Abstract
As an important aspect of computer-aided drug design, structure-based drug design brought a new horizon to pharmaceutical development. This in silico method permeates all aspects of drug discovery today, including lead identification, lead optimization, ADMET prediction and drug repurposing. Structure-based drug design has resulted in fruitful successes drug discovery targeting protein-ligand and protein-protein interactions. Meanwhile, challenges, noted by low accuracy and combinatoric issues, may also cause failures. In this review, state-of-the-art techniques for protein modeling (e.g. structure prediction, modeling protein flexibility, etc.), hit identification/optimization (e.g. molecular docking, focused library design, fragment-based design, molecular dynamic, etc.), and polypharmacology design will be discussed. We will explore how structure-based techniques can facilitate the drug discovery process and interplay with other experimental approaches.
Keywords: Structure-based drug design, protein modeling, focused library design, pharmacophore, flexible docking, high-throughput virtual screening, de novo design, protein-protein interaction, polypharmacology
Modern computational-aided drug design established a novel platform by which researchers perform in-depth in silico simulation prior to labor-extensive wet-lab validation [1]. It comprises of two major parts corresponding to the information of molecular source it utilizes: structure-based (or receptor-based) drug design and ligand-based drug design. Structure-based drug design, which relies on the knowledge of biological target structures, aims to discover small molecules/peptides leads with desired chemistry properties, and orchestrate the following experimental validation and lead optimization. Structure-based approach provides mechanism-based basis, where potential ligands are excavated using receptor-dependent parameters, while ligand-based approaches bypass the consideration of complex biomolecular “black box” in a living cell. This in silico method permeates all aspects of drug discovery today [2], and we expect it will draw more attentions with the unprecedented advances of computational power and modeling accuracy in this decade.
In this review, we will overview the state-of-the-art structure-based drug discovery techniques ranging from receptor modeling to lead identification and optimization. In each topic, we will also highlight the fruitful successes as well as challenges that obstructs or cause the direct failure in both preclinical and clinical stages.
Protein modeling
The development of techniques for macromolecular determination and the wide availability of these structures in repositories such as the PDB have provided essential knowledge for drug design. Rapid advances in structural biology, e.g. macromolecular crystallography, NMR and Cryo-EM, consequence a boosting of structural data deposited in Protein Data Bank (PDB), which contains 72104 structures nowadays. However, there are many macromolecules which are not amenable to structure determination, or have incompletely determined structures. As an example, membrane proteins are an important class of macromolecules which serve as important drug targets, but present technical challenges for structure determination [3]. Moreover, many structures are not necessarily static as they are determined. It is becoming increasingly acknowledged that the native state of a protein is not a static structure, but consists of an ensemble of structures which interconvert in a dynamic equilibrium. This dynamic equilibrium has functional importance, participating in such processes as the diffusion of oxygen into the heme cavity of myoglobin, the diffusion of ions across ion channels, substrate binding, and ion channel gating [4]. Here, we first highlighted the contribution of in silico receptor modeling, a critical process that aims to ease one of the major bottlenecks of structure-based drug discovery: limited availability of experimental protein structures [5, 6]. Despite the lower resolution than experimental counterparts, using protein models as the starting point has already achieved great successes in current pharmaceutical industry. They predict three dimensional protein models either by using such comparative protein structure prediction approaches, such as homology modeling, or sequence information in conjunction with physical principles (ab initio modeling). Second, we concentrated on the methods to model flexible proteins.
Homology modeling
Homology modeling is an established and widely documented process [7], a method based on the assumptions that proteins that possess similar sequences share similar three-dimensional structures, and only a limited number of protein folds exist in nature [8, 9]. Homology modeling has been stated as the best structure prediction method of homologous protein so far, and it was widely used in structure-based drug discovery projects [10]. In brief, homology modeling consists of 1) suitable homologous template selection, optimal target-template sequence alignment, crude model building stage and 2) model evaluation, refinement and validation stage. Continual research is advancing methods available for each stage. Methods for sequence alignment and target identification include bioinformatics-based approaches (MUSCLE [11], DIALIGN [12, 13], MuSiC [14], MANGO [15]), machine learning methods (hidden Markov models [16, 17], neural networks [18], support vector machines [19]), and threading methods where target sequence is compared and matched against known three dimensional folds [20]. Model construction includes the techniques based on satisfaction of spatial restraints (Modeller [21]), Cα positions from conserved segments in template structure(s) [22, 23]. Highly critical loop modeling part remains ambiguous, despite the approach such as matching protein structure segments from PDB and using them as templates [24]. Conformational search of unmatched sequence segments using energy-based selection [25, 26] and ab initio modeling [27] has shown promising results. However, the imperfection of the constructed models may limit their applicability in structure-based design methods. Several methods have been reported to refine crude homology models or at least the side-chain conformations of the crucial binding pocket residues [28–34]. Interested readers are encouraged to refer [10] for a comprehensive list of homology modeling programs and servers.
In the field of drug discovery, modeling ligand-bound structure is more crucial than modeling apo structure because complex structure offers a more reliable starting point for later virtual screening. Ligand-supported/ligand-steered homology modeling method shows exciting prospects in reproducing active site by introducing “induced-fit” theory [29]. By extending the basis of homology modeling that evolutionary-related proteins share similar conformations, the hypothesis that pertinent proteins should also bind similar ligands which share a common core substructure has been widely accepted [35, 36]. James and Richard recently developed two automated protein-ligand homology modeling approaches, both of which used 3D-Coffee for template alignment, Modeller for modeling construction, Omega for ligand conformer generation and LigMatch/Rocs for ligand superposition, in order to tackle the modeling of homologous protein with distinct but related native ligands. According to their assessment, both hybrid-template strategy (exchange ligands first) and original-template strategy (exchange ligands after homology modeling) outperformed Fred docking in absence of the information of new ligand by 1–2Å RMSD [36]. Instead of inputting new ligand by structural alignment, docking-based global energy minimization via stochastic side chain movement, force fields or user-defined scoring function has become another refinement routine. This “ligand-centric” approach has been widely employed in identification of novel inhibitors against GPCR targets including MCH-R1, mGluR and CCR2 [29, 34, 37–39]. Other applications include protein structural analysis and function prediction [40–45], understanding of protein-ligand interactions [46–53] and compound optimization [54–56]. Successful stories for drug design starting from homology models are listed in Table 4.
Table 4.
Recent development in drug development starting from homology modeling
| Target | Indication | Inhibitor | Reference |
|---|---|---|---|
| Renin | Hypertension | Aliskiren (Tekturna®) | [235] |
| EtCRK2 | Avian coccidiosis | BES062021, BES143551, BES241415, BES252034 | [238] |
| Dynamin receptor | Epileptic seizures | Pthaladyn scaffold | [237] |
| hIKK-2 | Chronic inflammatory diseases | ZINC03871389 | [239] |
| Cdc25A | Cancer | 1-phenyl-2-(5-phenyl-4H-[1,2,4]triazol-3-ylsulfanyl)ethanone scaffold | [240] |
| α-glucosidase | Diabetes | 13 inhibitors (< IC50 50μM) | [241] |
| PH domain leucine-rich protein phosphatase | Diabetes, heart disease | NCS45586, NCS117079 | [242] |
| NF-κB inducing kinase | Rheumatoid arthritis | 4H-isoquinoline-1,3-dione scaffold | [243] |
| Matriptase-2 | Iron homeostasis, breast/prostate cancer | Three N-protected dipeptide amides conjugated to 4-amidinobenzylamide | [244] |
| Chemokine receptor -2 (CCR2) | Anti-inflammation | G365-0350 | [245] |
Ab initio modeling
Ab initio structure prediction, or de novo structure prediction, is an energy-based method that predicts the 3D structure of protein from sequence alone. Ab initio approach was conceptualized based on the assumption that protein of specific sequence folds to a native/native-like conformation or ensemble that is close to/at the global free-energy minimum [57]. This protein structure modeling approach provides flexibility as well as uncertainty in absence of template as reference. The power of ab initio approach has been significantly improved within this decade under the community-based assessments by Critical Assessment of Structure Prediction (CASP), a blind-test platform for the evaluation of structure prediction methods. While it has been widely used in fold recognition or small protein modeling, ab initio modeling of large protein, especially proteins with over 150 amino acids, remains hard to cope with due to the exponentially growing computational expenses and accumulated errors.
There are two long-standing challenges involved: scoring function in solvent and vast conformational space of searching [58]. These can be severely problematic when applied to proteins with biologically relevant length (>150 aa) [59]. Rosetta ab initio module is the most famous and cited program, which employed Monte Carlo annealing algorithm for structural sampling [60, 61]. However, one of the bottlenecks of Rosetta was the limitation in sampling critical “linchpin” feature, which is characterized by irregular edge of β-strand frequently occurred in functional regions, which was evaluation by Baker’s group [62]. To overcome the ruggedness of energy potential surface in traditional Monte Carlo annealing algorithm, several advanced generalized ensemble Monte Carlo methods were developed, which includes Temperautre Replica Exchange Monte Carlo (TREM) and Hamiltonian Replica Exchange Monte Carlo (HREM) [63, 64]. According to Alena’s assessment, HREM outperforms other algorithms in its wide range of energy landscape and prediction accuracy when using Rosetta full atom predictive scoring function [59]. As for scoring function, Rosetta full-atom knowledge-based force field has shown encouraging results in CASPs [60, 61]. In addition, SimFold (solvent induced multibody force field) considered solvent-induced effects by introducing terms like residue hydrophobicity and preference of phi/psi dihedral angles, which were defined distinctly in Rosetta. SimFold achieved identical success rate (12/38 within RMSD 6.5Å) as Rosetta, and well-performed in CASP6 (http://predictioncenter.org/casp6) [65]. Interestingly, both Rosetta and SimFold groups indicated a significant contribution of hydrophobic effects upon protein folding, which was related to the bad performances on β-strand-only proteins. Readers interested in de novo scoring functions are recommended to read Shing-Chung’s review [66].
Since ab initio approach is computational expensive, to develop better algorithms and to run these algorithms on faster computing architecture are urgent requirements. Recent development in high-performance computing, such as supercomputers (i.e. Blue Gene), distributed computing projects (i.e. Rosetta@home) and crowdsourcing (i.e. Foldit) may make this approach applicable and computational affordable in nowadays translational research. TOUCHSTONE II is similar to Rosetta in conformational sampling and energy function but uses a new lattice representation of protein structure [67]. Cα, Cβ and side-chain center of mass are used so that this program explicitly reduced the computational cost. However, the accuracy of TOUCHSTONE II is still limited by the size of protein, which is less than 200 amino acids [67]. In addition, a number of established webservers, such as Robetta [68], provided user-friendly testing platform that can help improve the structural prediction algorithm. Even though there are few cases of drug discovery based on ab initio models, the algorithms and scoring functions developed by such template-free methods have undoubtedly exhibited great potentials in systems biology [69] and template-based model refinement [70]. Another study carried out recently observed a significant improvement in threading approach through employing ab initio energy function, in which 68 out of 127 were greatly improved in Prosup benchmark [71]. Christopher and Yu reported a rational combination of Rosetta ab inito structure prediction with I-SITES library of sequence motifs and HMMSTR model. In brief, a fragment moveset was generated by I-SITES, and then submitted to three separate HMMSTR models, which predicted backbone angles, secondary fold and supersecondary structure, respectively. These restraints were later applied in Rosetta Monte Carlo Metropolis conformational sampling [72]. I-SITES-HMMSTR-Rosetta approach resulted in 61% topologically corrected residues among 31 proteins in CASP4. Finally, de novo scoring functions may provide insights to guide polypeptide and antibody engineering in the future.
Modeling flexible active site
One of the earliest, and simplest, models of ligand binding is the lock-and-key paradigm [73], which views ligand-receptor binding as a rather rigid shape-matching process, akin to assembling a jigsaw puzzle. Modern models, such as the induced fit or conformational selection models, generally include flexibility in both ligand and substrate molecules [74]. While it has become common sense that biological receptors may subject to conformational rearrangements upon ligand binding [75], to model the bound-complex (or holo form) from unbound-receptor (or apo form) still remains quite challenging for years. Two distinct strategies offer potential solutions to this issue: direct modeling holo receptor prior to docking or ligand-dependent flexible docking. Recent developments have provided insights to modeling protein mobility by introducing receptor rearrangements in process of or prior to virtual screening. Several reviews are available in the previous literature [76–78]. In this section, we will briefly introduce the rationale to fulfill receptor flexibility and its related application (Table 1).
Table 1.
Major methods for flexible active site modeling
| Methods | Level of flexibility | Algorithm optimization | Reference |
|---|---|---|---|
| oft potentials | Side-chain | Toleration of minor steric clashes | [79–82] |
| Rotamer libraries random sampling | Side-chain, backbone | Discrete rotamer libraries, Monte Carlo minimization | [80, 81, 83–85, 99] |
| MD ensemble sampling | Side-chain, backbone, domain | Normal mode (low mode), temperature replica exchange | [88–90, 93] |
| 4D docking | Side-chain, backbone | Flexibility as the 4th discrete dimension of docking grids | [96] |
| QSiCR | Side-chain, backbone | Knowledge-based machine learning | [97, 98] |
Since the majority of movement upon ligand binding involves side-chain and loop rearrangement, flexible docking is a cost-effective means to consider small degrees of receptor flexibility during docking. Minor side-chain movement in protein can be addressed by soft potentials, which put “softer” penalties on non-polar van der Waals clashes [79]. Due to its relative computational efficiency, soft potential options have been routinely added in a large variety of docking packages [80–82]. Library-based rotamer exploration is another knowledge-based approach to explore side-chain variations. This strategy is most widely utilized to treat moderate side-chain and minor backbone movement. The rationale of using rotamer library is to model continuous rotametric changes by discrete amino acid side-chain rotamer libraries and circumvent computational-prohibitive exhaustive searching. Commercially available software, such as GOLD, ICM and Glide, implemented their own robust side-chain rotamer libraries. As for open-source ones, RosettaLigand developed by Backer’s lab aimed to save computational expenses by simultaneously searching protein side-chain, backbone and ligand rotamers using predetermined discrete rotamer libraries by Monte Carlo minimization [83, 84]. More recently, MedusaDock used stochastic rotamer library of ligands (STROLL) and was validated to sample ligand conformers in a more comprehensive fashion [85]. However, secondary structure rearrangements and domain movement still remain significantly challenging so far. As an efficient solution, normal-mode analysis based on protein elastic network model is able to estimate apo-holo transformation in which Cα movements are involved [86]. Still, it should be aware that flexible docking may offers both risks and prosperities in structure-based drug discovery [87], and expert knowledge on receptor and docking parameters are the bottom requirement.
Molecular dynamics (MD) can be used in various ways to model receptor flexibility. One of the most straightforward ways is to simply perform a simulation of the receptor structure prior to docking, and form an ensemble of structures by taking snapshots of structures during the simulation. This is the ensemble docking-based approach carried out by Sivanesan et al., who generated a 51-member ensemble of structures by performing 3ns of MD simulations of human estrogen receptor α [88]. These structures were used for screening a 3500 compound library, successfully identifying a handful of promising compounds. To sample large protein rearrangement that leads to apo-holo transition, including secondary structure/domain movement, normal modes or low modes are frequently used to model concerted motions observed in molecular dynamics. For instance, “low mode”, employed by MacroModel (Schrödinger), helped to validate novel inhibitors against such targets as farnesyltransferase [89] and Galectin-3 [90]. A slightly more sophisticated use of molecular dynamics is the research by McCammon and coworkers, who have shown that incorporating a short molecular dynamics refinement step into an iterative docking method gives improved results and can find alternative docking poses which otherwise would be missed [91]. However, due to the computational expense of standard molecular dynamics, its practical use in high-throughput virtual screening is limited to detailed studies of a limited number of selected compounds, late in the screening pipeline [92]. This may change in the future with improvements in computing hardware.
Standard molecular dynamics simulations remain too computationally expensive for virtual screening against a large compound database, but modifications to molecular dynamics have been developed to increase its efficiency. One such technique is the temperature replica exchange method [93], in which several replicas of the system of interest are run in parallel using molecular dynamics at different temperatures. Periodic exchanges are performed between replicas such that the correct sampling is achieved at each temperature. This allows for a faster search of configurational space. The parallelization of this algorithm is well-suited to modern cluster computing architecture. Examples of the application of replica exchange molecular dynamics include the work by Gerek and Ozkan who, in a ligand-binding replica exchange simulation study of PDZ domains, used dihedral restraints on the receptor to induce a bias towards binding site conformations [94], further reducing the simulation time, while still allowing for binding site flexibility. Of course, replica exchange can also be used to efficiently sample ligand populations as well [95]. Conceptually similar but more computational efficient than ensemble docking, in which compounds were docked iteratively to multiple receptor conformers (MRC), Abagyan’s group developed ICM four-dimensional docking (4D docking) on the basis of the idea that receptor flexibility can be reflected as the 4th discrete dimension on the docking grids [96]. 4D docking approach was tested against 99 proteins and 300 diverse ligands and achieved comparable accuracy in the benchmark (77.3%), but 4-fold efficiency in ligand conformational sampling compared to traditional MRC docking [96]. This innovative methodology, combined with receptor conformer sampling, may elicit promising speed and accuracy in flexible docking study of unfamiliar targets.
Interestingly, a ligand-based strategy may ease the flexible docking against some well-defined targets. Quantitative structure-induced conformation relationship (QSiCR) model [97, 98], sharing similar concept to structure-activity relationship (QSAR), predicts the displacements of key residues by statistical analysis of ligand-encoded properties [78]. In both CDK2 [97] and p38 MAP kinase [98] systems, QSiCR models derived from 2D descriptors of co-crystallized ligands were found sufficient to predict the variation in active sites. Take CDK2 as an example, the trans side-chain position in Lys33 was often induced by bulky negatively charged group, whereas Lys89 side-chain conformation was determined by the hydrophobicity of ligand [97]. This multivariate statistical method circumvents the combinatoric issue possessing by library-based rotamer searching and MD approaches, but the predictive power of QSiCR for non-cognate molecule still remains to be tested.
Basic methods for protein inhibitor design
The spirit of structure-based drug identification and optimization is to identify biologically active compounds from “compound forests” with higher efficiency and hit enrichment by utilizing the information from protein structures. This can be realized through different strategies. First, lead compounds or scaffolds can be identified from diversified compound pool and accelerated screening, such as high-throughput virtual screening (HTVS). Second, instead of making random screening pool, we build up a focused library based on the knowledge of bio-target, such as structure-based library design followed by screening. Third, we rationally shrink the screening pool based on the activity-associated patterns, such as structure-based/ligand-based pharmacophore modeling. Fourth, “conceptualize” the active compounds and search inversely for the compounds that may exist or synthesizable in the nature, such as de novo design/fragment-based design. Finally, we can augment the hit enrichment by directly estimating the binding free energy in a cost effective manner. We will discuss each strategy by the following part in more detail.
Molecular docking-based virtual screening
Generally speaking, molecular docking is a computational approach that predicts the orientation of a small molecule (ligand) in stable complex with a macromolecular target (receptor) using specific scoring functions. It has become the basis of structure-based virtual screening and has shown to be influential in current pharmaceutical design which is more economic than experimental screening. Docking is overly a “standard” procedure when protein-ligand complex structure is available. The advantages of structure-based virtual screening are obvious: quick and minimal manual interventions. Docking-based virtual screening has played pronounced roles in the lead identification and optimization in the history.
