Background: Interferon-induced transmembrane (IFITM) proteins are viral restriction factors with controversial topology.
Results: Ifitm3 with an intracellular N terminus and extracellular C terminus is the predominant form and exhibits viral restriction activity.
Conclusion: Ifitm3 is a type II transmembrane protein.
Significance: An understanding of the interaction of Ifitm3 with potential targets and cofactors is predicated on a correct understanding of its topology.
Keywords: Flow Cytometry, Interferon, Membrane Proteins, Microscopic Imaging, Virus Entry, Ifitm3
Abstract
The interferon-induced transmembrane (IFITM) proteins are a family of small membrane proteins that inhibit the cellular entry of several genera of viruses. These proteins had been predicted to adopt a two-pass, type III transmembrane topology with an intracellular loop, two transmembrane helices (TM1 and TM2), and extracellular N and C termini. Recent work, however, supports an intramembrane topology for the helices with cytosolic orientation of both termini. Here we determined the topology of murine Ifitm3. We found that the N terminus of Ifitm3 could be stained by antibodies at the cell surface but that this conformation was cell type-dependent and represented a minority of the total plasma membrane pool. In contrast, the C terminus was readily accessible to antibodies at the cell surface and extracellular C termini comprised most or all of those present at the plasma membrane. The addition of a C-terminal KDEL endoplasmic reticulum retention motif to Ifitm3 resulted in sequestration of Ifitm3 in the ER, demonstrating an ER-luminal orientation of the C terminus. C-terminal, but not N-terminal, epitope tags were also degraded within lysosomes, consistent with their luminal orientation. Furthermore, epitope-tagged Ifitm3 TM2 functioned as a signal anchor sequence when expressed in isolation. Collectively, our results demonstrate a type II transmembrane topology for Ifitm3 and will provide insight into its interaction with potential targets and cofactors.
Introduction
The interferon-induced transmembrane proteins are a family of small membrane proteins with immunologic and developmental roles. Humans have five IFITM loci, IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10 (1). IFITM1, -2, and -3, collectively termed immunity-related IFITMs,2 were recently recognized as viral restriction factors with an unusually broad activity against a wide variety of viruses (2). To date, IFITM proteins have been shown to restrict cellular entry of flaviviruses, filoviruses, influenza A virus, vesicular stomatitis virus, bunyaviruses, and reovirus. Other viruses, including alphaviruses, arenaviruses such as lymphocytic choriomeningitis virus, and the retrovirus murine leukemia virus (MLV), are unaffected (3–9). A few studies have reported HIV-1 restriction, but differ on which cell lines and IFITM proteins confer restriction, and uncertainty remains as to whether the activity is specifically entry-related (3, 10, 11). The immune-related IFITM proteins are, so far, unique among viral restriction factors in that they inhibit viral entry. In the case of enveloped viruses, they act prior to host/virus membrane fusion (12). Deleterious polymorphisms in human IFITM3 are associated with poorer outcomes for patients hospitalized with pneumonia (13, 14). IFITM5, which, despite its name, is not interferon-induced, plays a role in osteogenesis, with mutations resulting in osteogenesis imperfecta type V (15, 16) The function of IFITM10 is currently unknown.
Mice possess orthologs of all five human IFITM genes and two additional genes of unknown function: Ifitm6 and Ifitm7 (1). Knock-out mice with deletions of Ifitm3 alone or Ifitm1, -2, -3, -5, and -6 combined (IfitmDel) are viable and have known deficiencies in the control of influenza A viral infection (13, 17–19). Although most murine Ifitm proteins exhibit some degree of viral restriction activity when overexpressed in vitro, only Ifitm3 appears to contribute to the control of influenza in vivo (17). Ifitm3 is also unique among the murine Ifitm genes in that it, alone, is induced by interferons and acute phase cytokines (17).
