secretin receptor demonstrated by receptor mutagenesis. J Pharmacol Exp Ther. 1998;286:593–598. [PubMed] [Google Scholar]
- 39.Solano RM, et al. Two basic residues of the h-VPAC1 receptor second transmembrane helix are essential for ligand binding and signal transduction. J Biol Chem. 2001;276:1084–1088. doi: 10.1074/jbc.M007696200. [DOI] [PubMed] [Google Scholar]
- 40.Ceraudo E, et al. Spatial proximity between the VPAC1 receptor and the amino terminus of agonist and antagonist peptides reveals distinct sites of interaction. FASEB J. 2012;26:2060–2071. doi: 10.1096/fj.11-196444. [DOI] [PubMed] [Google Scholar]
- 41.Tan YV, Couvineau A, Laburthe M. Diffuse pharmacophoric domains of vasoactive intestinal peptide (VIP) and further insights into the interaction of VIP with the N-terminal ectodomain of human VPAC1 receptor by photoaffinity labeling with [Bpa6]-VIP. J Biol Chem. 2004;279:38889–38894. doi: 10.1074/jbc.M404460200. [DOI] [PubMed] [Google Scholar]
- 42.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Paolo E, et al. Role of charged amino acids conserved in the vasoactive intestinal polypeptide/secretin family of receptors on the secretin receptor functionality. Peptides. 1999;20:1187–1193. doi: 10.1016/s0196-9781(99)00122-9. [DOI] [PubMed] [Google Scholar]
- 44.Hanson MA, et al. Profiling of membrane protein variants in a baculovirus system by coupling cell-surface detection with small-scale parallel expression. Protein Expr Purif. 2007;56:85–92. doi: 10.1016/j.pep.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misquitta Y, Caffrey M. Detergents destabilize the cubic phase of monoolein: implications for membrane protein crystallization. Biophys J. 2003;85:3084–3096. doi: 10.1016/S0006-3495(03)74727-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 47.Xu F, Liu W, Hanson MA, Stevens RC, Cherezov V. Development of an Automated High Throughput LCP-FRAP Assay to Guide Membrane Protein Crystallization in Lipid Mesophases. Crystal growth & design. 2011;11:1193–1201. doi: 10.1021/cg101385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J R Soc Interface. 2009;6(Suppl 5):S587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terwilliger TC, et al. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarzenbacher R, Godzik A, Jaroszewski L. The JCSG MR pipeline: optimized alignments, multiple models and parallel searches. Acta Crystallogr D Biol Crystallogr. 2008;64:133–140. doi: 10.1107/S0907444907050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.BUSTER v. 2.8.0. Global Phasing Ltd; Cambridge, U.K: 2009. [Google Scholar]
- 56.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D. 2008;64:61–69. doi: 10.1107/S090744490705024x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhat T. Calculation of an OMIT map. J Appl Crystallogr. 1988;21:279–281. doi: 10.1107/S0021889887012755. [DOI] [Google Scholar]
- 59.ICM Manual v. 3.0. MolSoft LLC; La Jolla, CA: 2012. [Google Scholar]
- 60.Arnautova YA, Abagyan RA, Totrov M. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling. Proteins. 2011;79:477–498. doi: 10.1002/prot.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic acids research. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White JF, et al. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Day JW, et al. Charge inversion at position 68 of the glucagon and glucagon-like peptide-1 receptors supports selectivity in hormone action. J Pept Sci. 2011;17:218–225. doi: 10.1002/psc.1317. [DOI] [PubMed] [Google Scholar]
- 66.Mathi SK, Chan Y, Li X, Wheeler MB. Scanning of the glucagon-like peptide-1 receptor localizes G protein-activating determinants primarily to the N terminus of the third intracellular loop. Mol Endocrinol. 1997;11:424–432. doi: 10.1210/mend.11.4.9913. [DOI] [PubMed] [Google Scholar]
- 67.Dong M, Pinon DI, Miller LJ. Site of action of a pentapeptide agonist at the glucagon-like peptide-1 receptor. Insight into a small molecule agonist-binding pocket. Bioorg Med Chem Lett. 2012;22:638–641. doi: 10.1016/j.bmcl.2011.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.