As summarized by Elizabeth et al., the general issues in molecular docking that currently need to be addressed consist of: 1. bioinformatics and chemoinformatics of receptor and ligand representations (side-chain and backbone flexibility, protonation, structural water, tautomerism, rotamerism, efficiency of sampling, etc.) 2. sophisticated scoring function [100]. Besides, performing high-throughput virtual screening is faced with other critical obstructs such as combinatoric complexity and limited availability of computing resources. We made a list of popular docking software along with their respective algorithm in Table 2. Open-source software, such as Autodock (The Scripps Research Institute) [99], DOCK (UCSF) [101], RosettaLigand [83, 84] are widely used and exploited in several studies. The expandability of source code enables researches to understand, customize, improve and redistribute more robust docking packages [102, 103]. Commercial packages, such as Glide (Schrödinger) [81, 82], ICM (MolSoft LLC.) [104], GOLD (CCDC) [80], Surflex [105], FlexX (BioSolveIT) [106], Fred (OpenEye Inc.) [107], eHiTS (SimBioSys Inc.) [108] and LigandFit (Accelry Inc.) [109], are often considered more advanced. They usually outperform open-source competitors in atom typing, pocket representation, docking restraints setup, etc. Nonetheless, they are still far from perfection [110].
Table 2.
List of popular docking software and associated specifications
| Program | OSC | lFlex | pFlex | Parallel | Algorithm | SF | SF class | Owner |
|---|---|---|---|---|---|---|---|---|
| GOLD | √ | SP, RTML, MRC | CG | GA | GOLD_Fitness, Chemscore, | Empirical | CCDC | |
| ASP | Knowledge-based | |||||||
| eHiTS | √ | SP | CG, FG | Fragment | eHiTS SF | Knowledge-based | SimBioSys | |
| Surflex | √ | SP | CG, FG | Protomol | Surflex SF | Empirical | Tripos | |
| Glide | √ | SP, side-chain repackinga | CG | Energy funnel, MC | GlideScore | Empirical | Schrödinger | |
| Emodel | Empirical+Force field-based | |||||||
| LigandFit | √ | SP | CG, FG | MC | LigScore1/2 | Empirical | Accelry | |
| FlexX | √ | MRCb | CG | Fragment | FlexX score | Empirical | BioSolveIT | |
| ICM | √ | SP, MRC | CG | MC | ICM SF | Empirical | Molsoft | |
| ICM-PMF | Knowledge-based | |||||||
| Fred | CG | RB | Chemgauss3, Chemscore, OEChemscore | Empirical | OpenEye | |||
| Zapbindc | Force field-based | |||||||
| AutoDock | √ | √ | RTML | FG | GA, SA | AD4 | Empirical | Scripps |
| DOCK | √ | √ | Full-atom flexibility during AMBER score calculation | FG | Fragment | DOCK3.5 score, GB/SA score, PB/SA scorec, AMBER score | Force field-based | UCSF |
| RosettaLigand | √ | √ | Side-chain repacking | FG | MC | Rosetta energy function | Knowledge-based | University of Washington |
OSC=open source code; lFlex=ligand flexibility; pFlex=protein flexibility; SP=soft potentials; RTML=rotamer libraries; MRC=multiple receptor conformers; CG=coarse-grained parallelization; FG=fine-grained parallelization; SF=scoring function; GA=genetic algorithm; RB=rigid-body docking; SA=simulated annealling; Fragment=fragment-based algorithm; MC=Monte Carlo energy minimization; GB/SA=Generalized Boltzmann Surface Area; PB/SA=Poisson-Boltzmann Surface Area;
Prime (Schrödinger) is needed for induced fit docking.
FlexE (Tripos) is needed for MRC sampling.
Zap package (OpenEye) is needed for PB/SA calculation.
Here arises the paradox of using docking as an accelerator of drug discovery: predictor or liar?
Interestingly, although experimental high-throughput screening is known to considerably expensive and unreachable by most academic laboratories, the cost of establishment and maintenance of in silico virtual screening, including program licensing and purchase of high-performance computing facilities, are also considered significant [111]. It calls for the need to re-evaluate the docking accuracy before HTVS.
Conformational sampling and scoring are two sub-problems in docking programs, whereas accurate scoring remains more problematic than sampling [112]. A recent comprehensive comparison of 7 docking packages was carried out on a large dataset containing over 1300 protein-ligand complexes by Plewczynski et al. [113]. They proposed a general ranking regarding the pose prediction capability to be: GOLD ~ eHiTS > Surflex > Glide > LigandFit > FlexX ~ Autodock, yet the scoring functions for all 7 packages is less than satisfactory, with the best RMSD-rank correlations merely achieving 0.32 [113]. As to ranking and scoring decoys, stand-alone scoring packages, such as X-Score and DrugScoreCSD, are found to have better Spearman correlation around 0.66 based on another study using 195 diverse dataset [114]. According to another study carried out by Wang et al., 14 scoring functions were extensively evaluated on an 800 PBDbind database. X-Score, DrugScore, Sybyl::ChemScore and Cenius::PLP exhibited robustness based on consensus results [115]. Perola et al. found that GOLD and ICM are more binding site-dependent and docked poorer at the binding site where hydrophobic interactions are predominant, whereas Glide shares consistent performance across various pockets [110]. Rescoring poses by other knowledge-based scoring functions, such as ENTess which is based on interfacial atomic quadruplets and electronegativity, may offer solution to address this issue [116]. Moreover, the starting conformation of ligand may significantly impact the docking results in such software as Fred and AutoDock. However, since the full mastery of any docking package requires a high level of user intervention, these comparative studies may be only offered as a guideline for external users due to the differences in program settings and target characteristics. Stand-alone scoring function may also encounter overfitting issue. Therefore, to solve these behind-the-screen issues, one may need knowledge of the binding mode, multiple starting conformations, cross docking evaluation and consensus scoring.
Does docking-based virtual screening actually accelerate the drug discovery cycle? Honestly, yes, but only wisely programmed. Computer may also experience embarrassments as we human beings when faced with a screening task that involves millions of compounds. High-throughput technology has become a requirement for computer-aided drug discovery nowadays. Docking through parallelization or distributed computing experiences unprecedentedly advances than ever before. Parallel docking mode employed by such software as DOCK, GOLD and Glide has significantly enhanced the screening efficiency. As for open-source software, DOVIS, a HTVS program using AutoDock4 docking engine, enables researchers to perform massive virtual screening efficiently on Linux-based high performance clusters [102, 103]. Hyper-extensive file operation issue was optimized by automating load balancing. This software has been successfully employed during the development of PHT-427 which targets AKT1 pleckstrin homology (PH) domain [117, 118]. VSDocker, which is also relied on AutoDock engine, was the first high-performance virtual screening software under Microsoft Windows system [119]. In addition, several volunteer computing projects have been carried out recently that utilize unused computing resources to accelerate molecular discovery cycle [120]. For instance, developers of Rosetta Docking@home [121] and Docking@GRID aim to perform precise and effective docking; more importantly, they tried to explore the optimal searching pathway, maximize the simplicity of searching, and optimize distributed algorithms via metaheuristics methods or community sharing.
Structure-based ligand library design
Computational screening of large compound libraries have been characterized by shortfalls such as limited hit rates [122], poor biochemical quality of hits [123], high costs [124, 125] and quality of this process [126, 127]. One way of addressing the above mentioned shortcomings without compromising on success rates is the development of structure-based library design methods [128, 129]. Recent advances in the availability of protein structural information [130, 131], combinatorial chemistry [132] and computational methods (e.g. docking) have increased the feasibility of library design and enrichment methods. Fragment-based design and diversity oriented synthesis (DOS) [133] are two widely used approaches for library design and are discussed elsewhere [134–138]. On the other hand, ligand-based library design methods, e.g. using pharmacophores [139, 140] and QSAR [141], are also used and applied.
A protein-ligand complex structure serves as the foundation for subsequent structure-based library design. Protein-ligand interaction patterns (e.g. hydrogen bonds) are determined, and the compound core is retained. Next, either a library of compounds is generated by attaching relevant fragments satisfying the interaction patterns to the core, and then sequentially docked to identify top hits; or the core is placed in the binding pocket with fragments attached to generate target-specific library based on the docking scores. Docking programs e.g. CombiDock [142], DREAM++ [143], BUILDER v.2 [144], OptiDock [145], CombiGlide (Schrödinger), and CombiSMoG [146] have been specifically developed for structure-based library design. Here, we highlight a small set of applications for structure-based library design (Table 3).
Table 3.
List of structure-based library design method
| Library design method | Target: Disease | Reference |
|---|---|---|
| Optimized hit and scaffold based library design | NS5B Polymerase: Hepatitis C | [147] |
| Docking-based combinatorial library design | Soluble epoxide hydrolase: Cardiovascular disease | [148] |
| Anchor-based library tailoring approach | Receptor tyrosine kinase EphB4: Cancer | [150] |
| Combination of substructure-search and pharmacophore-based library design | Falcipain-2: Malaria | [151] |
For example, the complex structure consisting of a hit obtained from high-throughput screening and Hepatitis C NS5B polymerase was used to design a compound library for rational drug discovery [147]. The scaffold of this hit was used to intuitively design another compound to maximize interactions within the binding pocket while retaining a similar binding mode as the original hit. A focused library of 1175 compounds were docked and filtered based on the pharmacophore and revised scaffold, resulting in a 3μM hit amongst the 10 top ranking molecules. This compound laid the foundation of further focused library generation and low micromolar hit identification.
A group from Pfizer applied a structure-based library design methodology to identify hits for soluble epoxide hydrolase (sEH), a potential target against cardiovascular dysfunction and renal damage [148]. The crystal structure complex of sEH with a benzoxazole template compound was used to identify 2000 reagents. The compound library was docked and filtered using the presence of critical enzyme-compound hydrogen bonds in lieu of docking scores. 383 of the total of 591 compounds were synthesized. 102 compounds showed nanomolar activity (~26%) and 192 had submicromolar activity.
The hyperactivation of receptor tyrosine kinase EphB4 is responsible in many types of cancer [149]. Kolb et al. developed an anchor-based library tailoring (ALTA) approach to develop a structure-based focused library to identify inhibitors against EphB4 [150]. In brief, 35513 rigid fragments were identified from three commercial compound libraries (~782K). The fragments were rigidly docked against two homology models of EphB4 differing in the orientation of the hydroxyl group of Thr693, filtered by their ability to have a) a rigid moiety and b) hydrogen bonds with the residues Glu694 and Met696 in the hinge region, and finally ranked according to their binding energy. 21418 compounds containing the top ranked 1205 fragments were docked by keeping the fragment as an anchor in the two models and ranked using linear interaction energy with continuum electrostatics model. Top 40 hits were experimentally tested and 8 of them showed low micromolar activity in a florescence-based enzymatic assay.
Shah et al. developed a focused cysteine protease inhibitor library using a two-step process of substructure-search and pharmacophore based filter, prior to screening for anti-malarial inhibitors [151]. In step 1, compounds in 6 commercial libraries were shortlisted if they possessed soft electrophile substructures of α-heteroatom-substituted ketones and amides, azetidinones, α-keto amide, α-keto acid, and α-keto ester. A receptor-based pharmacophore was developed using the docked complex of the peptidic vinyl sulfone and Falcipain-2 (PDB ID: 2GHU), a related papain family cysteine protease. Two hydrophobic features corresponding to the S2 and S3 regions of the binding pocket, a hydrogen bond donor and hydrogen bond acceptor feature with Gly83 and Cys42 and Gln36 respectively formed the pharmacophore. Only those compounds from step 1 that satisfied three or four pharmacophore features were selected for the docking process. Subsequent docking, selection and experimental testing led to the discovery of 21 diverse non-peptidic inhibitors of FP-2 with a hit rate of 42%.
Structure-based pharmacophore modeling
A pharmacophore is defined as a spatial arrangement of functional groups and substructures common to active molecules and essential to biological activities [152]. The majority of pharmacophore models relied on the concept of “molecular similarity” of small molecules are derived from a series of active compounds and inactive ones [153]. Ligand-based 3D pharmacophore modeling, which is often used in the absence of target structure, has been reviewed in many publications [154–158]. On the other hand, various receptor-based pharmacophore approaches have been explored by probing predisposed interactions in the protein active site [159, 160]. The concept of structure-based pharmacophore has been widely introduced not only in drug discovery stage, but implemented in lead optimization and in silico ADME prediction, e.g. MetaSite [161], as well. The advantages of using pharmacophoric models over conventional docking scoring functions are that pharmacophore provides versatility for any type of protein of interest and options to accentuate target-specific interactions, such as π-orbital interactions and entropy effects [162].
LigandScout utilizes six types of chemical features and volume constraints to build pharmacophore models from ligand and its surrounding amino acids as the representation of protein-ligand interactions [152]. LigandScout was frequently used for in silico virtual screening against large-scale compound database [163]. This tool was first validated in context of human rhinovirus serotype 16 (HRV16) inhibitors [152]. Three dimensional pharmacophores were automatically created from three complex structures of HRV16, from which a common pharmacophore model was derived by applying the overlay algorithm of LigandScout. The combined HRV16 pharmacophore model was taken to search PDB and Maybridge database, which yielded four distinct hits, including such capsid-binding agents as pleconaril, all of which have been reported to be HRV16 hull protein binding agents [164]. More recent publications also implied the profound potentials of LigandScout in studying substrate binding specificity (e.g. Factor Xa), structure-guided pharmacophore-based virtual screening (e.g. novel sub-micromolar HIV-1 reverse transcriptase inhibitor and 11β-HSD1 inhibitors) [165, 166], and developing inhibitors targeting protein-protein interactions (e.g. CHIBA-3003 which interrupts HIV-1 Integrase-LEDGF/p75 interaction) [167].
Mason et al. developed a four-point pharmacophore method to perform molecular similarity search, which can be used for the design of combinatorial libraries [168]. This method calculated all potential pharmacophores/pharmacophoric shapes for a ligand and complementary site points in the binding site of a target protein. In combination with the ChemDiverse/Chem-X software, this pharmacophore method can be customized to enable a new measure of pharmacophore diversity. For example, Mason applied his method to design combinatorial libraries for 7-transmembrane GPCR targets, where “privileged” substructures were used as special features to internally referenced the pharmacophoric shapes [169]. The Mason’s method was used to consider up to 7 features and 15 distance ranges, which gave up to 350 million potential 4-point 3D pharmacophores/molecules. Subsequently, the resultant pharmacophore “key” served as a powerful measure for diversity or similarity, and provided a consistent frame of reference for comparing molecules, sets of molecules and protein sites.
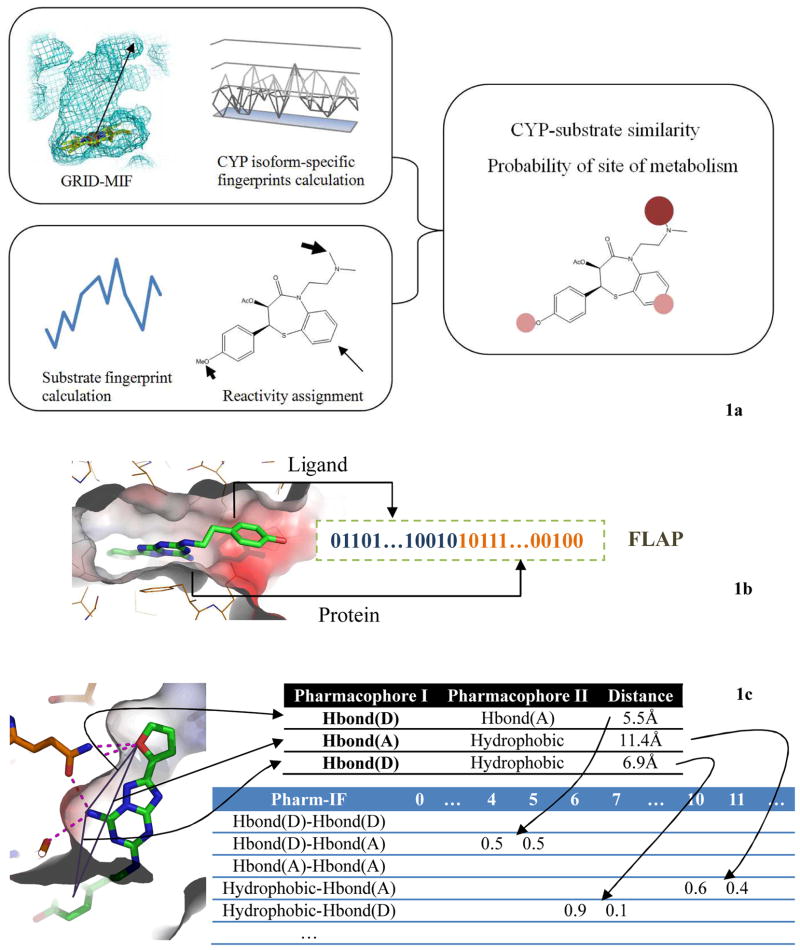
FLAP (Fingerprints for Ligands And Proteins) uses a common reference framework of four-point pharmacophore fingerprints derived from GRID analysis and a molecular-cavity shape to describe small molecules and protein structures [170]. The complementary description of the target and ligand generated by FLAP can firstly be implemented for selectivity or similarity analysis of macromolecules in a superimposition-free manner; secondly be used for ligand/structure-based virtual screening; and thirdly calculate chemometrics analysis and initiate a docking exercise [170]. Compare with DOCK and FieldScreen, better chemotype enrichment was achieved by both ligand-based and receptor-based screening by FLAP in an assessment using Directory of Useful Decoys (DUD) dataset [171]. Similar concept is embodied in MetaSite program, a robust software predicting the cytochrome P450 (CYP)-mediated metabolic site of given compound (Fig. 1a). Specifically, CYP-based GRID flexible molecular interaction fields (MIFs) are used to model the involved cytochrome, whereas GRID probe pharmacophore recognition is developed to characterize the substrate chemotypes [161]. The site of metabolism is predicted based on the hypothesis that the distance between the reactive center (e.g. oxene atom in protoporphyrin) on CYP and GRID-MIF points should correlate with the distance between the reactive center of the substrate and the positions of different chemotypes in the substrate. This state-of-the-art methodology facilitated the accurate prediction of the primary site of metabolism in 85% of the cases.
Fig. 1. Structure-based pharmacophore approaches in drug development.
1a. Workflow of prediction of the metabolism sites used by MetaSite. GRID-MIF of CYP is generated by chemotype probing technique. Then, similarity between GRID-MIF of CYPs and fingerprints of substrate is calculated to determine the substrate-specific CYPs. The site of metabolism is predicted as the degree of correlation based on the hypothesis that the distance between the reactive center on CYP and GRID-MIF points should correlate with the distance between the reactive center of the substrate and the positions of different chemotypes in the substrate. 1b. FLAP generation. Descriptors and fingerprints for both ligand and residues in the binding pockets are binned into FLAP fingerprints. Chemgenomic space is mined by machine learning method. 1c. Pharm-IF fingerprint generation. Only ligand atoms that participate in protein-ligand interaction are involved in fingerprints calculation. Pharm-IF fingerprints are derived from the distances of different pharmacophoric atoms. Target-specific Pharm-IF can be used to build pharmacophore models by machine learning method or similarity search.