Recent studies have begun to provide mechanistic insight into the antiviral activities of the IFITM proteins. In most cell lines, IFITM proteins concentrate in late endosomes and lysosomes where the viruses whose entry they restrict are thought to fuse (4, 12, 20). IFITM protein overexpression results in enlargement of perinuclear CD63 (cluster of differentiation 63) and lysosome-associated membrane protein 2 (LAMP2)-positive vesicles (4, 20, 21). Analysis of IFITM protein-enriched vesicles by fluorescence-lifetime imaging microscopy reveals that the membranes of these structures are less fluid than those of IFITM protein-free compartments (12). Amini-Bavil-Olyaee et al. (21) recently reported that these vesicles were highly enriched with cholesterol. They also identified a physical interaction between IFITM3 and VAMP (vesicle-associated membrane protein)-associated protein A (VAPA) and proposed that this interaction was responsible for the accumulation of cholesterol in IFITM protein-enriched endosomes and lysosomes by disrupting the association between VAMP-associated protein A and oxysterol-binding protein (21). Based on these findings, it appears that IFITM proteins may inhibit viral entry by altering the properties of endolysosomal limiting membranes, thus creating unfavorable energetics for viral membrane fusion.
IFITM proteins consist of a relatively long, hydrophilic N terminus, a conserved region consisting of a hydrophobic helix and cytosolic loop, a second hydrophobic helix, and a comparatively short, hydrophilic C terminus (1). Both helices are palmitoylated, and the palmitoylation is necessary for optimal viral restriction activity (22).
These proteins had been predicted to assume a type III, two-pass transmembrane topology with both N and C termini oriented toward the ER lumen or extracellular space (see Ref. 23 for a review of transmembrane protein topology nomenclature). Indeed, early investigators noted that antibodies against the N terminus of Leu-13 (IFITM1) caused aggregation of leukemia cells, suggesting exposure of extracellular IFITM1 epitopes (24, 25). Following enzymatic, cell surface radiolabeling, labeled IFITM1 could also be recovered by immunoprecipitation (26). Other groups have noted the presence of epitope-tagged IFITM protein N termini at the cell surface by flow cytometry (12).
Recently, however, two key studies provided substantial evidence in favor of a cytosolic N terminus. Yount et al. (27) demonstrated ubiquitylation of Lys-24 of the N terminus of IFITM3, implying access of this residue to cytosolic ubiquitin ligases, and also noted a lack of N-linked glycosylation of the NHT motif of the N terminus. Another critical piece of evidence was provided by Jia et al. (11), who revealed that tyrosine phosphorylation of IFITM3 Tyr-20 is necessary for its proper internalization into endolysosomal compartments. Furthermore, they were able to identify Fyn as the relevant kinase, again, consistent with the accessibility of the N terminus to cytosolic enzymes (11).
Yount et al. (27) also proposed a cytosolic orientation of the IFITM3 C terminus based on the ability of a prenylation reporter to bind an artificial C-terminal CLVL motif. The CLVL mutant, however, was studied only in conjunction with the introduction of an N-terminal myristoylation motif and simultaneous deletion of the palmitoylation sites, complicating interpretation of this finding (27). In another study, a cysteine near the C-terminal end of the predicted second transmembrane domain of the murine Ifitm1 was found to be palmitoylated, suggesting an intramembrane conformation for the C terminus of the molecule (28). A diagram of the two models for IFITM protein topology and a summary of the evidence for each are provided in Fig. 1.
FIGURE 1.
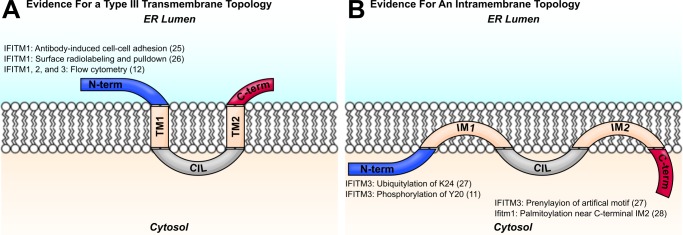
Two models of IFITM protein topology are shown. Evidence for the orientation of each terminus with respect to the membrane is shown with the relevant citation. A, IFITM proteins were predicted to assume a two-pass type III transmembrane topology, with two transmembrane domains (TM1 and TM2) flanking the conserved intracellular loop (CIL). Early studies identified IFITM1 N termini in the extracellular space, and other investigators have detected IFITM N termini at the plasma membrane by flow cytometry. (See figure for references.) B, more recent studies support a topology with two intramembrane domains (IM1 and IM2) and cytosolic N and C termini. Post-translational modifications of the IFITM3 N terminus imply access of the N terminus to cytosolic enzymes. Evidence for an intracellular C terminus comes from studies of lipidation motif reporters. (See figure for references.)
