Abstract
Escherichia coli DnaK acts as a molecular chaperone through its ATP-regulated binding and release of polypeptide substrates. Overexpressing a C-terminal fragment (CTF) of DnaK (Gly-384 to Lys-638) containing the polypeptide substrate binding domain is lethal in wild-type E. coli. This dominant-negative phenotype may result from the nonproductive binding of CTF to cellular polypeptide targets of DnaK. Mutations affecting DnaK substrate binding were identified by selecting noncytotoxic CTF mutants followed by in vitro screening. The clustering of such mutations in the three-dimensional structure of CTF suggests the model that loops L1,2 and L4,5 form a rigid core structure critical for interactions with substrate.
Full text
PDF

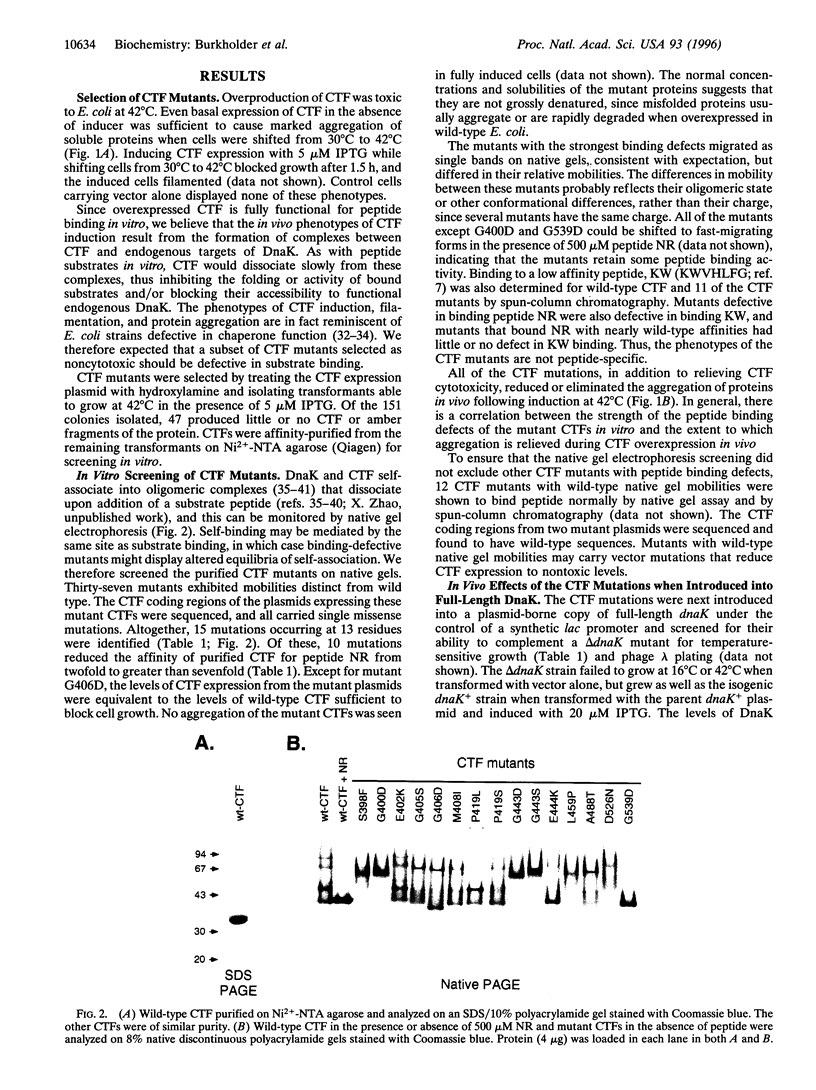
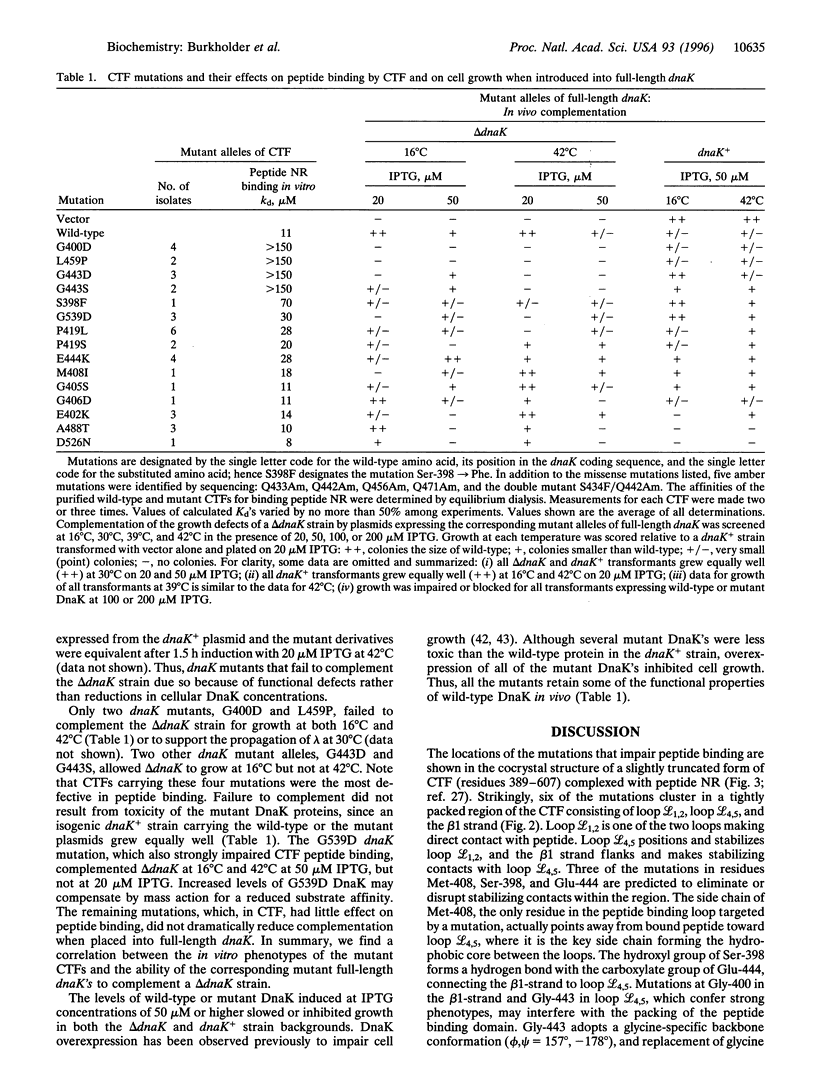


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banecki B., Zylicz M., Bertoli E., Tanfani F. Structural and functional relationships in DnaK and DnaK756 heat-shock proteins from Escherichia coli. J Biol Chem. 1992 Dec 15;267(35):25051–25058. [PubMed] [Google Scholar]
- Benaroudj N., Batelier G., Triniolles F., Ladjimi M. M. Self-association of the molecular chaperone HSC70. Biochemistry. 1995 Nov 21;34(46):15282–15290. doi: 10.1021/bi00046a037. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993 Nov 19;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S., Fourie A. M., Sambrook J. F., Gething M. J. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993 Jun 15;268(17):12730–12735. [PubMed] [Google Scholar]
- Blum P., Ory J., Bauernfeind J., Krska J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J Bacteriol. 1992 Nov;174(22):7436–7444. doi: 10.1128/jb.174.22.7436-7444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A., Theyssen H., Schröder H., McCarty J. S., Virgallita G., Milkereit P., Reinstein J., Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995 Jul 14;270(28):16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Buchberger A., Valencia A., McMacken R., Sander C., Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994 Apr 1;13(7):1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990 Dec;9(12):4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell T. G., Konforti B. B., Schmid S. L., Rothman J. E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987 Jan 15;262(2):746–751. [PubMed] [Google Scholar]