Weil and Rognan developed a novel protein-ligand fingerprint (PLFP) method to mine chemogenomic space (Fig. 1b) [172]. Their method combines standard fingerprint representations of ligand with a protein “cavity fingerprint (CavFP)” representing pharmacophore features of protein binding site residues. This concept was tested on GPCR family whose members share a homogeneous cavity description. GPCR’s CavFP is a composition of pharmacophoric properties retrieved from 30 cavity-lining residues, whereas ligand fingerprints are generated by combining MACCS, SHED descriptors and charge [172]. PLFP model was trained by known protein-ligand pairs, and in GPCR case, this combined model outperformed other corresponding ligand-fingerprint-only models in mining chemogenomic space. While ligand prediction (ligand screening) performed better than receptor prediction (target profiling), PLFP was already robust in terms of the high recall and precision, short hit list size and low hit rank in ligand screening [172]. This approach can be utilized in future polypharmacology design to directly predict chemogenomic protein-ligand pairs. Furthermore, Sato et al. reported a new interaction fingerprint (IF) based on ligand pharmacophore (Pharm-IF) (Fig. 1c) [162]. Similar to residue-based PLIF developed by MOE, Pharm-IF is calculated from the distances of pairs of pharmacophore features which form interactions with protein [162]. Trained with machine learning techniques, i.e. SVM, Pharm-IF can be used to post-process docking poses generated by Glide during virtual screening. Pharm-IF-SVM model was tested in various protein contexts (PKA, SRC, cathepsin K, carbonic anhydrase II and HIV-1 protease), and they demonstrated that Pharm-IF-SVM model achieved a higher enrichment factor at 10% (5.7) than those of Glide score (4.2) and PLIF (4.3). This structure-based pharmacophore modeling offers extra restraints on the interaction patterns so that it can effectively enhance the enrichment of active compound during virtual screening against well-known protein targets.
Fragment-based drug design (FBDD)
FBDD aims to discover novel chemical leads with expected pharmacological properties, namely tight binder, from small “efficient” binder [173]. FBDD approach has been successfully utilized to guide pharmaceutical design inhibitors against a variety of targets, such as CDK4 [174], estrogen receptor [175, 176], factor Xa [177], HIV protease [178], to name just a few [138]. Besides lead identification, FBDD also plays a significant role in lead optimization. Raf-1 kinase inhibitor, sorafenib, as a successful example, was derived from a 318Da fragment-like lead by FBDD [138, 179]. Building blocks in FBDD are typically the substructures from drug-like molecules in 140–400Da range [138]. To ensure a higher likelihood of obtaining chemicals with proper physiochemical properties, “Rule of 3” was introduced during the construction of fragment database [180]. Binding modes can be determined via either experimental (e.g. structure-activity relationship by NMR (SAR by NMR), X-ray crystallography and tethering) or in silico techniques (e.g. molecular docking). For instance, SAR by NMR was utilized to guide the synthesis of ABT-737 and SP-4206 [181, 182]. Higher-affinity binders are designed by either linking fragments bound in distinct parts of the binding pockets or growing initial scaffold based upon the concept of combinatorial chemistry [128]. From a computational perspective, initiating from the lead fragment, “seed and grow” method iteratively assemble suitable fragments binding site before onto its suitable sites pre-docked seed. In contrast, “dock and link” approach is to dock substituent groups based on the chemical environment, and then link based on chemistry constraints [128]. “Seed and grow” involves more combinatorics issues than “dock and link”. This issue has been partially addressed by a number of in silico tools that were developed to optimize the “privileged” fragment enumeration process. These computational tools have been reviewed [138, 173]. In brief, LEGEND [183], RASSE [184] and F-DycoBlock [185] were developed based on force field-based scoring functions. Other programs as LUDI [186, 187], LigBuilder [188], SPROUT [189] and CONCERTS [190] employed empirical scoring functions. SMoG implemented its own knowledge-based potential grounded from statistical analysis of ligand-protein interactions [146, 191]. Several programs developed combined scoring functions. GANDI scoring function, for example, is a weighted sum of force-field term and two knowledge-based similarity terms against known CDK2 inhibitors [192]. SYNOPSIS utilizes a hybrid scoring function which combines empirical scoring with force fields [178].
However, a number of challenges have been characterized in FBDD. Synthetic accessibility issue, for example, has been addressed by assigning restrictions on growing/linking site or penalties during fragment assembly. SYNOPSIS established “in silico reaction” that grow compounds by simulating organic synthesis steps [178]. Similarly, LigBuilder 2.0 also employed a synthesis accessibility analysis module which checks the synthetic feasibility and selects reasonable synthetic routes [193]. PRO_SELECT was implemented with template-substituent idea from combinatory chemistry when pre-docked seed and proposed fragment were linked. Of note, PRO_SELECT achieved great success in designing factor Xa protease inhibitor, LY-517717 by Eli Lilly [177]. Second, the binding mode of compound is not necessarily in a modular fashion as desired. High internal energy, abundance of abnormal torsions, unstable chemical bonds and tautomers may impede the designed molecule to maintain the proposed interactions. Third, since computational FBDD still largely depends on molecular docking, it inevitably inherits such unsolved issues as protein flexibility and solvation. The only report regarding incorporating residue rearrangement function was made by Ian et al. [194]. In summary, FBDD is a paradigm-shifting methodology which allows “privileged” chemicals to target “difficult” pockets and exercise lead optimization. Other than worldwide scoring function issue, more stringent ligand-based constraints await further developments.
Free energy calculation by molecular mechanics
Scoring function can be perceived as the representation of relative binding free energy, based on the assumption that the free energy difference upon different ligands binding is predominantly contributed by protein-ligand interactions as predicted in the end-point complex. From a recent perspective, formulating computational-efficient scoring functions by focusing on static molecular interactions may misleadingly neglect a large part of contribution of thermodynamics involved in binding free energy: solvation, long-range interactions and conformational changes [195]. Moreover, interaction pattern such as hydrogen bonding may not contribute equally under different chemical environments [196]. Protein-ligand binding energy is now perceived as a nonadditive effect, which is protein context-related, solvent-sensitive and involves protein-ligand cooperative dynamic processes [195, 197]. Therefore, a more sophisticated and context-flexible approach is essential in structure-based drug design.
Molecular dynamics simulations take on a vital role in calculation of binding free energy [2, 198–200]. Recent advances in coarse-graining techniques have allowed the probing of dynamics of large membrane-bound protein systems on the microsecond time scale [201]. Methods for deriving more accurate force fields are an intense area of current research involved in drug discovery, such as “polarizable” force fields [202, 203], in which atomic parameters (e.g. charge) may change in response to changes in environment. This can be expected to improve predictions of binding free energy, where the molecular environment of a ligand may change greatly during binding. For the interested reader, more details of the development of force fields and other aspects of molecular dynamics can be found in various references [204–206]. These knowledges will immensely facilitate free energy calculation. The difference in free energy between different states (such as the free and bound forms of a receptor-ligand system) or along a reaction path (such as the path for ligand binding) can be related to experimentally measurable quantities, such as dissociation constants. The methods for binding free energy computation are well-known [207, 208], but implementation has been hindered by the computational expense of accurate calculations. Nevertheless, binding free energy calculations are available in molecular dynamics packages such as GROMACS [209]. Arguably the most common type of free energy calculation involving molecular dynamics is the free energy perturbation (FEP) method. This method is often used to calculate relative binding free energy for two similar ligands to the same receptor. This method takes advantage of the possibility of simulating unphysical alchemical transformations of one ligand into another, and the free energy of binding is reconstructed using a thermodynamic cycle [209]. The transformation of ligands is accomplished by discretizing the path from one to the other. At each discrete point along the path, the system is simulating using a potential which is a combination of the potentials for the two different ligands. This can become very computationally intensive, especially if the ligands are very different. This limits the ligands studied to be a small set of chemically similar ligands, which lessens the attractiveness for use in high-throughput screening. Use of sheer computation is feasible if we limit ourselves to only a handful of system. Examples include the determination of the free energies of binding for eight ligands to FKBP using a massively parallel computing platform [210], or the calculation of free energies of binding for several sparso-mycin analogs to the bacterial ribosome [211]. The computational difficulty lies in the fact that free energies cannot be calculated from a single static structure, but requires sampling of a large configurational space.
As a result, for virtual screening of large numbers of ligands, binding free energy prediction during docking usually relies on fast approximation techniques, such as the use of simple empirical or knowledge-based scoring functions [212]. These approximations generally cannot accurately rank ligands in order of binding affinity, as we have discussed above [213, 214]. An attempt to quantify the errors introduced by various levels of approximation in calculating binding free energies was performed by Mitomo and coworkers [215], who used three methods, namely, in order of decreasing computational expense: explicit water molecular dynamics, an implicit solvent molecular mechanics generalized born surface area (MM-GBSA) model, and a docking scoring function. The test systems used were streptavidin and biotin. As expected results from the explicit water simulations most closely matched experimental measurements (error of 1.8 kcal/mol), with the MM-GBSA model overestimating (error of 17 kcal/mol) and the docking scoring function underestimating (error of 9.6 kcal/mol) the binding strength. Moreover, Noriaki et al. combined MM-PBSA with molecular docking in a virtual screening exercise, and yielded an improvement of 1.6–4.0 times that of enrichment performance with molecular docking alone [216]. This points the importance of correctly incorporating the solvent degrees of freedom, which are unfortunately often the most computationally expensive component of explicit solvent simulations. An attempt to generate an intermediate method which is faster than FEP but more accurate than simple scoring functions has been developed [217], called the linear interaction energy (LIE) model. Also, improvements in computational efficiency of free energy calculations can be obtained by using continuum solvation models, which treat water only implicitly, combined with molecular dynamics [218]. Research into fast implicit solvent methods which can match the accuracy of explicit solvent simulations is one of the most pressing problems which, if solved, would greatly enhance the effectiveness of computational drug design. High-throughput version of MM-PBSA, developed by Brown et al. from Abbott Laboratories, speeded up the relative binding free energy calculation to an order of 100 CPU min per structure by a rational combination of GB implicit solvation model and Zap (OpenEye) on an enterprise grid [219, 220]. To perform calculation more efficiently, Huang et al. incorporated the upstream docking (DOCK and Glide) to the free energy calculation, followed by energy minimization in GB implicit solvent, instead of commonly used MD conformer sampling. The speed has reached as fast as 1 min per structure [199]. Yet, this method may appear to be inaccurate because it bypassed the time-consuming entropy losses calculation and fully relied on GB semi-analytical approximation which contained documented drawbacks [221].
In conclusion, as computational costs continue to decline, we may expect molecular dynamics to play an increasing role in drug design. The power of molecular dynamics lies in its versatility. It is a general technique which can be readily adapted to specific needs, such as docking and free energy calculation. Concepts from molecular dynamics force field development have shown their influence in current pharmacology.
Targeting protein-ligand interaction
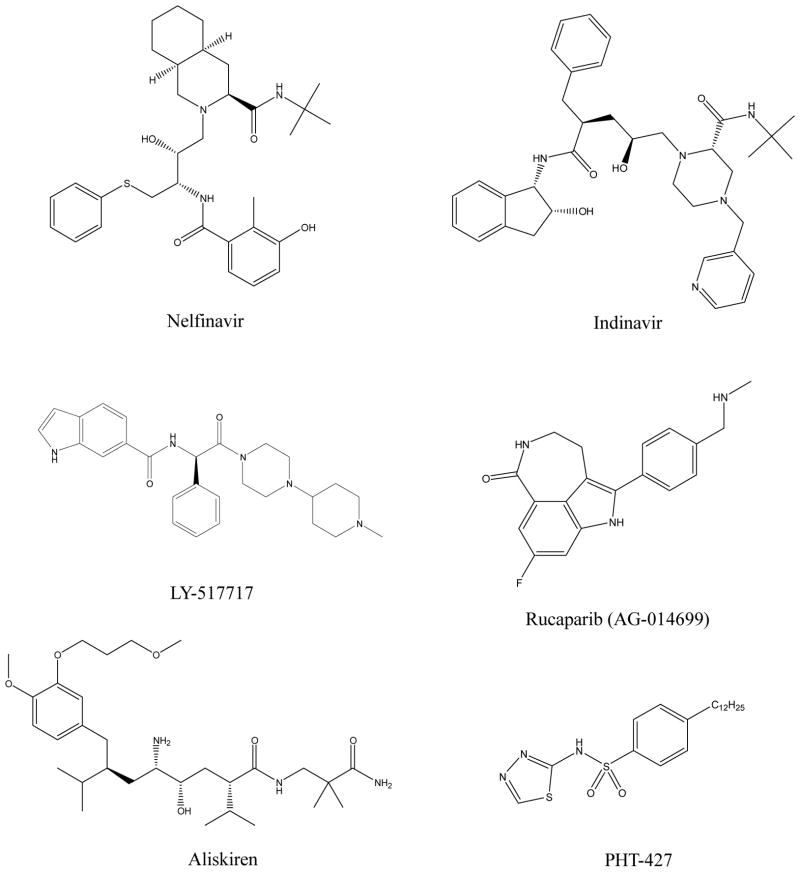
The discovery of a series of HIV-1 protease inhibitors, such as Ritonavir (Abbott laboratory), Indinavir (Merck) and Nelfi-navir (Agouron Pharmaceuticals, acquired by Pfizer), opened up an era of structure-based drug design scenario (Fig. 2) [222]. Previous successful development of protein inhibitors using free virtual screening packages, mainly AutoDock and DOCK, have been reviewed by Bruno et al. in their review [223]. Tanaji et al. reviewed the development of 12 small molecules that have entered late clinical trials or final approved by FDA for therapeutic use as direct consequences of structure-based drug design [224]. These drugs include Aliskiren (Tekturna®), NVP-AUY922 (Novartis), LY-517717 (Eli Lilly), etc. Van Montfort et al. discussed a subset of cancer-related drugs in which structure-based concept was involved in their development [225]. These targets involve oncogenic kinases (i.e. EGFR, Akt/PKB, PI3K, B-Raf, etc.), epigenomic target (i.e. HDACs), DNA repair target (i.e. PARP) and chaperone (i.e. Hsp90). In silico methods are frequently used in late lead optimization when target structure is accidentally available; however, in some cases, such as Aliskiren and LY-517717, computer models initiated the early lead identification. Here, combined with our lab’s experience in structure-based drug design, we picked up several typical successful stories in this field.
Fig. 2.
Example compounds that target protein-ligand interaction resulting from in silico structure-based design
From existing structures
The discovery of LY-517717 (Eli Lilly) is a direct consequence of computational de novo drug design approach. LY-517717 is an indole derivative (Fig. 2) in which indole replaces benzamidine moiety of 3a lead, and this inhibitor has just finished the phase II clinical trial in prevention of venous thromboembolism in patients with hip or knee replacement surgery (http://clinicaltrials.gov/ct2/show/NCT00074828). In identification of 3a, template-substituent idea of combinatorial chemistry was used by in silico program PRO_SELECT. Starting from benzamidine (Ki>200μM) as a seed, a series of factor Xa structure-based focused-libraries were iteratively constructed. The best benzamidine-based lead synthesized after virtual screening, 3a, obtained Ki 16nM [177]. To improve the oral bioavailability and other pharmacokinetic properties, benzamidine moiety was replaced by indole, and it resulted in LY-517717, which achieved Ki value at 5nM [226]. The success of LY-517717 in clinical trials is a proof-of-principle of de novo drug design and accentuates the importance of addressing synthetic issue in de novo design and library design.
Poly-ADP ribose polymerase-1 (PARP-1) involves in DNA single strand break pathway, which facilitates DNA damage repair by conjugating ADP-ribose to DNA and other DNA repair machineries [227]. PARP-1 inhibitor has been widely used in treatment of cancer, Parkinson’s disease and stroke [225]. Early developments of nicotinamide-based inhibitors were unsuccessful because poor specificity and pharmacokinetics were observed in spite of the high potency [227]. The breakthrough started from the development of tricyclic novel PARP-1 inhibitor, based on the crystal structure of conventional nicotinamide-based PARP inhibitors and chicken PARP complex [228]. To reach the pocket formed by two coplanar Tyr907 and Tyr896, amide edge was extended to [5,6,6]-tricyclic indole lactam or [5,6,7]-tricyclic indole lactam. Their model predicted a high binding affinity with [5,6,7]-tricyclic indole lactam, and later crystal structure showed that this crucial amide is locked in a cis conformation in this seven-member lactam ring, thus strengthening the interaction. In addition, the 2-phenyl group interacted with Tyr907 and Tyr896 via π-π stacking, as the design proposed [227]. Later minor optimizations led to Rucaparib (AG-014699), a potent and selective PARP-1 inhibitor with Ki <5nM [229]. Rucaparib has finished Phase I trial and submitted Phase II clinical trial in treatment of cancer with BRAC-1/2 mutation to achieve synthetic lethality (ClincialTrials.gov ID: NCT00664781) (Fig. 2).
A typical high-throughput docking-supported virtual screening was conducted in our lab which leaded to several promising sulfonamide inhibitors that targets AKT1 PH domain. AKT is known as oncogenic serine/threonine protein kinase that plays a key role in mediating signals for cell survival, proliferation, apoptosis, transcription and glucose metabolism [230]. AKT comprises three highly conserved isoforms, all of which have N-terminal pleckstrin homology (PH) domain, serine/threonine kinase domain and C-terminal hydrophobic motif [231]. PH domain is crucial for the translocation to the cellular membrane by attaching phosphatidylinositol (3,4,5)-triphosphate (PIP3). A ligand-based pharmacophore model identified 4 active compounds from a three million combined database. AutoDock was combined with GOLD to identify leads which contained sulfonamide moiety. After lead optimization and structure-activity relationship (SAR) investigations, PHT-427 was synthesized and exhibited Ki 2.7μm activity against AKT1 PH domain [117, 118, 232, 233] (Fig. 2). As we will further discuss in polypharmacology part, PHT-427, as a results of docking-based strategy, was also found to target phosphatidylinositide-dependent protein kinase 1 (PDPK1) PH domain at a slightly higher Ki (5.2μM) [118].
Starting from protein models
Strikingly, protein models, especially homology models, are widely used in structure-based drug discovery projects [10]. Some applications include protein structural analysis and function prediction [40–45], understanding of protein-ligand interactions [46–53] and compound optimization [54–56]. In order to compensate from the low-resolution models, additional target-specific constraints and/or more accurate scoring function are often applied. Here we will briefly discuss some successful hit identification stories starting from homology models (Table 4).
Renin is an aspartic peptidase which is able to cleave angiotensionogen and therefore trigger agiontensin II pathway, is an attractive target for the treatment of hypertension [234]. Homology model of renin played a critical role in developing Aliskiren (Tekturna®), an FDA approved drug in 2007 [235]. Aliskiren is the 3rd generation renin inhibitor, whereas both 1st generation peptide inhibitor CGP29287 and 2nd generation peptidomimetic inhibitor CGP38560 experienced disastrous failure in the clinical trials. The fact that CGP38560 exhibited very poor oral bioavailability (<1%) and half life (<7min) aroused the interests to develop small-molecule drugs that had better pharmacokinetic properties. Since no crystal structure available at that time, a homology model of renin completely orchestrated the structure-based drug designing project. The critical residues of binding were predicted by extensive docking of 2nd generation peptidomimetics, and four interaction pharmacophores (P1, P2, P3, P4) were determined. Then, a number of potent leads (<5nM) were rationally designed, including tetrahydroquinoline (THQ), phenoxy, indole and salicylamide, based on predicted docking poses and binding free energy. Aliskiren is a direct descendant of a phenxy lead extended by methoxyl-tert-butyl group, which has potent IC50 in a human cell line (0.6nM) (Fig. 2). The approval of Aliskiren raises two amazing facts: 1. The crystal structure for renin in complex with CGP38560 was solved in 1991, and this binding poses have already predicted in 1987 by homolog modeling, and 2. This 4-year-ahead advantage competed other pharmaceutical companies, such as Merck, Abbott, Roche, Pfizer, etc., in clinical trials and marketing of the drug targeting renin-angiotensin system (RAS) pathway [235].
Inhibiting uncontrolled cycles of endocytosis as in epileptic seizures represents a significant ($15 billion market) and largely unmet medical need [236]. Dynamin receptors are responsible for this disease. Lack of crystal structures for human dynamin has impeded rational drug discovery efforts, resulting in promiscuous compounds with varying side effects. Odell et al. have attempted to address this problem by developing a rational drug design study using homology modeling and docking-based protocol to identify selective dynamin I inhibitors. Two models of the GTPase domain of dynamin I were developed using the D. discoidum GDP-bound dynamin A GTPase domain, which were validated by their ability to reproduce low RMSD docking poses (<1Å) of GDP. The models were used in a virtual screening study against ~80K compounds and post experimental testing. A new class, namely pthaladyn, was identified to have micromolar potency. This hit and its protein-ligand interaction details were exploited to design several other potent GTP-competitive analogues [237].