Here we focused our studies on murine Ifitm3 because of the availability of knock-out murine embryonic fibroblasts (MEFs) and high affinity antisera. The antiviral activity of Ifitm3 in vitro (in both human and murine cells) and in vivo is well established (4, 13, 17), and the amino acid sequence is similar to that of human IFITM3 (1). We show that there exists a population of Ifitm3 molecules at the plasma membrane with extracellular N termini, consistent with earlier studies, but this conformation is apparently rare and possibly cell-type dependent. The Ifitm3 C terminus, however, is clearly extracellular at the plasma membrane and is also exposed to both the ER and the lysosomal lumina. We conclude that Ifitm3 is a type II transmembrane protein.
EXPERIMENTAL PROCEDURES
Generation of Stable Cells
HEK 293T cells were cultured in DMEM with 10% FBS and penicillin/streptomycin. MEFs derived from IfitmDel knock-out mice were grown in DMEM with 10% FBS, nonessential amino acid supplement (Life Technologies), and 55 μm β-mercaptoethanol. Cells were transduced with vesicular stomatitis virus glycoprotein-pseudotyped murine retrovirus particles as described previously (29). Retrovirus genomes were derived from vector pQCXIP (Clontech) containing variants of Ifitm3. Transduced cells were selected with either 2 μg/ml puromycin (293T) or 4 μg/ml puromycin (MEF), and all assays were performed within 1–2 weeks of drug selection to minimize potential artifacts of prolonged transgene expression.
Flow Cytometry
Surface staining for epitope tags was conducted on cells detached with nonenzymatic cell dissociation solution (Sigma) for 30 min at room temperature and blocked with 5% heat-inactivated goat serum on ice for 20 min prior to staining. Primary antibodies used were anti-Myc clone 9E10, anti-C9 clone 1D4 (Santa Cruz Biotechnology), anti-FLAG clone M2 (Sigma), or mouse IgG1 isotype control clone P3.6.2.8.1 (eBioscience). Secondary staining was performed with Alexa Fluor 488- or 633-labeled secondary antibodies of goat origin (Life Technologies). To assess membrane integrity, cells were incubated for 10 min on ice in the presence of 1 μg/ml propidium iodide and assayed in the same solution. Flow cytometry was performed with a BD Accuri C6 flow cytometer (BD Biosciences).
Western Blots
Cells were lysed in 1% Triton X-100 in PBS containing Complete protease inhibitor mixture (Roche Applied Science). Where indicated, peptide-N-glycosidase digestion was performed with enzyme and buffers from New England Biolabs according to the manufacturer's instructions. Immunoblotting for the native Ifitm3 N terminus was performed with goat anti-Ifitm3 (R&D Systems) and HRP-conjugated rabbit anti-goat (Sigma). Tubulin was stained with mouse clone SAP.4G5 (Sigma) and HRP-conjugated goat anti-mouse (Santa Cruz Biotechnology). Images were captured at 16-bit depth using an LAS4000 mini chemiluminescence reader (GE Healthcare) and windowed/leveled in ImageJ (30).
Fluorescence Microscopy
Cells were grown on 8-well glass chamber slides (LAB-TEK). For fixation and permeabilization experiments, fixation was performed using the FIX & PERM kit reagents (Life Technologies) at 1:1 and 1:3 dilutions in PBS, respectively, for 15 min each at room temperature. Otherwise, cells were fixed with a 1:1 mixture of methanol and acetone for 10 min at −20 °C. Serine palmitoyl transferase 1 (Sptlc1) cDNA was obtained from the Dana-Farber/Harvard Cancer Center DNA Resource Core (deposited by the Mammalian Genome Collection). Myc-tagged Ifitm3 was stained with clone 9E10 or 9E11, C9 tag was stained with clone 1D4, and FLAG-tagged Sptlc1 was stained with clone M2. α-Tubulin was detected with polyclonal rabbit antibodies (GeneTex). Alexa Fluor 488- or 568-conjugated antibodies of goat origin were used for secondary fluorescence staining (Life Technologies). Slides were coverslipped with ProLong Gold antifade medium with DAPI (Life Technologies) and imaged by wide-field epifluorescence. 16-bit grayscale images were pseudocolored, windowed, leveled, and composited in ImageJ.