- Craig E. A., Gambill B. D., Nelson R. J. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev. 1993 Jun;57(2):402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn G. C., Pohl J., Flocco M. T., Rothman J. E. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991 Oct 24;353(6346):726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Freeman B. C., Myers M. P., Schumacher R., Morimoto R. I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995 May 15;14(10):2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiden P. J., Gaut J. R., Hendershot L. M. Interconversion of three differentially modified and assembled forms of BiP. EMBO J. 1992 Jan;11(1):63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gragerov A., Nudler E., Komissarova N., Gaitanaris G. A., Gottesman M. E., Nikiforov V. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M. E. Specificity of DnaK-peptide binding. J Mol Biol. 1994 Jan 21;235(3):848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- Greene L. E., Zinner R., Naficy S., Eisenberg E. Effect of nucleotide on the binding of peptides to 70-kDa heat shock protein. J Biol Chem. 1995 Feb 17;270(7):2967–2973. doi: 10.1074/jbc.270.7.2967. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hartl F. U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol Gen Genet. 1976 Apr 23;145(1):101–108. doi: 10.1007/BF00331564. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee Y. J., Corry P. M. Constitutive HSP70: oligomerization and its dependence on ATP binding. J Cell Physiol. 1992 Nov;153(2):353–361. doi: 10.1002/jcp.1041530215. [DOI] [PubMed] [Google Scholar]
- Landry S. J., Jordan R., McMacken R., Gierasch L. M. Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature. 1992 Jan 30;355(6359):455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- Liberek K., Skowyra D., Zylicz M., Johnson C., Georgopoulos C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem. 1991 Aug 5;266(22):14491–14496. [PubMed] [Google Scholar]
- McCarty J. S., Buchberger A., Reinstein J., Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995 May 26;249(1):126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- McCarty J. S., Rüdiger S., Schönfeld H. J., Schneider-Mergener J., Nakahigashi K., Yura T., Bukau B. Regulatory region C of the E. coli heat shock transcription factor, sigma32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996 Mar 15;256(5):829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- McCarty J. S., Walker G. C. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Paek K. H., Walker G. C. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987 Jan;169(1):283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleros D. R., Reid K. L., McCarty J. S., Walker G. C., Fink A. L. DnaK, hsp73, and their molten globules. Two different ways heat shock proteins respond to heat. J Biol Chem. 1992 Mar 15;267(8):5279–5285. [PubMed] [Google Scholar]