Pharmacophore modeling can be used to promote the hit identification rate of homology model-based drug discovery. In an interesting approach involving homology models, researchers at Sanofi-Aventis developed sequence derived 3D pharmacophore models for class A GPCRs. A multi-step modeling and docking approach using known ligand-protein experimental data as restraints was used to model three-dimensional protein-ligand complexes. Key protein-ligand interactions (e.g. Asp332 interaction with positively charged nitrogen of ligands) were encoded as 2D chemoprints and were mapped to 35 single feature structure-based pharmacophores. As an application, the sequence of C3AR1 receptor identified four 2D chemoprints. Using the corresponding 4 pharmacophores, a ligand-based virtual screening identified the first small molecule inhibitors for C3AR1 [246]. A similar receptor-based pharmacophore was successful applied by Sala et al. to a virtual screening study against the homology model of hIKK-2 protein to increase the identification rate of novel chemotype inhibitor [239].
Targeting protein-protein interaction
In order to function properly, most proteins do not act as isolated units; they often form complexes with other macromolecules, usually proteins. The formation of these complexes relies on the protein interactions (PPI). PPIs are ubiquitous in all biological processes, and cellular dysfunction via faulty protein-protein interactions is the root cause of a plethora of diseases, including cancer and neurological disorders [247, 248]. Similar to protein-ligand interaction, recent structural analyses of protein-protein surfaces refined the definition of PPI as locally-optimized and stable protein-protein association, dominantly contributed by clustered, networked, highly packed, cooperative and structurally conserved residues [249]. However, in contrast to classic protein inhibitors that target well-defined small molecule’s binding grooves or channels [250, 251], PPIs are usually mediated by flat and large interfaces. The rapidly increasing knowledge of thermodynamic basis of PPI as well as the quality and diversity of lead compound have allowed multiple studies of drugs indeed targeting interfaces [252, 253]. Hence, this new class of novel, druggable interactions greatly expands the target space available for a myriad of diseases.
Hot spots identification and prediction
It has been well documented that for all PPI interface, the energy distribution is not uniform across a particular interaction; a small set of residues contribute more significantly to the binding free energy than other residues, so-called hot spots [254–261]. Bogan and Thorn defined a hot spot as a residue whose mutation to alanine results in a binding free energy change of at least 2.0 kcal/mol [262]. As one might expect, the residues at protein interfaces [263] and functional sites [264] are mutating at a slower rate when compared to other surface residues. Furthermore, hot-spot residues are complementary, interfacial residues that are usually surrounded by weaker interactions providing specificity [265].
Experimental identification of hot spot residues in PPIs is primarily performed by alanine scanning, whereby hot spot is defined as the residue that, when mutated to an alanine, results in a remarkable drop in the binding constant, typically tenfold or higher [266]. Alternatively, we can consider a hot spot mutation as a mutation that destabilizes the bound state ensemble relative to the unbound one if we consider the insights of free-energy surfaces and conformational ensembles from protein folding studies [267, 268]. Alanine scanning allows one to determine the specific contribution of residues to the stability of the ensemble, and protein’s resulting function. Results from alanine scanning are deposited in the Alanine Scanning Energetics Database (ASEdb), and the Binding Interface Database (BID) contains experimentally verified hot spots from the literature [262, 269]. These repositories, while useful, may have drawbacks such as limited complexes [266]. Experimental mutagenesis of target proteins for elucidation of hot spots is not applicable on a large scale since it is very time consuming and expensive. People are seeking effective ways to accelerate the laborious alanine scanning, such as using reflectometric interference spectroscopy [270], “shotgun scanning” [271] and in silico prediction. There are many computational tools available to the user that can accelerate this process. Three primary types of predictive approaches exist for computational hot spot prediction: energy-based, machine learning-based and empirical-based methods. Energy-based method estimates the energetic contribution of each residue to the total binding energy via either virtual alanine scanning (e.g. FOLDEF [272], Robetta [68, 273], PP_SITE [274]) or MD simulations (e.g. snapshot-based ΔΔGbinding estimation [275], anchor residue analysis [276, 277]). Machine learning-based methods can overcome the blockroad of energy-based method-high computational cost-through both pure machine-learning models (e.g. decision tree-based KFC and SVM-based model [278]) and hybrid models where energetic terms are partially involved (e.g. KFCA [279] and HSPred [280]). While machine learning methods hold great promise, simpler empirical models may better their predictive power with the introduction of additional terms, such as residue conservation. For example, both HotPoint webserver [281] and HotSprint database [282] were established based on empirical models which combined residue conservation term with other terms such as buried/complex accessible surface area and knowledge-based pairwise residue potentials of the interface residues.
Towards potent PPI inhibitors
PPI networks have been studied extensively in multiple species, and their complexity and diversity are unrivaled among other macromolecular interactions [283–286]. The hub proteins (ones with multiple binding partners) in disease-related pathways, such as p53, have proved to be crucial to signaling networks; these proteins gain much interests as they can interact with multiple binding partners through the same interface (promiscuous proteins), as well as simultaneously through different binding regions [287–289]. A number of studies have been carried out through targeting these critical PPIs (Table 5).
Table 5.
Strategies and mechanisms of protein-protein interactions inhibitors
| Molecule | Mechanism of action | Identification method | Stage in clinical trials |
|---|---|---|---|
| Nutlin-1/-2/-3 | MDM2 binding | High-throughput screening | Phase I |
| MI-219 | MDM2 binding | Structure-based de novo design | Phase I |
| benzodiazepine-based inhibitors | MDM2 binding | High-throughput screening | Preclinical |
| ABT-737 | Bcl-2, Bcl-XL and Bcl-w binding | Structure-activity relationship by NMR | Phase I |
| ABT-263 (Navitoclax) | Bcl-2, Bcl-XL and Bcl-w binding | Lead optimization from ABT-737 | Phase I/II |
| HA14-1 | Bcl-2 binding | Virtual screening by DOCK | Preclinical |
| J042 | Bcl-2 binding | Receptor-based pharmacophore modeling | Preclinical |
| Embelin | XIAP binding | Virtual screening by DOCK | Preclinical |
| Azabicyclooctane derivatives | XIAP binding | Computational de novo optimization from peptide using CAVEAT program | Preclinical |
| Enfuvirtide | gp41 protein folding | Peptide mimetics | Approved |
| ADS-J1/ADS-J2 | gp41 protein folding | Virtual screening by DOCK | Preclinical |
| NB-64, NB-2 and derivatives | gp41 protein folding | Sandwich ELISA screening combined with molecular docking | Preclinical |
| NSC23766 | Rac-1 binding | Virtual screening by UNITY | Preclinical |
| ZINC08010136 | Rac-1 binding | Pharmacophore filtering, virtual screening by AutoDock and MOE | Preclinical |
| Ro26-4550 | IL-2 and IL-2Rα binding | Peptidomimetic of IL-2 | Preclinical |
| SP-4206 | IL-2Rα binding | Fragment-based design from Ro26-4550 | Preclinical |
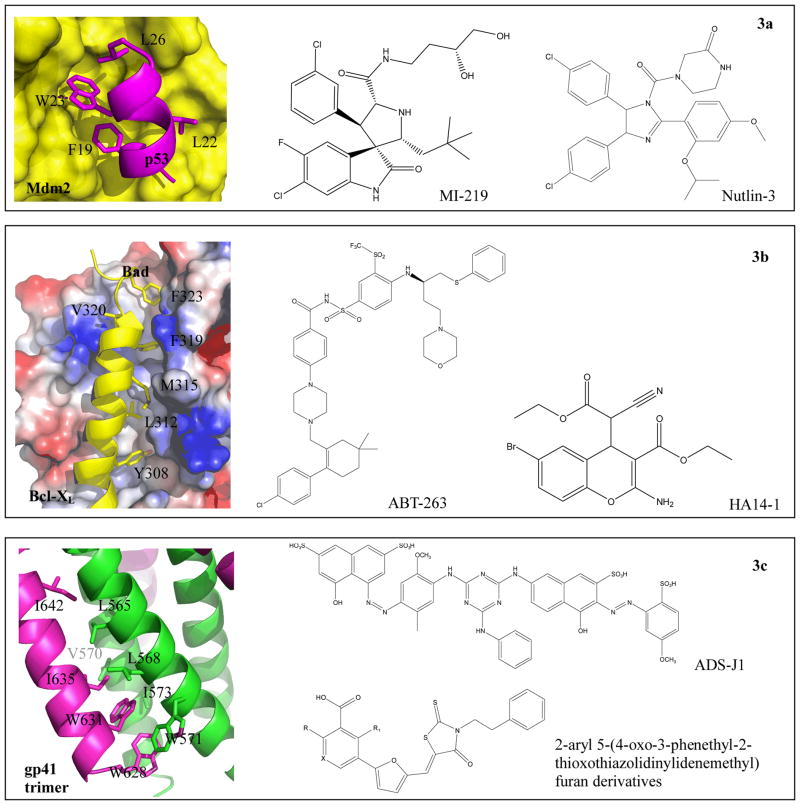
The most successful PPI inhibition target up till now is Mdm2-p53. Mdm2, a ubiquitin E3 ligase, targets p53 for 26S pro-teosome-mediated proteolytic pathway. The overexpression of Mdm2 in some tumor cell line was found correlated with the inactivation of p53-dependent apoptosis, and thereby “awakening” p53 by inhibiting Mdm2 binding has been highlighted in previous researches [290]. Mdm2 binds to a 15-residue α-helical region in the transactivation domain of p53, where three contact points (Leu26, Trp23 and Phe19) on this helix confer most of the binding energy (Fig. 3a) [291, 292]. Three classes of inhibitors have been reported as small-molecule potent inhibitors against Mdm2-mediated ubiquitylation: Nutlins [293], benzodiazepine-based inhibitors [294] and spiro-oxindole based inhibitors [295]. Of note, Nutlins and benzodiazepine derivatives were discovered through high-throughput screening, whereas spiro-oxindole scaffold through structure-guided de novo design. Seeded with Trp23 residue, a number of spiro-oxindole scaffold containing natural compounds, such as Alstonisine and spirotryprostatin A, were discovered. Further de novo lead optimization was performed and guided by GOLD docking conformations to achieve maximal binding affinities. A series of spiro-oxindole based low nanomolar inhibitors, namely MI-63 (Ki=3nM) [296] and a more bioavailable derivative MI-219 (Ki=5nM) (Fig. 3a) [297], were designed by expanding the binding pocket to the proximal Leu22. Particularly, MI-219 was fully capable of inhibiting cancer cell proliferation by induction of p53-dependent apoptosis both in vitro and in vivo. Both Nutlin and MI-219 had entered phase I clinical trial for safety testing [298]. Other active compounds with novel chemical cores, such as NSC66811, were recently identified by a combination of pharmacophore filtering and structure-based virtual screening against NCI database [299]. The resulted NSC66811 acquired binding affinity Ki 120nM that was 3.3 times less potent than Nutlin-3 (Fig. 3a) (36nM), yet exhibited functional restoration of p53 and p21 in HCT-116 p53+/+ colon cancer cell lines [299].
Fig. 3. Successful protein-protein interaction targets for therapeutic developments.
3a. Four hot-spot residues of p53 (magenta)-MDM2 (yellow) complex and two inhibitors (MI-219 and Nutlin-3). Complex structure was got from (PDB ID: 1YCR). 3b. Interactions between Bcl-XL and Bad (PDB ID: 1G5J) and structures of two inhibitors (ABT-263 and HA14-1). Peptide of Bad are colored in yellow, and the electrostatic potential surface of Bcl-XL is displayed. 3c. Hydrophobic groove of C-terminal heptad repeats from HIV-1 gp41 trimer (PDB ID: 1AIK). Two representatives of inhibitors (ADS-J1 and 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl) furan derivatives) are in right side. All critical residues are displayed in sticks.
B-cell lymphoma 2 (Bcl-2) family of proteins are pronounced regulators of apoptotic cell death, which include both anti-apoptotic factors, such as Bcl-2 and Bcl-XL, and death agonists such as Bak, Bax, Bim, Bad, etc. [300, 301]. These proteins can form homodimers and heterodimers with other family members, particularly the pro-apoptotic molecule Bak containing BH3 domain (Bcl-2-antagonist/killer) [302]. Many research groups have produced α-helical mimics targeting the hydrophobic binding pocket which mediates Bcl-2/Bak interaction [303–306]. The most distinguished of these are ABT-737 and ABT-263 (Fig. 3b) (Abbott Laboratories), inhibitors of Bcl-2, Bcl-XL and Bcl-w that were discovered by NMR-based high-throughput screening of a chemical library [307]. These molecules mimics the Bak-derived peptide in binding region on Bcl-XL, and binds deeper in cavities with more puckered grooves at subnanomolar level [307]. ABT-737 exhibits profound inhibitory potency against lymphoma and leukemia cell lines, and synergizes with other cytotoxic chemotherapeutic agents. ABT-737 has been in phase I clinical trial [308]. Bioavailable ABT-263 (Navitoclax) was developed upon ABT-737 lead and is currently in Phase I/II clinical trial [309]. Besides ABT-737 derivatives, Jia-Lun et al. identified a nonpeptidic Bcl-2 inhibitor, HA14-1, after a virtual screening exercise using Bcl-2 homology model and DOCK for screening MDL/ACD database (Fig. 3b). This compound achieved ~9μM IC50 against Flu-BakBH3 and exhibited in vitro pro-apoptotic efficacy in an Apaf-1-dependent manner [310]. Other research groups, such as, applied similar approaches and identified hits against this target from NCI 3D database. 7 compounds out of 35 selected hits were tested to obtain anti-proliferative effects varying from IC50 1.6 to 14.0 μM. Michele et al. conducted structure-based virtual screening targeting BH3 binding pocket using the crystal structure of Bcl-XL in complex with Bak derived peptide. A total of 320 hits, selected from Maybridge according to FlexX docking poses and H-bonding to Bcl-XL, [311]. J042 was identified from ZINC database using a combination of receptor-based pharmacophore filtering and cross-docking, and achieved 2.58 μM activity [312]. All of the above active compounds were assessed based on several apoptosis-related parameters, such as swelling, ΔΨm loss and cytochrome c release, in which ABT-737 still outperformed other compounds that were designed computationally [313].
As another promising prosurvival target, XIAP (X-linked inhibitor of apoptosis), was demonstrated to bind and specifically inhibit caspase-9 activity via BIR3 domain [314]. XIAP-caspase-9 interaction also shows its direct role in resistance to radiation therapy in cancer cells by blocking both intrinsic and extrinsic apoptosis pathway. Furthermore, the existence of endogenous XIAP inhibitors, such as SMAC and HTRA2, provides proof-of-principle to exploit XIAP as a novel therapeutic target. An incomplete list of XIAP inhibitor is available in Nature Cell and Differentiation Review [314]. As an example of XIAP inhibitors screened from Traditional Chinese Medicinal herbs 3D database, embelin was first discovered by a computational virtual screening using DOCK program [315]. Embelin, along with its derivatives [316], has been proved to be robust apoptotic-inducing agents at low micromolar IC50 by overcoming protective effects of XIAP overexpression [315]. The other strategy of targeting XIAP is to block its interaction with caspase-9 by the compounds mimicking SMAC-XIAP N-terminal interactive motif, NH2-AVPI [317]. Frederick et al. performed an in silico optimization by CAVEAT program on ML-IAP-binding peptide, ALP-2,2-diphenethylamine. They discovered bioavailable azabicyclooctane derivatives as a novel antagonist of apoptosis inhibitor that suppressed BIR3 domain binding at Ki 140nM [318]. Starting from NH2-AVPI peptide, Huang et al. introduced a fragment-based approach by replacing amino acids by drug-like and synthetically accessible scaffolds [319]. Optimizing the lead compound guided by docking, they discovered and synthesized cell-permeable inhibitor BI-75D2, which has anti-IAP cellular activity at IC50 16.4 μM.
Recent anti-HIV researches shed more spotlights on targeting early stage of HIV-1 infection, such as fusion and entry, other than maturation. Two critical protein, gp120 and gp41, were found to mediate the CD4+ cell recognition and membrane fusion, in which gp41 forms a six-helix bundle core upon surface gp120 binding to CD4 and co-receptors (e.g. CCR5 and CXCR4). Massive efforts have been exerted on CCR5 targets in order to block gp120/CCR5 interactions. Maraviroc (Pfizer) defeated aplaviroc (GlaxoSmithKline) and vicriviroc (Schering-Plough) for final approval after racing for the first CCR5-targeted entry inhibitor. As to gp41 target, the stabilization of six-helical bundle requires the antiparallel packing of three C-terminal heptad repeats (HR1-3) into the hydrophobic grooves on the central trimeric core (Fig. 3c), making such hydrophobic cavity attractive as a potential drug-binding pocket [320, 321]. Although great successes have been made by Enfuvirtide [322], the first polypeptidic fusion inhibitor derived from HR2 and approved by FDA, people are still seeking more bioavail-able drugs that circumvents the frequent intravenous injection. The first nonpeptidic gp41 inhibitor identified by structure-based method was ADS-J1 and ADS-J2 (Fig. 3c), two large and hydrophobic compounds that suppressed the fusion-active gp41 core at IC50 4.95 μM and 21.85 μM respectively [323]. XTT formazan has similar anti-fusion activity against gp41 according to their ELISA and docking results [324]. Further drug-like inhibitors explorations, including the primary identification of NB-2 and NB-64 micromolar leads [325] and docking-guided optimization to derivatives of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl) furan (Fig. 3c), have been carried out recently by the same group [326, 327]. The most potent inhibitors synthesized so far acquired 14nM IC50 against the formation of gp41.
Ras GTPase family proteins are well-known notorious in its oncogenic mutations that lead to constitutively active growth signals and mutants-associated drug-resistances [328]. Efforts have been done to silence Ras (or other oncogenic Ras family members e.g. Rho and Rac) signaling by inhibiting 1) post-translational modification, such as farnesylation 2) Ras-effector interaction 3) Ras-GEF interaction 4) GTPase activity restoration [329]. However, little progress was made in designing potent PPI inhibitors probably due to the chemical property of Ras-binding pocket and its associated binding modes. In sharp contrast to other targets discussed above, the interactions observed from Ras-effector complex are mainly composed of antiparallel β-strands hydrogen bonds and electrostatic attractions. Early developments of Ras-Raf1 inhibitor elicited some high micromolar hits, such as MCP derivatives [330], radicicol [331], and effector-derived synthetic peptides [332, 333]. Yuan et al. made breakthrough on Rac1-GEF interaction by targeting Trp56 region from comparative and mutagenesis analysis. NSC23766 was identified by structure-based virtual screening using UNITY (Tripos Associtates) [334]. Similar docking-based virtual screening was utilized by Nicola et al. in their study, and the best inhibitor they discovered was ZINC08010136 with IC50 12.2 μM, which was slightly better than NSC23766 [335]. Other less potent allosteric inhibitor (e.g. cyclen-metal) that can stabilize the weak binding state for downstream effectors is still in early stage development [336]. In summary, hot topic as Ras-associated disease is nowadays, silencing mutant Ras signaling by interrupting Ras-effector interactions still confronts great challenges.