Pseudovirus Entry Assay
Pseudotyped retroviruses encoding eGFP were prepared as described previously (29) using the indicated viral entry proteins. Envelope proteins included hemagglutinins derived from influenza virus A/Thailand/2(SP-33)/2004 (H5), neuraminidase from influenza virus A/PR/8/34 (N1), and glycoproteins from vesicular stomatitis virus, lymphocytic choriomeningitis virus, and MLV. Infection of MEFs was carried out with 0.45 μm-filtered supernatants containing 5 μg/ml Polybrene at 4000 × g at 4 °C for 1 h. MEFs were then returned to the 37 °C, 5% CO2 incubator for 2 h followed by removal of the inoculum and return to growth medium. 24 h later, cells were harvested by trypsinization, and the percentage of eGFP-positive cells was determined by flow cytometry.
RESULTS
Localization of Ifitm3 N and C Termini to the Extracellular Space
We first determined whether N- or C-terminal epitope tags added to Ifitm3 could be detected extracellularly at the plasma membrane. We included critical controls to rule out potential artifactual positive results. Specifically, we took measures to exclude 1) positive staining resulting from membrane permeabilization and access of antibodies to intracellular epitopes and 2) artifactual staining due to membrane-adherent exosomes or cellular debris.
We first created stable cell lines from HEK 293T cells and Ifitm knock-out murine embryonic fibroblasts (IfitmDel MEFs) expressing various epitope-tagged Ifitm3 constructs. Cells expressing N- or C-terminal FLAG-tagged Ifitm3 (termed FLAG-Ifitm3 and Ifitm3-FLAG, respectively) were stained by indirect immunofluorescence.
To ensure staining of only extracellular epitope tags, we incubated cells in propidium iodide (PI) prior to flow cytometry. PI is a membrane-impermeant, nucleic acid-binding dye with a molecular mass of ∼0.7 kDa. PI-negative cells, therefore, are cells whose membranes are intact and would therefore be expected to block access of antibodies (molecular mass ∼150 kDa) to intracellular epitopes.
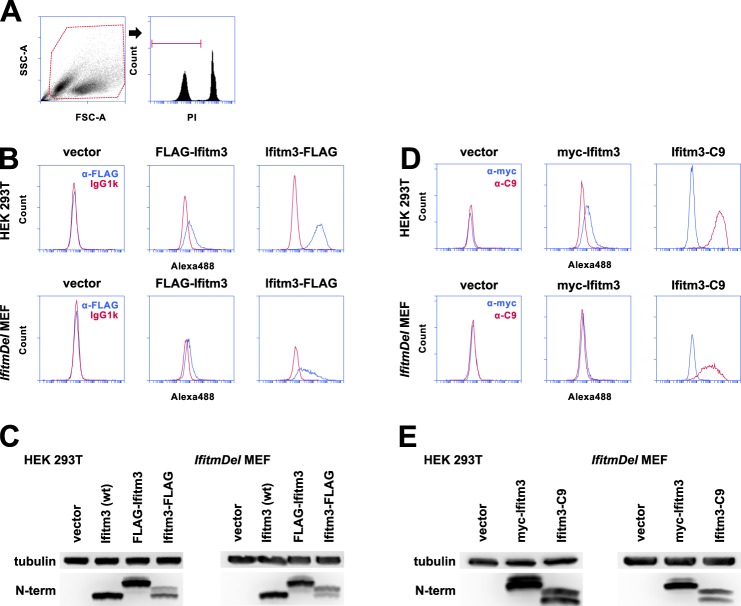
Both N-terminal and C-terminal FLAG tags could be detected on the surface of PI− 293T cells, albeit with a much greater intensity for staining of the C-terminal tag (Fig. 2A). Staining of the Ifitm3 N terminus in MEFs was very weak despite a robust signal for the C terminus (Fig. 2B). Total cellular expression of the N- and C-terminal tagged constructs was assessed by Western blot (Fig. 2C). These results were not specific to the FLAG epitope; the same staining pattern was observed in both cell lines when the FLAG tags were replaced by Myc and C9 on the N and C termini, respectively (Fig. 2, D and E).