- Palleros D. R., Reid K. L., Shi L., Welch W. J., Fink A. L. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993 Oct 14;365(6447):664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- Palleros D. R., Shi L., Reid K. L., Fink A. L. hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J Biol Chem. 1994 May 6;269(18):13107–13114. [PubMed] [Google Scholar]
- Palleros D. R., Welch W. J., Fink A. L. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E., Moreno C., Young D. Mapping of Hsp70-binding sites on protein antigens. Eur J Biochem. 1994 May 15;222(1):65–73. doi: 10.1111/j.1432-1033.1994.tb18842.x. [DOI] [PubMed] [Google Scholar]
- Schmid D., Baici A., Gehring H., Christen P. Kinetics of molecular chaperone action. Science. 1994 Feb 18;263(5149):971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- Sharff A. J., Rodseth L. E., Szmelcman S., Hofnung M., Quiocho F. A. Refined structures of two insertion/deletion mutants probe function of the maltodextrin binding protein. J Mol Biol. 1995 Feb 10;246(1):8–13. doi: 10.1006/jmbi.1994.0059. [DOI] [PubMed] [Google Scholar]
- Shi L., Kataoka M., Fink A. L. Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry. 1996 Mar 12;35(10):3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- Szabo A., Langer T., Schröder H., Flanagan J., Bukau B., Hartl F. U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka I. M., Leung S. M., McAndrew S. J., Brown J. P., Hightower L. E. Hsc70-binding peptides selected from a phage display peptide library that resemble organellar targeting sequences. J Biol Chem. 1995 Aug 25;270(34):19839–19844. doi: 10.1074/jbc.270.34.19839. [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Wang C. Uncoupling of peptide-stimulated ATPase and clathrin-uncoating activity in deletion mutant of hsc70. J Biol Chem. 1994 Feb 25;269(8):5958–5962. [PubMed] [Google Scholar]
- Ueguchi C., Shiozawa T., Kakeda M., Yamada H., Mizuno T. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J Bacteriol. 1995 Jul;177(13):3894–3896. doi: 10.1128/jb.177.13.3894-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. F., Chang J. H., Wang C. Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J Biol Chem. 1993 Dec 15;268(35):26049–26051. [PubMed] [Google Scholar]
- Wawrzynów A., Zylicz M. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem. 1995 Aug 18;270(33):19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- Wilbanks S. M., Chen L., Tsuruta H., Hodgson K. O., McKay D. B. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995 Sep 26;34(38):12095–12106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996 Jun 14;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]