It is worthwhile to notice that most successful pockets of PPI inhibitor development are characterized with massive hydrophobicity. The core scaffolds that target Bcl-XL-Bad, gp41, Mdm2-p53, XIAP-caspase-9, CD4-MHC II [337] and IL-2-IL-2Rα interactions [338, 339] are overwhelmingly hydrophobic. In contrast, such interactions as Ras-effector and PTEN-MAGI3 [340, 341] are mainly mediated by β-strands hydrogen bonds and electrostatic attractions, and those targets have unavoidably experienced obstacles to achieve low IC50. These facts imply the poorly-understood but pivotal roles of entropy and desolvation effects that trigger protein-protein association. Furthermore, targeting PPI also experiences common challenges such as inherent flexibility. From the lesson of developing IL-2 inhibitors, we know that Ro26-4550 (Roche) induced significant conformational changes compared to unbound IL-2 [339]. Hence, novel insights are needed for future drug design and scoring function improvement.
Polypharmacology: Selectivity or promiscuity? Or both?
Modern drug development projects primarily aim to deliver target-specific active compounds. The genesis of this approach was the “one disease, one target and one drug” Mendelian model. However, retrospective analysis proved that approved drugs are often promiscuous and bind to several protein targets [342, 343]. This property of an active compound binding to multiple proteins is termed as polypharmacology. Polypharmacology of bioactive compounds elicit either beneficial (e.g. Sorafenib: A Raf inhibitor originally developed against lung or pancreatic cancer [344] proved effective against renal cell cancer by its action on VEGFR2 receptors [345]) or adverse effects (e.g. Paxil, a serotonin uptake inhibitor, also binds to beta-adrenergic receptors thus offering plausible explanation for increased heart rate [346]). Several other examples can be found are available in previous review [347]. Moreover, increasing evidence proving the complexity of diseases that void the Mendelian model (e.g. multiple targets implicated for one disease state [342]) and the robust or compensatory behaviour of biological systems [348] suggest that designing selectively promiscuous compounds may offer better therapeutic outcomes [349]. Likewise, prediction of compound polypharmacology has the potential to identify possible adverse effects [350, 351] which are implicated in about 30% of drug failures.
Several methods have been developed for the computational prediction of compound polypharmacology. Network-based methods integrate chemoinformatics, bioinformatics, and systems biology techniques to investigate compound polypharmacology by mapping drug-target associations [352–355]. Our laboratory developed a network that allows users to intuitively interrogate drug-target relationships based on functional annotation (e.g. biological targets, reported side effects, toxicities or metabolism) and chemical structural similarities [352]. At present over 5000 drugs, ~10 million virtual library compounds, and ~56K biological macromolecules are available for exploration via a web-portal which will be publicly accessible in the near future. Our novel network-based prediction of quercetin’s chemical structure similarity with Dasatanib and thus its possible use as a tyrosine kinase inhibitor has been independently validated [356]. Chen et al. combined networks obtained from experimental assay information in PubChem, protein-protein interaction, drug-target interaction and biological pathways to map polypharmacology within PubChem database [357].
Structure-based methods are based on the premise that proteins with similar binding sites will attract similar compounds. Examples include the similarity ensemble approach developed by Keiser et al. [346], three-dimensional shape based approach for identifying diverse targets [358], structural similarities of molecular scaffolds to map drug promiscuity [359]. Vulpetti et al. developed a shape-based binding site similarity method to elucidate polypharmacology in the kinase family [360]. Weill et al. described the binding site of GPCRs by encoding both, ligand descriptors and protein pharmacophoric properties, in a low dimensional fingerprint for chemogenomic screening applications [172]. Text mining approaches [361] and semantic framework based methods [362] have been applied to map and predict polypharmacology using widely available biochemical information contained in publicly available resources. Although current structure-based drug design hardly considers multi-targeting issue, efficient polyphamacology prediction may be helpful in the future to facilitate drug repurposing and to discover more potent drug with less off-target toxicity.
Acknowledgments
This work was supported in part by the US Department of Defense Concept Awards (BC085871), US National Institute of Health P41 grant (5P41GM079588-03), Grant # IRG-08-061-01 from the American Cancer Society, and MDACC-UT Austin CTT-TI3D grants.
Footnotes
Conflict of interest
The authors declare that they do not have competing interests.
References
- 1.Zhang S. Structure/Ligand-based Drug Design and Structure Bioinformatics: Basics, Concepts, Methods, and Applications. VDM Verlag; 2009. [Google Scholar]
- 2.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004 Mar 19;303(5665):1813–8. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 3.Cherezov V, Abola E, Stevens RC. Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol Biol. 2010;654:141–68. doi: 10.1007/978-1-60761-762-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahar I, Lezon TR, Bakan A, Shrivastava IH. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem Rev. 2009 Mar 10;110(3):1463–97. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt J. Structural genomics-impact on biomedicine and drug discovery. Exp Cell Res. May 1;316(8):1332–8. doi: 10.1016/j.yexcr.2010.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Grabowski M, Chruszcz M, Zimmerman MD, Kirillova O, Minor W. Benefits of structural genomics for drug discovery research. Infect Disord Drug Targets. 2009 Nov;9(5):459–74. doi: 10.2174/187152609789105704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavasotto CN, Phatak SS. Homology modeling in drug discovery: current trends and applications. Drug Discov Today. 2009 Jul;14(13–14):676–83. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Daga PR, Patel RY, Doerksen RJ. Template-based protein modeling: recent methodological advances. Curr Top Med Chem. 10(1):84–94. doi: 10.2174/156802610790232314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malmstrom L, Goodlett DR. Protein structure modeling. Methods Mol Biol. 673:63–72. doi: 10.1007/978-1-60761-842-3_5. [DOI] [PubMed] [Google Scholar]
- 10.Grant MA. Protein structure prediction in structure-based ligand design and virtual screening. Comb Chem High Throughput Screen. 2009 Dec;12(10):940–60. doi: 10.2174/138620709789824718. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004 Aug 19;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999 Mar;15(3):211–8. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 13.Morgenstern B. Alignment of genomic sequences using DIALIGN. Methods Mol Biol. 2007;395:195–204. doi: 10.1007/978-1-59745-514-5_12. [DOI] [PubMed] [Google Scholar]
- 14.Tsai YT, Huang YP, Yu CT, Lu CL. MuSiC: a tool for multiple sequence alignment with constraints. Bioinformatics. 2004 Sep 22;20(14):2309–11. doi: 10.1093/bioinformatics/bth220. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Lin H, Li M. MANGO: a new approach to multiple sequence alignment. Comput Syst Bioinformatics Conf. 2007;6:237–47. [PubMed] [Google Scholar]
- 16.Bystroff C, Krogh A. Hidden Markov Models for prediction of protein features. Methods Mol Biol. 2008;413:173–98. doi: 10.1007/978-1-59745-574-9_7. [DOI] [PubMed] [Google Scholar]
- 17.Karplus K. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 2009 Jul 1;37(Web Server issue):W492–7. doi: 10.1093/nar/gkp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooney C, Pollastri G. Beyond the Twilight Zone: automated prediction of structural properties of proteins by recursive neural networks and remote homology information. Proteins. 2009 Oct;77(1):181–90. doi: 10.1002/prot.22429. [DOI] [PubMed] [Google Scholar]
- 19.Fariselli P, Rossi I, Capriotti E, Casadio R. The WWWH of remote homolog detection: the state of the art. Brief Bioinform. 2007 Mar;8(2):78–87. doi: 10.1093/bib/bbl032. [DOI] [PubMed] [Google Scholar]
- 20.Wan XF, Xu D. Computational methods for remote homolog identification. Curr Protein Pept Sci. 2005 Dec;6(6):527–46. doi: 10.2174/138920305774933231. [DOI] [PubMed] [Google Scholar]
- 21.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006 Oct;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bystroff C, Baker D. Prediction of local structure in proteins using a library of sequence-structure motifs. J Mol Biol. 1998 Aug 21;281(3):565–77. doi: 10.1006/jmbi.1998.1943. [DOI] [PubMed] [Google Scholar]
- 23.Levitt M. Accurate modeling of protein conformation by automatic segment matching. J Mol Biol. 1992 Jul 20;226(2):507–33. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Lee D, Park H, Coutsias EA, Seok C. Protein loop modeling by using fragment assembly and analytical loop closure. Proteins. Dec;78(16):3428–36. doi: 10.1002/prot.22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto CS, Fasnacht M, Zhu J, Forrest L, Honig B. Loop modeling: Sampling, filtering, and scoring. Proteins. 2008 Feb 15;70(3):834–43. doi: 10.1002/prot.21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnautova YA, Abagyan RA, Totrov M. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling. Proteins. Feb;79(2):477–98. doi: 10.1002/prot.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruccoleri RE. Ab initio loop modeling and its application to homology modeling. Methods Mol Biol. 2000;143:247–64. doi: 10.1385/1-59259-368-2:247. [DOI] [PubMed] [Google Scholar]
- 28.Phatak SS, Gatica EA, Cavasotto CN. Ligand-steered modeling and docking: a benchmarking study in class a g-protein-coupled receptors. J Chem Inf Model. Dec 27;50(12):2119–28. doi: 10.1021/ci100285f. [DOI] [PubMed] [Google Scholar]
- 29.Vilar S, Ferino G, Phatak SS, Berk B, Cavasotto CN, Costanzi S. Docking-based virtual screening for ligands of G protein-coupled receptors: not only crystal structures but also in silico models. J Mol Graph Model. Feb;29(5):614–23. doi: 10.1016/j.jmgm.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neves MA, Simoes S, Sae Melo ML. Ligand-guided optimization of CXCR4 homology models for virtual screening using a multiple chemotype approach. J Comput Aided Mol Des. Dec;24(12):1023–33. doi: 10.1007/s10822-010-9393-x. [DOI] [PubMed] [Google Scholar]
- 31.Michino M, Chen J, Stevens RC, Brooks CL., 3rd FoldGPCR: structure prediction protocol for the transmembrane domain of G protein-coupled receptors from class A. Proteins. Aug 1;78(10):2189–201. doi: 10.1002/prot.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Fan H, Periole X, Honig B, Mark AE. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins. 2008 Sep;72(4):1171–88. doi: 10.1002/prot.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannan S, Zacharias M. Application of biasing-potential replica-exchange simulations for loop modeling and refinement of proteins in explicit solvent. Proteins. Oct;78(13):2809–19. doi: 10.1002/prot.22796. [DOI] [PubMed] [Google Scholar]
- 34.Kimura SR, Tebben AJ, Langley DR. Expanding GPCR homology model binding sites via a balloon potential: A molecular dynamics refinement approach. Proteins. 2008 Jun;71(4):1919–29. doi: 10.1002/prot.21906. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell JBO. The relationship between the sequence identities of alpha helical proteins in the PDB and the molecular similarities of their ligands. Journal of Chemical Information and Computer Sciences. 2001 Nov-Dec;41(6):1617–22. doi: 10.1021/ci010364q. [DOI] [PubMed] [Google Scholar]
- 36.Dalton JA, Jackson RM. Homology-modelling protein-ligand interactions: allowing for ligand-induced conformational change. J Mol Biol. 2010 Jun 18;399(4):645–61. doi: 10.1016/j.jmb.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 37.Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the beta2-adrenergic receptor. J Med Chem. 2008 May 22;51(10):2907–14. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radestock S, Weil T, Renner S. Homology model-based virtual screening for GPCR ligands using docking and target-biased scoring. Journal of Chemical Information and Modeling. 2008 May;48(5):1104–17. doi: 10.1021/ci8000265. [DOI] [PubMed] [Google Scholar]
- 39.Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, et al. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J Med Chem. 2008 Feb 14;51(3):581–8. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi H, Akitaya T, Yu T, Kidachi Y, Kamiie K, Noshita T, et al. Homology modeling and structural analysis of 11beta-hydroxysteroid dehydrogenase type 2. Eur J Med Chem. Apr;46(4):1325–30. doi: 10.1016/j.ejmech.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 41.Williams AL, Bielopolski N, Meroz D, Lam AD, Passmore DR, Ben-Tal N, et al. Structural and functional analysis of tomosyn identifies domains important in exocytotic regulation. J Biol Chem. Feb 17; doi: 10.1074/jbc.M110.215624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meirelles GV, Silva JC, de Mendonca YA, Ramos CH, Torriani IL, Kobarg J. Human Nek6 is a monomeric mostly globular kinase with an unfolded short N-terminal domain. BMC Struct Biol. 11:12. doi: 10.1186/1472-6807-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costanzi S. Modeling G Protein-Coupled Receptors: a Concrete Possibility. Chim Oggi. 28(3):26–31. [PMC free article] [PubMed] [Google Scholar]
- 44.Ode H, Yokoyama M, Kanda T, Sato H. Identification of folding preferences of cleavage junctions of HIV-1 precursor proteins for regulation of cleavability. J Mol Model. Feb;17(2):391–9. doi: 10.1007/s00894-010-0739-z. [DOI] [PubMed] [Google Scholar]
- 45.Rutledge RM, Esser L, Ma J, Xia D. Toward understanding the mechanism of action of the yeast multidrug resistance transporter Pdr5p: a molecular modeling study. J Struct Biol. Feb;173(2):333–44. doi: 10.1016/j.jsb.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang JL, Zheng QC, Zhang HX. Theoretical improvement of the specific inhibitor of human carbonic anhydrase VII. Comput Biol Chem. Feb;35(1):50–6. doi: 10.1016/j.compbiolchem.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Tschammer N, Elsner J, Goetz A, Ehrlich K, Schuster S, Ruberg M, et al. Highly Potent 5-Aminotetrahydropyrazolopyridines: Enantioselective Dopamine D(3) Receptor Binding, Functional Selectivity, and Analysis of Receptor-Ligand Interactions. J Med Chem. Mar 9; doi: 10.1021/jm101639t. [DOI] [PubMed] [Google Scholar]
- 48.Wang Q, Mach RH, Luedtke RR, Reichert DE. Subtype selectivity of dopamine receptor ligands: insights from structure and ligand-based methods. J Chem Inf Model. Nov 22;50(11):1970–85. doi: 10.1021/ci1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA. Dual role of the second extracellular loop of the cannabinoid receptor 1: ligand binding and receptor localization. Mol Pharmacol. 2009 Oct;76(4):833–42. doi: 10.1124/mol.109.057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz P, Phatak SS, Xu J, Fronczek FR, Astruc-Diaz F, Thompson CM, et al. 2,3-Dihydro-1-benzofuran derivatives as a series of potent selective cannabinoid receptor 2 agonists: design, synthesis, and binding mode prediction through ligand-steered modeling. ChemMedChem. 2009 Oct;4(10):1615–29. doi: 10.1002/cmdc.200900226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diaz P, Phatak SS, Xu J, Astruc-Diaz F, Cavasotto CN, Naguib M. 6-Methoxy-N-alkyl isatin acylhydrazone derivatives as a novel series of potent selective cannabinoid receptor 2 inverse agonists: design, synthesis, and binding mode prediction. J Med Chem. 2009 Jan 22;52(2):433–44. doi: 10.1021/jm801353p. [DOI] [PubMed] [Google Scholar]
- 52.Ryu H, Jin MK, Kim SY, Choi HK, Kang SU, Kang DW, et al. Stereospecific high-affinity TRPV1 antagonists: chiral N-(2-benzyl-3-pivaloyloxypropyl) 2-[4-(methylsulfonylamino)phenyl]propionamide analogues. J Med Chem. 2008 Jan 10;51(1):57–67. doi: 10.1021/jm701049p. [DOI] [PubMed] [Google Scholar]
- 53.Sander T, Frolund B, Bruun AT, Ivanov I, McCammon JA, Balle T. New insights into the GABA(A) receptor structure and orthosteric ligand binding: Receptor modeling guided by experimental data. Proteins. Jan 4; doi: 10.1002/prot.22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levoin N, Calmels T, Poupardin-Olivier O, Labeeuw O, Danvy D, Robert P, et al. Refined docking as a valuable tool for lead optimization: application to histamine H3 receptor antagonists. Arch Pharm (Weinheim) 2008 Oct;341(10):610–23. doi: 10.1002/ardp.200800042. [DOI] [PubMed] [Google Scholar]
- 55.Sheng C, Wang W, Che X, Dong G, Wang S, Ji H, et al. Improved model of lanosterol 14alpha-demethylase by ligand-supported homology modeling: validation by virtual screening and azole optimization. ChemMedChem. Mar 1;5(3):390–7. doi: 10.1002/cmdc.200900468. [DOI] [PubMed] [Google Scholar]
- 56.Nowak P, Cole DC, Brooijmans N, Bursavich MG, Curran KJ, Ellingboe JW, et al. Discovery of potent and selective inhibitors of the mammalian target of rapamycin (mTOR) kinase. J Med Chem. 2009 Nov 26;52(22):7081–9. doi: 10.1021/jm9012642. [DOI] [PubMed] [Google Scholar]
- 57.Bonneau R, Baker D. Ab initio protein structure prediction: progress and prospects. Annu Rev Biophys Biomol Struct. 2001;30:173–89. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- 58.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005 Sep 16;309(5742):1868–71. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 59.Shmygelska A, Levitt M. Generalized ensemble methods for de novo structure prediction. Proc Natl Acad Sci U S A. 2009 Feb 3;106(5):1415–20. doi: 10.1073/pnas.0812510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins-Structure Function and Genetics. 1999:171–6. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- 61.Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69( Suppl 8):118–28. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 62.Kim DE, Blum B, Bradley P, Baker D. Sampling bottlenecks in de novo protein structure prediction. J Mol Biol. 2009 Oct 16;393(1):249–60. doi: 10.1016/j.jmb.2009.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamoto Y. Generalized-ensemble algorithms: enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J Mol Graph Model. 2004 May;22(5):425–39. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Fukunishi H, Watanabe O, Takada S. On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: Application to protein structure prediction. Journal of Chemical Physics. 2002 May 22;116(20):9058–67. [Google Scholar]
- 65.Fujitsuka Y, Chikenji G, Takada S. SimFold energy function for de novo protein structure prediction: consensus with Rosetta. Proteins. 2006 Feb 1;62(2):381–98. doi: 10.1002/prot.20748. [DOI] [PubMed] [Google Scholar]
- 66.Ngan SC, Hung LH, Liu T, Samudrala R. Scoring functions for de novo protein structure prediction revisited. Methods Mol Biol. 2008;413:243–81. doi: 10.1007/978-1-59745-574-9_10. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: a new approach to ab initio protein structure prediction. Biophys J. 2003 Aug;85(2):1145–64. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W526–31. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonneau R, Baliga NS, Deutsch EW, Shannon P, Hood L. Comprehensive de novo structure prediction in a systems-biology context for the archaea Halobacterium sp. NRC-1. Genome Biol. 2004;5(8):R52. doi: 10.1186/gb-2004-5-8-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77( Suppl 9):89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao M, Wang S, Wang C, Yuan X, Li S, Zheng W, et al. Incorporating Ab Initio energy into threading approaches for protein structure prediction. BMC Bioinformatics. 2011;12(Suppl 1):S54. doi: 10.1186/1471-2105-12-S1-S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bystroff C, Shao Y. Fully automated ab initio protein structure prediction using I-SITES, HMMSTR and ROSETTA. Bioinformatics. 2002;18( Suppl 1):S54–61. doi: 10.1093/bioinformatics/18.suppl_1.s54. [DOI] [PubMed] [Google Scholar]