FIGURE 2.
FLAG-tagged Ifitm3 N and C termini can be detected at the surface of intact cells. A, the gating strategy used to produce subsequent histograms is shown here. To confirm staining of only extracellular FLAG antigen, PI-positive cells were excluded from analysis. SSC-A, side scatter; FSC-A, forward scatter. B, staining for the FLAG epitopes on 293T cells and MEFs is shown in blue. Fluorescence staining from the isotype control is shown in red. Alexa488, Alexa Fluor 488. C, expression levels of wild type and N- and C-terminally FLAG-tagged Ifitm3 were determined by Western blot with an antibody against the native Ifitm3 N terminus. D, the experiment shown in A was repeated with N-terminal Myc and C-terminal C9 tags in place of the FLAG epitopes. Myc staining is shown in blue, and C9 staining is shown in red. E, total expression of the constructs was determined by Western blot. The double band pattern for C-terminally tagged Ifitm3 constructs is explored in detail in Fig. 6.
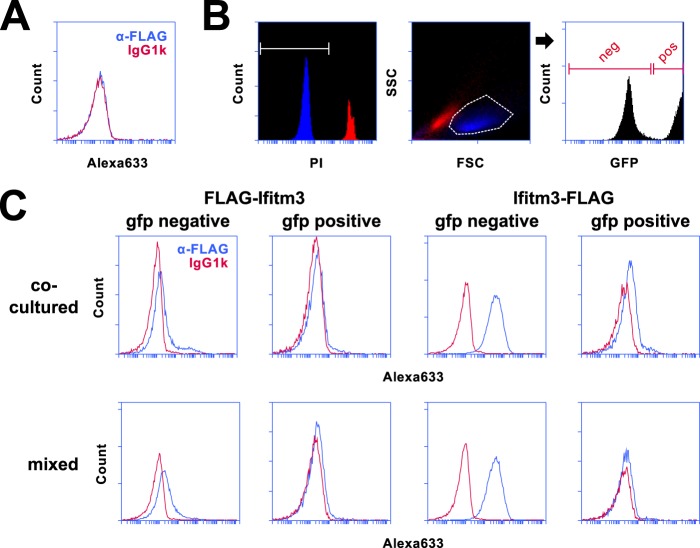
We next ruled out the possibility that the surface staining observed in Fig. 2 was the result of adherent cellular debris (derived from exosomes or dead cells) deposited on the plasma membrane. We generated a line of 293T cells stably expressing eGFP to see whether these cells could acquire the FLAG epitope from Ifitm3 stable cells. First, we verified the lack of any nonspecific FLAG staining on the surface of the eGFP+ cells (Fig. 3A). Because eGFP fluorescence spilled over into the PI channel, complicating our ability to identify intact cells by vital staining, we verified that intact 293T cells could be effectively identified by morphology (i.e. forward and side scatter gating) alone. In Fig. 3B, we show both PI-positive and PI-negative 293T cells plotted against the forward and side scatter axes. Intact (PI−) and permeabilized (PI+) 293T cells could be clearly distinguished on the basis of scatter gating alone. Therefore, by scatter gating followed by eGFP gating, we could isolate populations of intact eGFP+ or eGFP− cells. Furthermore, by indirect immunofluorescence for Ifitm3 epitope tags, we could distinguish cells expressing tagged Ifitm3 (Ifitm3+/eGFP−) or acquiring Ifitm3 from neighboring cells (Ifitm3−/eGFP+).
FIGURE 3.