- 73.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Berichte der deutschen chemischen Gesellschaft. 1894;27(3):2985–93. [Google Scholar]
- 74.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends in Biochemical Sciences. 2010 Oct;35(10):539–46. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003 Jul;2(7):527–41. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 76.Kroemer RT. Structure-based drug design: docking and scoring. Curr Protein Pept Sci. 2007 Aug;8(4):312–28. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- 77.Carlson HA, McCammon JA. Accommodating protein flexibility in computational drug design. Mol Pharmacol. 2000 Feb;57(2):213–8. [PubMed] [Google Scholar]
- 78.Chandrika BR, Subramanian J, Sharma SD. Managing protein flexibility in docking and its applications. Drug Discov Today. 2009 Apr;14(7–8):394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 79.Jiang F, Kim SH. “Soft docking”: matching of molecular surface cubes. J Mol Biol. 1991 May 5;219(1):79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- 80.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of Molecular Biology. 1995;245(1):43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 81.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring 1. Method and assessment of docking accuracy. J Med Chem. 2004 Mar 25;47(7):1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 82.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, et al. Glide: a new approach for rapid, accurate docking and scoring 2. Enrichment factors in database screening. J Med Chem. 2004 Mar 25;47(7):1750–9. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 83.Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol. 2009 Jan 16;385(2):381–92. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 84.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006 Nov 15;65(3):538–48. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 85.Ding F, Yin S, Dokholyan NV. Rapid Flexible Docking Using a Stochastic Rotamer Library of Ligands. Journal of Chemical Information and Modeling. 2010;50(9):1623–32. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran HT, Zhang S. Accurate prediction of the bound form of the akt pleckstrin homology domain using normal mode analysis to explore structural flexibility. J Chem Inf Model. 2011 Sep 26;51(9):2352–60. doi: 10.1021/ci2001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barril X, Morley SD. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. J Med Chem. 2005 Jun 30;48(13):4432–43. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 88.Sivanesan D, Rajnarayanan RV, Doherty J, Pattabiraman N. In-silico screening using flexible ligand binding pockets: a molecular dynamics-based approach. J Comput Aided Mol Des. 2005 Apr;19(4):213–28. doi: 10.1007/s10822-005-4788-9. [DOI] [PubMed] [Google Scholar]
- 89.Guida WC, Hamilton AD, Crotty JW, Sebti SM. Protein farnesyltransferase: flexible docking studies on inhibitors using computational modeling. J Comput Aided Mol Des. 2005 Dec;19(12):871–85. doi: 10.1007/s10822-005-9030-2. [DOI] [PubMed] [Google Scholar]
- 90.Salameh BA, Cumpstey I, Sundin A, Leffler H, Nilsson UJ. 1H-1,2,3-triazol-1-yl thiodigalactoside derivatives as high affinity galectin-3 inhibitors. Bioorg Med Chem. 2010 Jul 15;18(14):5367–78. doi: 10.1016/j.bmc.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 91.Lin JH, Perryman AL, Schames JR, McCammon JA. Computational drug design accommodating receptor flexibility: the relaxed complex scheme. J Am Chem Soc. 2002 May;124:5632–3. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 92.Okimoto N, Futatsugi N, Fuji H, Suenaga A, Morimoto G, Yanai R, et al. High-performance drug discovery: computational screening by combining docking and molecular dynamics simulations. PLoS Comput Biol. 2009 Oct;5:1000528. doi: 10.1371/journal.pcbi.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314(1–2):141–51. [Google Scholar]
- 94.Gerek ZN, Ozkan SB. A flexible docking scheme to explore the binding selectivity of PDZ domains. Protein Sci. May;19:914–28. doi: 10.1002/pro.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Okumura H, Gallicchio E, Levy RM. Conformational populations of ligand-sized molecules by replica exchange molecular dynamics and temperature reweighting. J Comput Chem. May;31:1357–67. doi: 10.1002/jcc.21419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J Med Chem. 2009 Jan 22;52(2):397–406. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Subramanian J, Sharma S, CBR A novel computational analysis of ligand-induced conformational changes in the ATP binding sites of cyclin dependent kinases. J Med Chem. 2006 Sep 7;49(18):5434–41. doi: 10.1021/jm060172s. [DOI] [PubMed] [Google Scholar]
- 98.Subramanian J, Sharma S, CBR Modeling and selection of flexible proteins for structure-based drug design: backbone and side chain movements in p38 MAPK. ChemMedChem. 2008 Feb;3(2):336–44. doi: 10.1002/cmdc.200700255. [DOI] [PubMed] [Google Scholar]
- 99.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19(14):1639–62. [Google Scholar]
- 100.Yuriev E, Agostino M, Ramsland PA. Challenges and advances in computational docking: 2009 in review. J Mol Recognit. Oct 23; doi: 10.1002/jmr.1077. [DOI] [PubMed] [Google Scholar]
- 101.Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992 Aug 21;257(5073):1078–82. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 102.Jiang X, Kumar K, Hu X, Wallqvist A, Reifman J. DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chem Cent J. 2008;2:18. doi: 10.1186/1752-153X-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang S, Kumar K, Jiang X, Wallqvist A, Reifman J. DOVIS: an implementation for high-throughput virtual screening using AutoDock. BMC Bioinformatics. 2008;9:126. doi: 10.1186/1471-2105-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abagyan R, Totrov M, Kuznetsov D. Icm - a New Method for Protein Modeling and Design - Applications to Docking and Structure Prediction from the Distorted Native Conformation. Journal of Computational Chemistry. 1994 May;15(5):488–506. [Google Scholar]
- 105.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003 Feb 13;46(4):499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 106.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996 Aug 23;261(3):470–89. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 107.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Gaussian docking functions. Biopolymers. 2003 Jan;68(1):76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 108.Zsoldos Z, Reid D, Simon A, Sadjad BS, Johnson AP. eHiTS: an innovative approach to the docking and scoring function problems. Curr Protein Pept Sci. 2006 Oct;7(5):421–35. doi: 10.2174/138920306778559412. [DOI] [PubMed] [Google Scholar]
- 109.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model. 2003 Jan;21(4):289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 110.Perola E, Walters WP, Charifson PS. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins. 2004 Aug 1;56(2):235–49. doi: 10.1002/prot.20088. [DOI] [PubMed] [Google Scholar]
- 111.Miteva MA, Lee WH, Montes MO, Villoutreix BO. Fast structure-based virtual ligand screening combining FRED, DOCK, and Surflex. J Med Chem. 2005 Sep 22;48(19):6012–22. doi: 10.1021/jm050262h. [DOI] [PubMed] [Google Scholar]
- 112.Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Docking: successes and challenges. Curr Pharm Des. 2005;11(3):323–33. doi: 10.2174/1381612053382106. [DOI] [PubMed] [Google Scholar]
- 113.Plewczynski D, ŁaŸniewski M, Augustyniak R, Ginalski K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. Journal of Computational Chemistry. 2010;32(4):742–55. doi: 10.1002/jcc.21643. [DOI] [PubMed] [Google Scholar]
- 114.Cheng T, Li X, Li Y, Liu Z, Wang R. Comparative Assessment of Scoring Functions on a Diverse Test Set. Journal of Chemical Information and Modeling. 2009;49(4):1079–93. doi: 10.1021/ci9000053. [DOI] [PubMed] [Google Scholar]
- 115.Wang R, Lu Y, Fang X, Wang S. An extensive test of 14 scoring functions using the PDBbind refined set of 800 protein-ligand complexes. J Chem Inf Comput Sci. 2004 Nov-Dec;44(6):2114–25. doi: 10.1021/ci049733j. [DOI] [PubMed] [Google Scholar]
- 116.Zhang S, Golbraikh A, Tropsha A. Development of quantitative structure-binding affinity relationship models based on novel geometrical chemical descriptors of the protein-ligand interfaces. J Med Chem. 2006 May 4;49(9):2713–24. doi: 10.1021/jm050260x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moses SA, Ali MA, Zuohe S, Du-Cuny L, Zhou LL, Lemos R, et al. In vitro and in vivo activity of novel small-molecule inhibitors targeting the pleckstrin homology domain of protein kinase B/AKT. Cancer Res. 2009 Jun 15;69(12):5073–81. doi: 10.1158/0008-5472.CAN-08-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meuillet EJ, Zuohe S, Lemos R, Ihle N, Kingston J, Watkins R, et al. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol Cancer Ther. 2010 Mar;9(3):706–17. doi: 10.1158/1535-7163.MCT-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Prakhov ND, Chernorudskiy AL, Gainullin MR. VSDocker: a tool for parallel high-throughput virtual screening using AutoDock on Windows-based computer clusters. Bioinformatics. May 15;26(10):1374–5. doi: 10.1093/bioinformatics/btq149. [DOI] [PubMed] [Google Scholar]
- 120.Buch I, Harvey MJ, Giorgino T, Anderson DP, De Fabritiis G. High-throughput all-atom molecular dynamics simulations using distributed computing. J Chem Inf Model. 2010 Mar 22;50(3):397–403. doi: 10.1021/ci900455r. [DOI] [PubMed] [Google Scholar]
- 121.Estrada T, Armen R, Taufer M. Automatic selection of near-native protein-ligand conformations using a hierarchical clustering and volunteer computing. Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology; Niagara Falls, New York: ACM; 2010. [Google Scholar]
- 122.Keseru GM, Makara GM. The influence of lead discovery strategies on the properties of drug candidates. Nat Rev Drug Discov. 2009 Mar;8(3):203–12. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- 123.Lipkin MJ, Stevens AP, Livingstone DJ, Harris CJ. How large does a compound screening collection need to be? Comb Chem High Throughput Screen. 2008 Jul;11(6):482–93. doi: 10.2174/138620708784911492. [DOI] [PubMed] [Google Scholar]
- 124.Schnecke V, Bostrom J. Computational chemistry-driven decision making in lead generation. Drug Discov Today. 2006 Jan;11(1–2):43–50. doi: 10.1016/S1359-6446(05)03703-7. [DOI] [PubMed] [Google Scholar]
- 125.Davies JW, Glick M, Jenkins JL. Streamlining lead discovery by aligning in silico and high-throughput screening. Curr Opin Chem Biol. 2006 Aug;10(4):343–51. doi: 10.1016/j.cbpa.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 126.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009 Oct;9(5):580–8. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 127.Janzen WP, Popa-Burke IG. Advances in improving the quality and flexibility of compound management. J Biomol Screen. 2009 Jun;14(5):444–51. doi: 10.1177/1087057109335262. [DOI] [PubMed] [Google Scholar]
- 128.Cavasotto CN, Phatak SS. Docking methods for structure-based library design. Methods Mol Biol. 685:155–74. doi: 10.1007/978-1-60761-931-4_8. [DOI] [PubMed] [Google Scholar]
- 129.Schnur DM. Recent trends in library design: ‘rational design’ revisited. Curr Opin Drug Discov Devel. 2008 May;11(3):375–80. [PubMed] [Google Scholar]
- 130.Manjasetty BA, Turnbull AP, Panjikar S, Bussow K, Chance MR. Automated technologies and novel techniques to accelerate protein crystallography for structural genomics. Proteomics. 2008 Feb;8(4):612–25. doi: 10.1002/pmic.200700687. [DOI] [PubMed] [Google Scholar]
- 131.Gileadi O, Knapp S, Lee WH, Marsden BD, Muller S, Niesen FH, et al. The scientific impact of the Structural Genomics Consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J Struct Funct Genomics. 2007 Sep;8(2–3):107–19. doi: 10.1007/s10969-007-9027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moos WH, Hurt CR, Morales GA. Combinatorial chemistry: oh what a decade or two can do. Mol Divers. 2009 May;13(2):241–5. doi: 10.1007/s11030-009-9127-y. [DOI] [PubMed] [Google Scholar]
- 133.Hajduk PJ, Galloway WR, Spring DR. Drug discovery: A question of library design. Nature. Feb 3;470(7332):42–3. doi: 10.1038/470042a. [DOI] [PubMed] [Google Scholar]
- 134.Hartenfeller M, Schneider G. De novo drug design. Methods Mol Biol. 672:299–323. doi: 10.1007/978-1-60761-839-3_12. [DOI] [PubMed] [Google Scholar]
- 135.Schulz MN, Hubbard RE. Recent progress in fragment-based lead discovery. Curr Opin Pharmacol. 2009 Oct;9(5):615–21. doi: 10.1016/j.coph.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 136.Dandapani S, Marcaurelle LA. Current strategies for diversity-oriented synthesis. Curr Opin Chem Biol. Jun;14(3):362–70. doi: 10.1016/j.cbpa.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 137.Spandl RJ, Bender A, Spring DR. Diversity-oriented synthesis; a spectrum of approaches and results. Org Biomol Chem. 2008 Apr 7;6(7):1149–58. doi: 10.1039/b719372f. [DOI] [PubMed] [Google Scholar]
- 138.Loving K, Alberts I, Sherman W. Computational approaches for fragment-based and de novo design. Curr Top Med Chem. 2010;10(1):14–32. doi: 10.2174/156802610790232305. [DOI] [PubMed] [Google Scholar]
- 139.Chen H, Engkvist O, Blomberg N. Combinatorial library design from reagent pharmacophore fingerprints. Methods Mol Biol. 685:135–52. doi: 10.1007/978-1-60761-931-4_7. [DOI] [PubMed] [Google Scholar]
- 140.Chen H, Borjesson U, Engkvist O, Kogej T, Svensson MA, Blomberg N, et al. ProSAR: a new methodology for combinatorial library design. J Chem Inf Model. 2009 Mar;49(3):603–14. doi: 10.1021/ci800231d. [DOI] [PubMed] [Google Scholar]
- 141.Ebalunode JO, Zheng W, Tropsha A. Application of QSAR and shape pharmacophore modeling approaches for targeted chemical library design. Methods Mol Biol. 685:111–33. doi: 10.1007/978-1-60761-931-4_6. [DOI] [PubMed] [Google Scholar]
- 142.Sun Y, Ewing TJ, Skillman AG, Kuntz ID. CombiDOCK: structure-based combinatorial docking and library design. J Comput Aided Mol Des. 1998 Nov;12(6):597–604. doi: 10.1023/a:1008036704754. [DOI] [PubMed] [Google Scholar]
- 143.Makino S, Ewing TJ, Kuntz ID. DREAM++: flexible docking program for virtual combinatorial libraries. J Comput Aided Mol Des. 1999 Sep;13(5):513–32. doi: 10.1023/a:1008066310669. [DOI] [PubMed] [Google Scholar]
- 144.Roe DC, Kuntz ID. BUILDER v.2: improving the chemistry of a de novo design strategy. J Comput Aided Mol Des. 1995 Jun;9(3):269–82. doi: 10.1007/BF00124457. [DOI] [PubMed] [Google Scholar]
- 145.Sprous DG, Lowis DR, Leonard JM, Heritage T, Burkett SN, Baker DS, et al. OptiDock: virtual HTS of combinatorial libraries by efficient sampling of binding modes in product space. J Comb Chem. 2004 Jul-Aug;6(4):530–9. doi: 10.1021/cc034068x. [DOI] [PubMed] [Google Scholar]
- 146.Grzybowski BA, Ishchenko AV, Kim CY, Topalov G, Chapman R, Christianson DW, et al. Combinatorial computational method gives new picomolar ligands for a known enzyme. Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1270–3. doi: 10.1073/pnas.032673399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yan S, Selliah R. Structure-based library design in efficient discovery of novel inhibitors. Methods Mol Biol. 685:175–90. doi: 10.1007/978-1-60761-931-4_9. [DOI] [PubMed] [Google Scholar]
- 148.Xing L, McDonald JJ, Kolodziej SA, Kurumbail RG, Williams JM, Warren CJ, et al. Discovery of potent inhibitors of soluble epoxide hydrolase by combinatorial library design and structure-based virtual screening. J Med Chem. Mar 10;54(5):1211–22. doi: 10.1021/jm101382t. [DOI] [PubMed] [Google Scholar]
- 149.Cheng N, Brantley DM, Chen J. The ephrins and Eph receptors in angiogenesis. Cytokine Growth Factor Rev. 2002 Feb;13(1):75–85. doi: 10.1016/s1359-6101(01)00031-4. [DOI] [PubMed] [Google Scholar]
- 150.Kolb P, Kipouros CB, Huang D, Caflisch A. Structure-based tailoring of compound libraries for high-throughput screening: discovery of novel EphB4 kinase inhibitors. Proteins. 2008 Oct;73(1):11–8. doi: 10.1002/prot.22028. [DOI] [PubMed] [Google Scholar]
- 151.Shah F, Mukherjee P, Gut J, Legac J, Rosenthal PJ, Tekwani BL, et al. Identification of Novel Malarial Cysteine Protease Inhibitors Using Structure-Based Virtual Screening of a Focused Cysteine Protease Inhibitor Library. J Chem Inf Model. Mar 23; doi: 10.1021/ci200029y. [DOI] [PubMed] [Google Scholar]
- 152.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005 Jan-Feb;45(1):160–9. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 153.Lajiness MS, Johnson MA, Maggiora GM. Implementing drug screening programs using molecular similarity methods. Prog Clin Biol Res. 1989;291:173–6. [PubMed] [Google Scholar]
- 154.Yang SY. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today. 2011 Jun;15(11–12):444–50. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 155.Hou T, Xu X. Recent development and application of virtual screening in drug discovery: an overview. Curr Pharm Des. 2004;10(9):1011–33. doi: 10.2174/1381612043452721. [DOI] [PubMed] [Google Scholar]
- 156.Gao Q, Yang L, Zhu Y. Pharmacophore based drug design approach as a practical process in drug discovery. Curr Comput Aided Drug Des. 2010 Mar;6(1):37–49. doi: 10.2174/157340910790980151. [DOI] [PubMed] [Google Scholar]
- 157.Schuster D, Wolber G. Identification of bioactive natural products by pharmacophore-based virtual screening. Curr Pharm Des. 2010 May;16(15):1666–81. doi: 10.2174/138161210791164072. [DOI] [PubMed] [Google Scholar]
- 158.Mason JS, Good AC, Martin EJ. 3-D pharmacophores in drug discovery. Curr Pharm Des. 2001 May;7(7):567–97. doi: 10.2174/1381612013397843. [DOI] [PubMed] [Google Scholar]
- 159.Wolber G, Seidel T, Bendix F, Langer T. Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov Today. 2008 Jan;13(1–2):23–9. doi: 10.1016/j.drudis.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 160.Leach AR, Gillet VJ, Lewis RA, Taylor R. Three-dimensional pharmacophore methods in drug discovery. J Med Chem. 2010 Jan 28;53(2):539–58. doi: 10.1021/jm900817u. [DOI] [PubMed] [Google Scholar]
- 161.Cruciani G, Carosati E, De Boeck B, Ethirajulu K, Mackie C, Howe T, et al. MetaSite: understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem. 2005 Nov 3;48(22):6970–9. doi: 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- 162.Sato T, Honma T, Yokoyama S. Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J Chem Inf Model. Jan;50(1):170–85. doi: 10.1021/ci900382e. [DOI] [PubMed] [Google Scholar]
- 163.Steindl TM, Schuster D, Wolber G, Laggner C, Langer T. High-throughput structure-based pharmacophore modelling as a basis for successful parallel virtual screening. J Comput Aided Mol Des. 2006 Dec;20(12):703–15. doi: 10.1007/s10822-006-9066-y. [DOI] [PubMed] [Google Scholar]
- 164.Hadfield AT, Diana GD, Rossmann MG. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci U S A. 1999 Dec 21;96(26):14730–5. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Barreca ML, De Luca L, Iraci N, Rao A, Ferro S, Maga G, et al. Structure-based pharmacophore identification of new chemical scaffolds as non-nucleoside reverse transcriptase inhibitors. J Chem Inf Model. 2007 Mar-Apr;47(2):557–62. doi: 10.1021/ci600320q. [DOI] [PubMed] [Google Scholar]
- 166.Yang H, Shen Y, Chen J, Jiang Q, Leng Y, Shen J. Structure-based virtual screening for identification of novel 11beta-HSD1 inhibitors. Eur J Med Chem. 2009 Mar;44(3):1167–71. doi: 10.1016/j.ejmech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 167.De Luca L, Barreca ML, Ferro S, Christ F, Iraci N, Gitto R, et al. Pharmacophore-based discovery of small-molecule inhibitors of protein-protein interactions between HIV-1 integrase and cellular cofactor LEDGF/p75. ChemMedChem. 2009 Aug;4(8):1311–6. doi: 10.1002/cmdc.200900070. [DOI] [PubMed] [Google Scholar]