Surface staining of N and C termini is not an artifact of membrane adherent cellular debris. A, staining of Ifitm3−/eGFP+ with anti-FLAG (blue) and isotype control (red) antibodies is shown. Alexa633, Alexa Fluor 633. B, PI− (intact) and PI+ (permeabilized) 293T cell populations were mapped onto a forward/side scatter plot (FSC/SSC). Intact 293T cells (blue) could be clearly distinguished from cells with compromised membrane integrity (red) by scatter gating alone. Scatter gating was used in lieu of PI staining to ensure analysis of only intact cells for during subsequent experiments. Cells were further sorted based on eGFP expression as shown in the rightmost histogram. C, cells expressing either FLAG-Ifitm3 or Ifitm3-FLAG were co-cultured with Ifitm3−/eGFP+ cells for 18 h (top panels) or mixed with Ifitm3−/eGFP+ cells immediately prior to surface staining (bottom panels). FLAG staining is shown in blue, and isotype control staining is shown in red.
We performed two variations of this experiment. In the first, Ifitm3−/eGFP+ cells were co-cultured with Ifitm3+/eGFP− cells for 18 h prior to flow cytometry. In the second, Ifitm3−/eGFP+ cells were cultured separately from Ifitm3+/eGFP− cells but mixed prior to surface staining (Fig. 3C). We found that the Ifitm3−/eGFP+ cells did, in fact, acquire FLAG epitope from their Ifitm3+/eGFP− counterparts during co-culture. The amount of FLAG acquired was minor, however, and not appreciably different from the amount transferred by simply mixing cells prior to staining.
Determination of the Relative Abundance of Intracellular and Extracellular Epitope Tags
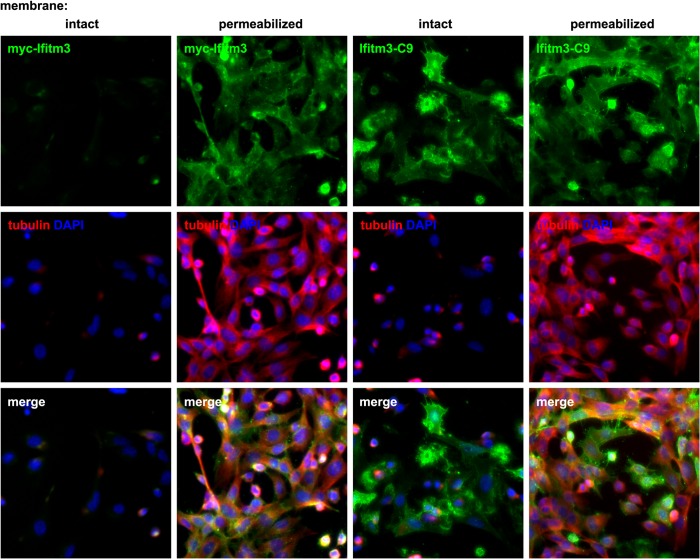
Flow cytometry confirmed the presence of extracellular N- and C-terminal epitopes at the plasma membrane. Flow cytometry, however, cannot exclude the possibility of intracellular epitopes at this location. Therefore, to determine the relative amounts of intracellular and extracellular Ifitm3 termini, we performed fluorescence microscopy with staining for N-terminal Myc and C-terminal C9 epitope tags in fixed or fixed and permeabilized IfitmDel MEFs. We expected that staining for primarily intracellular epitopes would show a marked increase in intensity following permeabilization, whereas staining for primarily extracellular epitopes should be equally bright under both conditions. As a control for permeabilization, co-staining was performed with an antibody against the cytoskeletal protein, α-tubulin (Fig. 4). In the absence of permeabilization, no N-terminal Myc staining was evident. With permeabilization, however, intense plasma membrane staining was observed. In contrast, C-terminal C9 staining was equally intense in both permeabilized and nonpermeabilized cells. Therefore, we conclude that Ifitm3 N termini are intracellular but that the C termini are mostly or entirely extracellular.
FIGURE 4.
In contrast to Ifitm3 N termini, the C termini are mostly or entirely extracellular. MEFs expressing either Myc-Ifitm3 or Ifitm3-C9 were first fixed and then either permeabilized with detergent or left with membranes intact prior to indirect immunofluorescence. Ifitm3 epitope tag staining is shown in green (top panels). Membrane integrity was assessed by co-staining for α-tubulin (middle panels, red). Nuclei were counterstained with DAPI (blue). Exposure times for each channel were identical, and contrast adjustment was applied uniformly to all panels for each color channel.