- 168.Mason JS, Morize I, Menard PR, Cheney DL, Hulme C, Labaudiniere RF. New 4-point pharmacophore method for molecular similarity and diversity applications: overview of the method and applications, including a novel approach to the design of combinatorial libraries containing privileged substructures. J Med Chem. 1999 Aug 26;42(17):3251–64. doi: 10.1021/jm9806998. [DOI] [PubMed] [Google Scholar]
- 169.Evans BE, Rittle KE, Bock MG, DiPardo RM, Freidinger RM, Whitter WL, et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem. 1988 Dec;31(12):2235–46. doi: 10.1021/jm00120a002. [DOI] [PubMed] [Google Scholar]
- 170.Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J Chem Inf Model. 2007 Mar-Apr;47(2):279–94. doi: 10.1021/ci600253e. [DOI] [PubMed] [Google Scholar]
- 171.Cross S, Baroni M, Carosati E, Benedetti P, Clementi S. FLAP: GRID molecular interaction fields in virtual screening. validation using the DUD data set. J Chem Inf Model. 2010 Aug 23;50(8):1442–50. doi: 10.1021/ci100221g. [DOI] [PubMed] [Google Scholar]
- 172.Weill N, Rognan D. Development and validation of a novel protein-ligand fingerprint to mine chemogenomic space: application to G protein-coupled receptors and their ligands. J Chem Inf Model. 2009 Apr;49(4):1049–62. doi: 10.1021/ci800447g. [DOI] [PubMed] [Google Scholar]
- 173.Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat Rev Drug Discov. 2005 Aug;4(8):649–63. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 174.Honma T, Hayashi K, Aoyama T, Hashimoto N, Machida T, Fukasawa K, et al. Structure-based generation of a new class of potent Cdk4 inhibitors: new de novo design strategy and library design. J Med Chem. 2001 Dec 20;44(26):4615–27. doi: 10.1021/jm0103256. [DOI] [PubMed] [Google Scholar]
- 175.Firth-Clark S, Kirton SB, Willems HM, Williams A. De novo ligand design to partially flexible active sites: application of the ReFlex algorithm to carboxypeptidase A, acetylcholinesterase, and the estrogen receptor. J Chem Inf Model. 2008 Feb;48(2):296–305. doi: 10.1021/ci700282u. [DOI] [PubMed] [Google Scholar]
- 176.Firth-Clark S, Willems HM, Williams A, Harris W. Generation and selection of novel estrogen receptor ligands using the de novo structure-based design tool, SkelGen. J Chem Inf Model. 2006 Mar-Apr;46(2):642–7. doi: 10.1021/ci0502956. [DOI] [PubMed] [Google Scholar]
- 177.Liebeschuetz JW, Jones SD, Morgan PJ, Murray CW, Rimmer AD, Roscoe JM, et al. PRO_SELECT: combining structure-based drug design and array-based chemistry for rapid lead discovery 2. The development of a series of highly potent and selective factor Xa inhibitors. J Med Chem. 2002 Mar 14;45(6):1221–32. doi: 10.1021/jm010944e. [DOI] [PubMed] [Google Scholar]
- 178.Vinkers HM, de Jonge MR, Daeyaert FF, Heeres J, Koymans LM, van Lenthe JH, et al. SYNOPSIS: SYNthesize and OPtimize System in Silico. J Med Chem. 2003 Jun 19;46(13):2765–73. doi: 10.1021/jm030809x. [DOI] [PubMed] [Google Scholar]
- 179.Lowinger TB, Riedl B, Dumas J, Smith RA. Design and discovery of small molecules targeting raf-1 kinase. Curr Pharm Des. 2002;8(25):2269–78. doi: 10.2174/1381612023393125. [DOI] [PubMed] [Google Scholar]
- 180.Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Discov Today. 2003 Oct 1;8(19):876–7. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 181.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005 Jun 2;435(7042):677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 182.Braisted AC, Oslob JD, Delano WL, Hyde J, McDowell RS, Waal N, et al. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J Am Chem Soc. 2003 Apr 2;125(13):3714–5. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 183.Nishibata Y, Itai A. Automatic creation of drug candidate structures based on receptor structure. Starting point for artificial lead generation. Tetrahedron. 1991;47(43):8985–90. [Google Scholar]
- 184.Luo Z, Wang R, Lai L. RASSE: a new method for structure-based drug design. J Chem Inf Comput Sci. 1996 Nov-Dec;36(6):1187–94. doi: 10.1021/ci950277w. [DOI] [PubMed] [Google Scholar]
- 185.Zhu J, Fan H, Liu H, Shi Y. Structure-based ligand design for flexible proteins: application of new F-DycoBlock. J Comput Aided Mol Des. 2001 Nov;15(11):979–96. doi: 10.1023/a:1014817911249. [DOI] [PubMed] [Google Scholar]
- 186.Bohm HJ. LUDI: rule-based automatic design of new substituents for enzyme inhibitor leads. J Comput Aided Mol Des. 1992 Dec;6(6):593–606. doi: 10.1007/BF00126217. [DOI] [PubMed] [Google Scholar]
- 187.Bohm HJ. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J Comput Aided Mol Des. 1992 Feb;6(1):61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 188.Wang R, Gao Y, Lai L. LigBuilder: A Multi-Purpose Program for Structure-Based Drug Design. Journal of Molecular Modeling. 2000;6(7):498–516. [Google Scholar]
- 189.Gillet V, Johnson AP, Mata P, Sike S, Williams P. SPROUT: a program for structure generation. J Comput Aided Mol Des. 1993 Apr;7(2):127–53. doi: 10.1007/BF00126441. [DOI] [PubMed] [Google Scholar]
- 190.Pearlman DA, Murcko MA. CONCERTS: dynamic connection of fragments as an approach to de novo ligand design. J Med Chem. 1996 Apr 12;39(8):1651–63. doi: 10.1021/jm950792l. [DOI] [PubMed] [Google Scholar]
- 191.DeWitte R, Shakhnovich E. SMoG: de Novo Design Method Based on Simple, Fast, and Accurate Free Energy Estimates 1. Methodology and Supporting Evidence. Journal of the American Chemical Society. 1996;118(47):11733–44. [Google Scholar]
- 192.Dey F, Caflisch A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J Chem Inf Model. 2008 Mar;48(3):679–90. doi: 10.1021/ci700424b. [DOI] [PubMed] [Google Scholar]
- 193.Ni S, Yuan Y, Huang J, Mao X, Lv M, Zhu J, et al. Discovering potent small molecule inhibitors of cyclophilin A using de novo drug design approach. J Med Chem. 2009 Sep 10;52(17):5295–8. doi: 10.1021/jm9008295. [DOI] [PubMed] [Google Scholar]
- 194.Alberts IL, Todorov NP, Dean PM. Receptor flexibility in de novo ligand design and docking. J Med Chem. 2005 Oct 20;48(21):6585–96. doi: 10.1021/jm050196j. [DOI] [PubMed] [Google Scholar]
- 195.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010 Jul 22;53(14):5061–84. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Scatena LF, Brown MG, Richmond GL. Water at hydrophobic surfaces: Weak hydrogen bonding and strong orientation effects. Science. 2001 May 4;292(5518):908–12. doi: 10.1126/science.1059514. [DOI] [PubMed] [Google Scholar]
- 197.Dill KA. Additivity principles in biochemistry. J Biol Chem. 1997 Jan 10;272(2):701–4. doi: 10.1074/jbc.272.2.701. [DOI] [PubMed] [Google Scholar]
- 198.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 199.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. Phys Chem Chem Phys. 2006 Nov 28;8(44):5166–77. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- 200.Noy E, Senderowitz H. Molecular Simulations for the Evaluation of Binding Free Energies in Lead Optimization. Drug Develop Res. 2011 Feb;72(1):36–44. [Google Scholar]
- 201.Lindahl E, Sansom MS. Membrane proteins: molecular dynamics simulations. Curr Opin Struct Biol. 2008 Aug;18(4):425–31. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 202.Jiao D, Golubkov PA, Darden TA, Ren P. Calculation of protein-ligand binding free energy by using a polarizable potential. Proc Natl Acad Sci USA. 2008 Apr;105:6290–5. doi: 10.1073/pnas.0711686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jiao D, Zhang J, Duke RE, Li G, Schnieders MJ, Ren P. Trypsin-ligand binding free energies from explicit and implicit solvent simulations with polarizable potential. J Comput Chem. 2009 Aug;30:1701–11. doi: 10.1002/jcc.21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Leach AR. Molecular Modelling: Principles and Applications. Dorchester, Dorset, UK: Pearson Education Limited; 2001. [Google Scholar]
- 205.Frenkel D, Smit B. Understanding Molecular Simulation, Second Edition: From Algorithms to Applications. San Diego, California, USA: Academic Press; 2002. [Google Scholar]
- 206.Allen MP, Tildesley DJ. Computer Simulation of Liquids. New York, USA: Oxford University Press; 1989. [Google Scholar]
- 207.Shirts MR, Pande VS. Comparison of efficiency and bias of free energies computed by exponential averaging, the Bennett acceptance ratio, and thermodynamic integration. J Chem Phys. 2005 Apr;122:144107. doi: 10.1063/1.1873592. [DOI] [PubMed] [Google Scholar]
- 208.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009 Feb;113:2234–46. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hess B, Kutzner C, Spoel Dvd, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4(3):435–47. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 210.Fujitani H, Tanida Y, Ito M, Jayachandran G, Snow CD, Shirts MR, et al. Direct calculation of the binding free energies of FKBP ligands. J Chem Phys. 2005 Aug;123:084108. doi: 10.1063/1.1999637. [DOI] [PubMed] [Google Scholar]
- 211.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008 Apr;108(4):1171–224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 212.Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed Engl. 2002 Aug;41(15):2644–76. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 213.Jorgensen WL. Efficient drug lead discovery and optimization. Acc Chem Res. 2009 Jun;42(6):724–33. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tirado-Rives J, Jorgensen WL. Contribution of conformer focusing to the uncertainty in predicting free energies for protein-ligand binding. J Med Chem. 2006 Oct;49(20):5880–4. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 215.Mitomo D, Fukunishi Y, Higo J, Nakamura H. Calculation of protein-ligand binding free energy using smooth reaction path generation (SRPG) method: a comparison of the explicit water model, gb/sa model and docking score function. Genome Inform. 2009 Oct;23(1):85–97. [PubMed] [Google Scholar]
- 216.Okimoto N, Futatsugi N, Fuji H, Suenaga A, Morimoto G, Yanai R, et al. High-performance drug discovery: computational screening by combining docking and molecular dynamics simulations. PLoS Comput Biol. 2009 Oct;5(10):e1000528. doi: 10.1371/journal.pcbi.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Aqvist J, Luzhkov VB, Brandsdal BO. Ligand binding affinities from MD simulations. Acc Chem Res. 2002 Jun;35(6):358–65. doi: 10.1021/ar010014p. [DOI] [PubMed] [Google Scholar]
- 218.Hou T, Wang J, Li Y, Wang W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J Chem Inf Model. 2010 Nov; doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Brown SP, Muchmore SW. High-throughput calculation of protein-ligand binding affinities: modification and adaptation of the MM-PBSA protocol to enterprise grid computing. J Chem Inf Model. 2006 May-Jun;46(3):999–1005. doi: 10.1021/ci050488t. [DOI] [PubMed] [Google Scholar]
- 220.Brown SP, Muchmore SW. Rapid estimation of relative protein-ligand binding affinities using a high-throughput version of MM-PBSA. J Chem Inf Model. 2007 Jul-Aug;47(4):1493–503. doi: 10.1021/ci700041j. [DOI] [PubMed] [Google Scholar]
- 221.Nymeyer H, Garcia AE. Simulation of the folding equilibrium of alpha-helical peptides: a comparison of the generalized Born approximation with explicit solvent. Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):13934–9. doi: 10.1073/pnas.2232868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: A major success of structure-assisted drug design. Annual Review of Biophysics and Biomolecular Structure. 1998;27:249–84. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 223.Villoutreix BO, Renault N, Lagorce D, Sperandio O, Montes M, Miteva MA. Free resources to assist structure-based virtual ligand screening experiments. Curr Protein Pept Sci. 2007 Aug;8(4):381–411. doi: 10.2174/138920307781369391. [DOI] [PubMed] [Google Scholar]
- 224.Talele TT, Khedkar SA, Rigby AC. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr Top Med Chem. 2009;10(1):127–41. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- 225.van Montfort RL, Workman P. Structure-based design of molecular cancer therapeutics. Trends Biotechnol. 2009 May;27(5):315–28. doi: 10.1016/j.tibtech.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 226.Jones SD, Liebeschuetz JW, Morgan PJ, Murray CW, Rimmer AD, Roscoe JM, et al. The design of phenylglycine containing benzamidine carboxamides as potent and selective inhibitors of factor Xa. Bioorg Med Chem Lett. 2001 Mar 12;11(5):733–6. doi: 10.1016/s0960-894x(01)00042-7. [DOI] [PubMed] [Google Scholar]
- 227.Canan Koch SS, Thoresen LH, Tikhe JG, Maegley KA, Almassy RJ, Li J, et al. Novel tricyclic poly(ADP-ribose) polymerase-1 inhibitors with potent anticancer chemopotentiating activity: design, synthesis, and X-ray cocrystal structure. J Med Chem. 2002 Nov 7;45(23):4961–74. doi: 10.1021/jm020259n. [DOI] [PubMed] [Google Scholar]
- 228.Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD(+) binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998 Mar 17;37(11):3893–900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 229.Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007 Mar;6(3):945–56. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 230.Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004 May;29(5):233–42. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 231.Morrow JK, Du-Cuny L, Chen L, Meuillet EJ, Mash EA, Powis G, et al. Recent development of anticancer therapeutics targeting Akt. Recent Pat Anticancer Drug Discov. 2011 Jan;6(1):146–59. doi: 10.2174/157489211793980079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Du-Cuny L, Song Z, Moses S, Powis G, Mash EA, Meuillet EJ, et al. Computational modeling of novel inhibitors targeting the Akt pleckstrin homology domain. Bioorg Med Chem. 2009 Oct 1;17(19):6983–92. doi: 10.1016/j.bmc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ahad AM, Zuohe S, Du-Cuny L, Moses SA, Zhou LL, Zhang S, et al. Development of sulfonamide AKT PH domain inhibitors. Bioorg Med Chem. 2011 Mar 15;19(6):2046–54. doi: 10.1016/j.bmc.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Azizi M, Guyene TT, Chatellier G, Menard J. Blood pressure effects of acute intravenous renin or oral angiotensin converting enzyme inhibition in essential hypertension. J Hypertens. 1994 Apr;12(4):419–27. [PubMed] [Google Scholar]
- 235.Cohen NC. Structure-based drug design and the discovery of aliskiren (Tekturna): perseverance and creativity to overcome a R&D pipeline challenge. Chem Biol Drug Des. 2007 Dec;70(6):557–65. doi: 10.1111/j.1747-0285.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- 236.Perucca E, Yasothan U, Clincke G, Kirkpatrick P. Lacosamide. Nat Rev Drug Discov. 2008 Dec;7(12):973–4. doi: 10.1038/nrd2764. [DOI] [PubMed] [Google Scholar]
- 237.Odell LR, Howan D, Gordon CP, Robertson MJ, Chau N, Mariana A, et al. The pthaladyns: GTP competitive inhibitors of dynamin I and II GTPase derived from virtual screening. J Med Chem. 2010 Jul 22;53(14):5267–80. doi: 10.1021/jm100442u. [DOI] [PubMed] [Google Scholar]
- 238.Engels K, Beyer C, Suarez Fernandez ML, Bender F, Gassel M, Unden G, et al. Inhibition of Eimeria tenella CDK-related kinase 2: From target identification to lead compounds. ChemMedChem. Aug 2;5(8):1259–71. doi: 10.1002/cmdc.201000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sala E, Guasch L, Iwaszkiewicz J, Mulero M, Salvado MJ, Pinent M, et al. Identification of Human IKK-2 Inhibitors of Natural Origin (Part I): Modeling of the IKK-2 Kinase Domain, Virtual Screening and Activity Assays. PLoS One. 6(2):e16903. doi: 10.1371/journal.pone.0016903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Park H, Bahn YJ, Jung SK, Jeong DG, Lee SH, Seo I, et al. Discovery of novel Cdc25 phosphatase inhibitors with micromolar activity based on the structure-based virtual screening. J Med Chem. 2008 Sep 25;51(18):5533–41. doi: 10.1021/jm701157g. [DOI] [PubMed] [Google Scholar]
- 241.Park H, Hwang KY, Oh KH, Kim YH, Lee JY, Kim K. Discovery of novel alpha-glucosidase inhibitors based on the virtual screening with the homology-modeled protein structure. Bioorg Med Chem. 2008 Jan 1;16(1):284–92. doi: 10.1016/j.bmc.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 242.Sierecki E, Sinko W, McCammon JA, Newton AC. Discovery of small molecule inhibitors of the PH domain leucine-rich repeat protein phosphatase (PHLPP) by chemical and virtual screening. J Med Chem. Oct 14;53(19):6899–911. doi: 10.1021/jm100331d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mortier J, Masereel B, Remouchamps C, Ganeff C, Piette J, Frederick R. NF-kappaB inducing kinase (NIK) inhibitors: identification of new scaffolds using virtual screening. Bioorg Med Chem Lett. Aug 1;20(15):4515–20. doi: 10.1016/j.bmcl.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 244.Sisay MT, Steinmetzer T, Stirnberg M, Maurer E, Hammami M, Bajorath J, et al. Identification of the first low-molecular-weight inhibitors of matriptase-2. J Med Chem. 2010 Aug 12;53(15):5523–35. doi: 10.1021/jm100183e. [DOI] [PubMed] [Google Scholar]
- 245.Kim JH, Lim JW, Lee SW, Kim K, No KT. Ligand supported homology modeling and docking evaluation of CCR2: docked pose selection by consensus scoring. J Mol Model. Jan 7; doi: 10.1007/s00894-010-0943-x. [DOI] [PubMed] [Google Scholar]
- 246.Klabunde T, Giegerich C, Evers A. Sequence-derived three-dimensional pharmacophore models for G-protein-coupled receptors and their application in virtual screening. J Med Chem. 2009 May 14;52(9):2923–32. doi: 10.1021/jm9001346. [DOI] [PubMed] [Google Scholar]
- 247.White AW, Westwell AD, Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:e8. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- 248.Blazer LL, Neubig RR. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009 Jan;34(1):126–41. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 249.Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13(22):2607–25. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 250.Siehler S. Cell-based assays in GPCR drug discovery. Biotechnol J. 2008 Apr;3(4):471–83. doi: 10.1002/biot.200800001. [DOI] [PubMed] [Google Scholar]
- 251.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002 Sep;1(9):727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 252.Sharma SK, Ramsey TM, Bair KW. Protein-protein interactions: lessons learned. Curr Med Chem Anticancer Agents. 2002 Mar;2(2):311–30. doi: 10.2174/1568011023354191. [DOI] [PubMed] [Google Scholar]
- 253.Keskin O, Gursoy A, Ma B, Nussinov R. Towards drugs targeting multiple proteins in a systems biology approach. Curr Top Med Chem. 2007;7(10):943–51. doi: 10.2174/156802607780906690. [DOI] [PubMed] [Google Scholar]
- 254.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995 Jan 20;267(5196):383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 255.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998 Jul 3;280(1):1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 256.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 257.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995 Apr 28;248(2):478–86. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 258.Stites WE. Proteinminus signProtein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chem Rev. 1997 Aug 5;97(5):1233–50. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 259.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol. 1998 Apr 17;277(5):1111–28. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 260.Hu Z, Ma B, Wolfson H, Nussinov R. Conservation of polar residues as hot spots at protein interfaces. Proteins. 2000 Jun 1;39(4):331–42. [PubMed] [Google Scholar]
- 261.Kouadio JL, Horn JR, Pal G, Kossiakoff AA. Shotgun alanine scanning shows that growth hormone can bind productively to its receptor through a drastically minimized interface. J Biol Chem. 2005 Jul 8;280(27):25524–32. doi: 10.1074/jbc.M502167200. [DOI] [PubMed] [Google Scholar]
- 262.Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001 Mar;17(3):284–5. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- 263.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002 Apr 26;296(5568):750–2. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 264.Panchenko AR, Kondrashov F, Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004 Apr;13(4):884–92. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007 Sep 1;68(4):803–12. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 266.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002 Feb;12(1):14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 267.Dinner AR, Sali A, Smith LJ, Dobson CM, Karplus M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem Sci. 2000 Jul;25(7):331–9. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
- 268.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000 Jan;9(1):10–9. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fischer TB, Arunachalam KV, Bailey D, Mangual V, Bakhru S, Russo R, et al. The binding interface database (BID): a compilation of amino acid hot spots in protein interfaces. Bioinformatics. 2003 Jul 22;19(11):1453–4. doi: 10.1093/bioinformatics/btg163. [DOI] [PubMed] [Google Scholar]
- 270.Piehler J, Schreiber G. Fast transient cytokine-receptor interactions monitored in real time by reflectometric interference spectroscopy. Anal Biochem. 2001 Feb 15;289(2):173–86. doi: 10.1006/abio.2000.4920. [DOI] [PubMed] [Google Scholar]
- 271.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000 Aug 1;97(16):8950–4. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002 Jul 5;320(2):369–87. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 273.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14116–21. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gao Y, Wang R, Lai L. Structure-based method for analyzing protein-protein interfaces. J Mol Model. 2004 Feb;10(1):44–54. doi: 10.1007/s00894-003-0168-3. [DOI] [PubMed] [Google Scholar]
- 275.Huo S, Massova I, Kollman PA. Computational alanine scanning of the 1:1 human growth hormone-receptor complex. J Comput Chem. 2002 Jan 15;23(1):15–27. doi: 10.1002/jcc.1153. [DOI] [PubMed] [Google Scholar]
- 276.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci U S A. 2004 Aug 3;101(31):11287–92. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kimura SR, Brower RC, Vajda S, Camacho CJ. Dynamical view of the positions of key side chains in protein-protein recognition. Biophys J. 2001 Feb;80(2):635–42. doi: 10.1016/S0006-3495(01)76044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Cho KI, Kim D, Lee D. A feature-based approach to modeling protein-protein interaction hot spots. Nucleic Acids Res. 2009 May;37(8):2672–87. doi: 10.1093/nar/gkp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004 Feb;Oct;2004(219):pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 280.Lise S, Buchan D, Pontil M, Jones DT. Predictions of hot spot residues at protein-protein interfaces using support vector machines. PLoS One. 2011;6(2):e16774. doi: 10.1371/journal.pone.0016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009 Jun 15;25(12):1513–20. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 282.Guney E, Tuncbag N, Keskin O, Gursoy A. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008 Jan;36(Database issue):D662–6. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005 Nov 3;438(7064):103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 284.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006 Mar 30;440(7084):637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 285.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007 Apr 12;446(7137):806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 286.Komurov K, White M. Revealing static and dynamic modular architecture of the eukaryotic protein interaction network. Mol Syst Biol. 2007;3:110. doi: 10.1038/msb4100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Tuncbag N, Kar G, Gursoy A, Keskin O, Nussinov R. Towards inferring time dimensionality in protein-protein interaction networks by integrating structures: the p53 example. Mol Biosyst. 2009 Dec;5(12):1770–8. doi: 10.1039/b905661k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009 Dec;5(12):e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006 Dec 22;314(5807):1938–41. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 290.Zhang, Wang H. MDM2 oncogene as a novel target for human cancer therapy. Curr Pharm Des. 2000 Mar;6(4):393–416. doi: 10.2174/1381612003400911. [DOI] [PubMed] [Google Scholar]
- 291.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996 Nov 8;274(5289):948–53. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 292.Popowicz GM, Domling A, Holak TA. The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew Chem Int Ed Engl. 2011 Mar 14;50(12):2680–8. doi: 10.1002/anie.201003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004 Feb 6;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 294.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005 Feb 24;48(4):909–12. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 295.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, et al. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc. 2005 Jul 27;127(29):10130–1. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 296.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006 Jun 15;49(12):3432–5. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 297.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009 Dec;9(12):862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 299.Lu Y, Nikolovska-Coleska Z, Fang X, Gao W, Shangary S, Qiu S, et al. Discovery of a nanomolar inhibitor of the human murine double minute 2 (MDM2)-p53 interaction through an integrated, virtual database screening strategy. J Med Chem. 2006 Jun 29;49(13):3759–62. doi: 10.1021/jm060023+. [DOI] [PubMed] [Google Scholar]
- 300.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998 Aug 28;281(5381):1322–6. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 301.Petros AM, Nettesheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000 Dec;9(12):2528–34. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997 Feb 14;275(5302):983–6. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 303.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004 Sep 3;305(5689):1466–70. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Sadowsky JD, Fairlie WD, Hadley EB, Lee HS, Umezawa N, Nikolovska-Coleska Z, et al. (alpha/beta+alpha)-peptide antagonists of BH3 domain/Bcl-x(L) recognition: toward general strategies for foldamer-based inhibition of protein-protein interactions. J Am Chem Soc. 2007 Jan 10;129(1):139–54. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]
- 305.Sadowsky JD, Murray JK, Tomita Y, Gellman SH. Exploration of backbone space in foldamers containing alpha- and beta-amino acid residues: developing protease-resistant oligomers that bind tightly to the BH3-recognition cleft of Bcl-xL. Chembiochem. 2007 May 25;8(8):903–16. doi: 10.1002/cbic.200600546. [DOI] [PubMed] [Google Scholar]
- 306.Yin H, Lee GI, Sedey KA, Kutzki O, Park HS, Orner BP, et al. Terphenyl-Based Bak BH3 alpha-helical proteomimetics as low-molecular-weight antagonists of Bcl-xL. J Am Chem Soc. 2005 Jul 27;127(29):10191–6. doi: 10.1021/ja050122x. [DOI] [PubMed] [Google Scholar]
- 307.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007 Dec 13;450(7172):1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 308.Hann CL, Daniel VC, Sugar EA, Dobromilskaya I, Murphy SC, Cope L, et al. Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer. Cancer Res. 2008 Apr 1;68(7):2321–8. doi: 10.1158/0008-5472.CAN-07-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008 May 1;68(9):3421–8. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 310.Wang JL, Liu D, Zhang ZJ, Shan S, Han X, Srinivasula SM, et al. Structure-based discovery of an organic compound that binds Bcl-2 protein and induces apoptosis of tumor cells. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7124–9. doi: 10.1073/pnas.97.13.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rega MF, Leone M, Jung D, Cotton NJ, Stebbins JL, Pellecchia M. Structure-based discovery of a new class of Bcl-xL antagonists. Bioorg Chem. 2007 Aug;35(4):344–53. doi: 10.1016/j.bioorg.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Mukherjee P, Desai P, Zhou YD, Avery M. Targeting the BH3 domain mediated protein-protein interaction of Bcl-xL through virtual screening. J Chem Inf Model. 2010 May 24;50(5):906–23. doi: 10.1021/ci1000373. [DOI] [PubMed] [Google Scholar]
- 313.Buron N, Porceddu M, Brabant M, Desgue D, Racoeur C, Lassalle M, et al. Use of Human Cancer Cell Lines Mitochondria to Explore the Mechanisms of BH3 Peptides and ABT-737-Induced Mitochondrial Membrane Permeabilization. Plos One. 2010 Mar 31;5(3) doi: 10.1371/journal.pone.0009924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Schimmer AD, Dalili S, Batey RA, Riedl SJ. Targeting XIAP for the treatment of malignancy. Cell Death Differ. 2006 Feb;13(2):179–88. doi: 10.1038/sj.cdd.4401826. [DOI] [PubMed] [Google Scholar]
- 315.Nikolovska-Coleska Z, Xu L, Hu ZJ, Tomita Y, Li P, Roller PP, et al. Discovery of embelin as a cell-permeable, small-molecular weight inhibitor of XLAP through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. Journal of Medicinal Chemistry. 2004 May 6;47(10):2430–40. doi: 10.1021/jm030420+. [DOI] [PubMed] [Google Scholar]
- 316.Chen J, Nikolovska-Coleska Z, Wang G, Qiu S, Wang S. Design, synthesis, and characterization of new embelin derivatives as potent inhibitors of X-linked inhibitor of apoptosis protein. Bioorg Med Chem Lett. 2006 Nov 15;16(22):5805–8. doi: 10.1016/j.bmcl.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 317.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003 Feb;11(2):519–27. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 318.Cohen F, Alicke B, Elliott LO, Flygare JA, Goncharov T, Keteltas SF, et al. Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold. J Med Chem. 2009 Mar 26;52(6):1723–30. doi: 10.1021/jm801450c. [DOI] [PubMed] [Google Scholar]
- 319.Huang JW, Zhang Z, Wu B, Cellitti JF, Zhang X, Dahl R, et al. Fragment-based design of small molecule X-linked inhibitor of apoptosis protein inhibitors. J Med Chem. 2008 Nov 27;51(22):7111–8. doi: 10.1021/jm8006992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995 Dec;2(12):1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 321.Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proceedings of the National Academy of Sciences of the United States of America. 1998 Dec 22;95(26):15613–7. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Hardy N, Skolnik PR. Enfuvirtide, a new fusion inhibitor for therapy of human immunodeficiency virus infection. Pharmacotherapy. 2004 Feb;24(2):198–211. doi: 10.1592/phco.24.2.198.33141. [DOI] [PubMed] [Google Scholar]
- 323.Debnath AK, Radigan L, Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J Med Chem. 1999 Aug 26;42(17):3203–9. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- 324.Zhao Q, Ernst JT, Hamilton AD, Debnath AK, Jiang SB. XTT formazan widely used to detect cell viability inhibits HIV type 1 infection in vitro by targeting gp41. Aids Research and Human Retroviruses. 2002 Sep 20;18(14):989–97. doi: 10.1089/08892220260235353. [DOI] [PubMed] [Google Scholar]
- 325.Jiang SB, Lu H, Liu SW, Zhao Q, He YX, Debnath AK. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrobial Agents and Chemotherapy. 2004 Nov;48(11):4349–59. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Jiang SB, Tala SR, Lu H, Abo-Dya NE, Avan I, Gyanda K, et al. Design, Synthesis, and Biological Activity of Novel 5-((Arylfuran/1H-pyrrol-2-yl)methylene)-2-thioxo-3-(3-(trifluoromethyl)phenyl)thiazolidin-4-ones as HIV-1 Fusion Inhibitors Targeting gp41. Journal of Medicinal Chemistry. 2011 Jan 27;54(2):572–9. doi: 10.1021/jm101014v. [DOI] [PubMed] [Google Scholar]
- 327.Katritzky AR, Tala SR, Lu H, Vakulenko AV, Chen QY, Sivapackiam J, et al. Design, synthesis, and structure-activity relationship of a novel series of 2-aryl 5-(4-oxo-3-phenethyl-2-thioxothiazolidinylidenemethyl)furans as HIV-1 entry inhibitors. J Med Chem. 2009 Dec 10;52(23):7631–9. doi: 10.1021/jm900450n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Dempke WCM, Heinemann V. Ras Mutational Status Is a Biomarker for Resistance to EGFR Inhibitors in Colorectal Carcinoma. Anticancer Research. 2010 Nov;30(11):4673–7. [PubMed] [Google Scholar]
- 329.Spoerner M, Graf T, Konig B, Kalbitzer HR. A novel mechanism for the modulation of the Ras-effector interaction by small molecules. Biochemical and Biophysical Research Communications. 2005 Aug 26;334(2):709–13. doi: 10.1016/j.bbrc.2005.06.144. [DOI] [PubMed] [Google Scholar]
- 330.Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, et al. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2002 Oct 29;99(22):14398–403. doi: 10.1073/pnas.222222699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Ki SW, Kasahara K, Kwon HJ, Eishima J, Takesako K, Cooper JA, et al. Identification of radicicol as an inhibitor of in vivo Ras/Raf interaction with the yeast two-hybrid screening system. Journal of Antibiotics. 1998 Oct;51(10):936–44. doi: 10.7164/antibiotics.51.936. [DOI] [PubMed] [Google Scholar]
- 332.Ohnishi M, Yamawaki-Kataoka Y, Kariya K, Tamada M, Hu CD, Kataoka T. Selective inhibition of ras interaction with its particular effector by synthetic peptides corresponding to the ras effector region. Journal of Biological Chemistry. 1998 Apr 24;273(17):10210–5. doi: 10.1074/jbc.273.17.10210. [DOI] [PubMed] [Google Scholar]
- 333.Zeng J, Nheu T, Zorzet A, Catimel B, Nice E, Maruta H, et al. Design of inhibitors of Ras-Raf interaction using a computational combinatorial algorithm. Protein Engineering. 2001 Jan;14(1):39–45. doi: 10.1093/protein/14.1.39. [DOI] [PubMed] [Google Scholar]
- 334.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004 May 18;101(20):7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ferri N, Corsini A, Bottino P, Clerici F, Contini A. Virtual screening approach for the identification of new Rac1 inhibitors. J Med Chem. 2009 Jul 23;52(14):4087–90. doi: 10.1021/jm8015987. [DOI] [PubMed] [Google Scholar]
- 336.Rosnizeck IC, Graf T, Spoerner M, Trankle J, Filchtinski D, Herrmann C, et al. Stabilizing a Weak Binding State for Effectors in the Human Ras Protein by Cyclen Complexes. Angewandte Chemie-International Edition. 2010;49(22):3830–3. doi: 10.1002/anie.200907002. [DOI] [PubMed] [Google Scholar]
- 337.Xiao H, Feng JN, Yu ZY, Zhang L, Yu M, He XH, et al. A5, a new small-molecule inhibitor of CD4 D1 obtained from a computer-aided screening method, contributes to the inhibition of CD4(+) T-cell function. Journal of Biomolecular Screening. 2007 Sep;12(6):800–8. doi: 10.1177/1087057107305505. [DOI] [PubMed] [Google Scholar]
- 338.Tilley JW, Chen L, Fry DC, Emerson SD, Powers GD, Biondi D, et al. Identification of a small molecule inhibitor of the IL-2/IL-2R alpha receptor interaction which binds to IL-2. Journal of the American Chemical Society. 1997 Aug 13;119(32):7589–90. [Google Scholar]
- 339.Wilson CGM, Arkin MR. Small-Molecule Inhibitors of IL-2/IL-2R: Lessons Learned and Applied. Small-Molecule Inhibitors of Protein-Protein Interactions. 2011;348:25–59. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- 340.Fujii N, Haresco JJ, Novak KA, Stokoe D, Kuntz ID, Guy RK. A selective irreversible inhibitor targeting a PDZ protein interaction domain. J Am Chem Soc. 2003 Oct 8;125(40):12074–5. doi: 10.1021/ja035540l. [DOI] [PubMed] [Google Scholar]
- 341.Fujii N, Haresco JJ, Novak KA, Gage RM, Pedemonte N, Stokoe D, et al. Rational design of a nonpeptide general chemical scaffold for reversible inhibition of PDZ domain interactions. Bioorg Med Chem Lett. 2007 Jan 15;17(2):549–52. doi: 10.1016/j.bmcl.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 342.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007 Oct;25(10):1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 343.Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness--the Achilles heel of drug-target networks. Nat Biotechnol. 2008 Sep;26(9):983–4. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 344.Hauschild A, Agarwala SS, Trefzer U, Hogg D, Robert C, Hersey P, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J Clin Oncol. 2009 Jun 10;27(17):2823–30. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 345.Strumberg D, Voliotis D, Moeller JG, Hilger RA, Richly H, Kredtke S, et al. Results of phase I pharmacokinetic and pharmacodynamic studies of the Raf kinase inhibitor BAY 43-9006 in patients with solid tumors. Int J Clin Pharmacol Ther. 2002 Dec;40(12):580–1. doi: 10.5414/cpp40580. [DOI] [PubMed] [Google Scholar]
- 346.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, et al. Predicting new molecular targets for known drugs. Nature. 2009 Nov 12;462(7270):175–81. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Boran AD, Iyengar R. Systems approaches to polypharmacology and drug discovery. Curr Opin Drug Discov Devel. May;13(3):297–309. [PMC free article] [PubMed] [Google Scholar]
- 348.Barabasi AL. Scale-free networks: a decade and beyond. Science. 2009 Jul 24;325(5939):412–3. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 349.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol. 2008 Nov;4(11):691–9. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Durrant JD, Amaro RE, Xie L, Urbaniak MD, Ferguson MA, Haapalainen A, et al. A multidimensional strategy to detect polypharmacological targets in the absence of structural and sequence homology. PLoS Comput Biol. Jan;6(1):e1000648. doi: 10.1371/journal.pcbi.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Wallach I, Jaitly N, Lilien R. A structure-based approach for mapping adverse drug reactions to the perturbation of underlying biological pathways. PLoS One. 5(8):e12063. doi: 10.1371/journal.pone.0012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Tian L, Zhang S. Mapping drug-target interaction networks. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:2336–9. doi: 10.1109/IEMBS.2009.5335053. [DOI] [PubMed] [Google Scholar]
- 353.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006 Jul;24(7):805–15. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 354.Morrow JK, Tian L, Zhang S. Molecular networks in drug discovery. Crit Rev Biomed Eng. 38(2):143–56. doi: 10.1615/critrevbiomedeng.v38.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Metz JT, Hajduk PJ. Rational approaches to targeted polypharmacology: creating and navigating protein-ligand interaction networks. Curr Opin Chem Biol. Aug;14(4):498–504. doi: 10.1016/j.cbpa.2010.06.166. [DOI] [PubMed] [Google Scholar]
- 356.Winger JA, Hantschel O, Superti-Furga G, Kuriyan J. The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2) BMC Struct Biol. 2009;9:7. doi: 10.1186/1472-6807-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Chen B, Wild D, Guha R. PubChem as a source of polypharmacology. J Chem Inf Model. 2009 Sep;49(9):2044–55. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- 358.Cleves AE, Jain AN. Robust ligand-based modeling of the biological targets of known drugs. J Med Chem. 2006 May 18;49(10):2921–38. doi: 10.1021/jm051139t. [DOI] [PubMed] [Google Scholar]
- 359.Hu Y, Bajorath J. Molecular scaffolds with high propensity to form multi-target activity cliffs. J Chem Inf Model. Apr 26;50(4):500–10. doi: 10.1021/ci100059q. [DOI] [PubMed] [Google Scholar]
- 360.Milletti F, Vulpetti A. Predicting polypharmacology by binding site similarity: from kinases to the protein universe. J Chem Inf Model. Aug 23;50(8):1418–31. doi: 10.1021/ci1001263. [DOI] [PubMed] [Google Scholar]
- 361.Plake C, Schroeder M. Computational polypharmacology with text mining and ontologies. Curr Pharm Biotechnol. Mar 1;12(3):449–57. doi: 10.2174/138920111794480624. [DOI] [PubMed] [Google Scholar]
- 362.Chen B, Dong X, Jiao D, Wang H, Zhu Q, Ding Y, et al. Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinformatics. 11:255. doi: 10.1186/1471-2105-11-255. [DOI] [PMC free article] [PubMed] [Google Scholar]