Localization of the Ifitm3 C Terminus to the ER Lumen
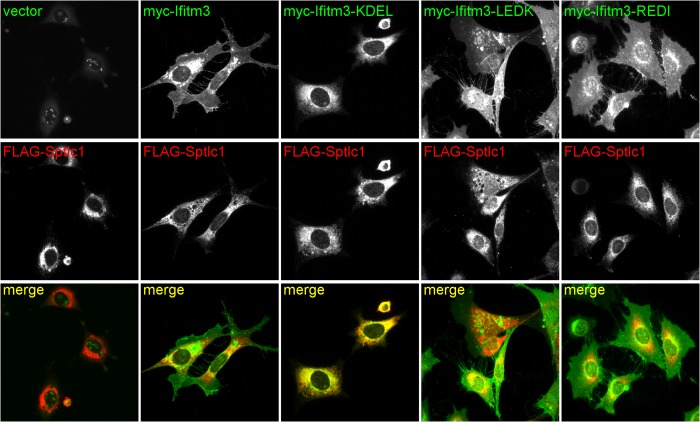
We next sought to determine whether the Ifitm3 C terminus was exposed to the lumen of the endoplasmic reticulum. We created an N-terminally Myc-tagged Ifitm3 with the addition of a C-terminal GSSG linker and a KDEL motif. C-terminal KDEL motifs are bound by chaperones that sequester proteins in the ER lumen. Normally, KDEL prevents secretion of soluble proteins but has been used experimentally to force ER retention of type II transmembrane proteins as well (31). We anticipated that if the Ifitm3 C terminus were oriented toward the ER lumen, a C-terminal KDEL motif would block egress of Ifitm3 from the ER. As controls, we created two additional Ifitm3 mutants. One was terminated with the reverse motif (LEDK), and the other was terminated with a motif consisting of similar charge and hydrophobicity (REDI). MEFs stably expressing these Ifitm3 mutants were transduced a second time with FLAG-tagged Sptlc1 (FLAG-Sptlc1) to serve as an ER marker. Cells were stained for both Myc (Ifitm3) and FLAG (Sptlc1) epitopes and examined by epifluorescence microscopy (Fig. 5). Myc-Ifitm3, the LEDK, and the REDI control showed identical localization patterns: a combination of cytoplasmic puncta and plasma membrane staining. Ifitm3-KDEL, however, co-localized nearly entirely with the ER marker and exhibited no punctate or plasma membrane localization. ER retention of Ifitm3-KDEL, but not Ifitm3-REDI or Ifitm3-LEDK implies a specific interaction between the KDEL motif and ER-luminal proteins, demonstrating the ER-luminal orientation of the Ifitm3 C terminus.
FIGURE 5.
The addition of a C-terminal KDEL motif to Myc-Ifitm3 results in its retention in the ER. Top panels show Myc (N-terminal) staining of cells expressing Ifitm3 with or without the indicated C-terminal additions. Middle panels show staining for FLAG-tagged Sptlc1 (an ER marker). Myc-Ifitm3-KDEL was poorly expressed in comparison with the other constructs, so the brightness of this panel was adjusted independently to better visualize its co-localization with Sptlc1. The nucleolar Myc staining is an artifact of the 9E11 antibody and has been previously described (17).
Localization of the Ifitm3 C Terminus to the Lysosomal Lumen
In Fig. 2, C and E, we noted that the addition of a C-terminal epitope tag to Ifitm3 resulted in a double band on the Western blot when probed with an N-terminal antibody. This suggested that the C terminus may, at some frequency, be cleaved or degraded. To further explore this phenomenon, we created an Ifitm3 construct with both an N-terminal Myc tag and a C-terminal C9 tag (Myc-Ifitm3-C9). Staining this protein with antibodies against the N-terminal Myc tag or native N terminus revealed a double band. When stained for the C9 epitope, however, only the higher molecular weight band was observed, consistent with removal of the C terminus of the molecule (Fig. 6A).
FIGURE 6.
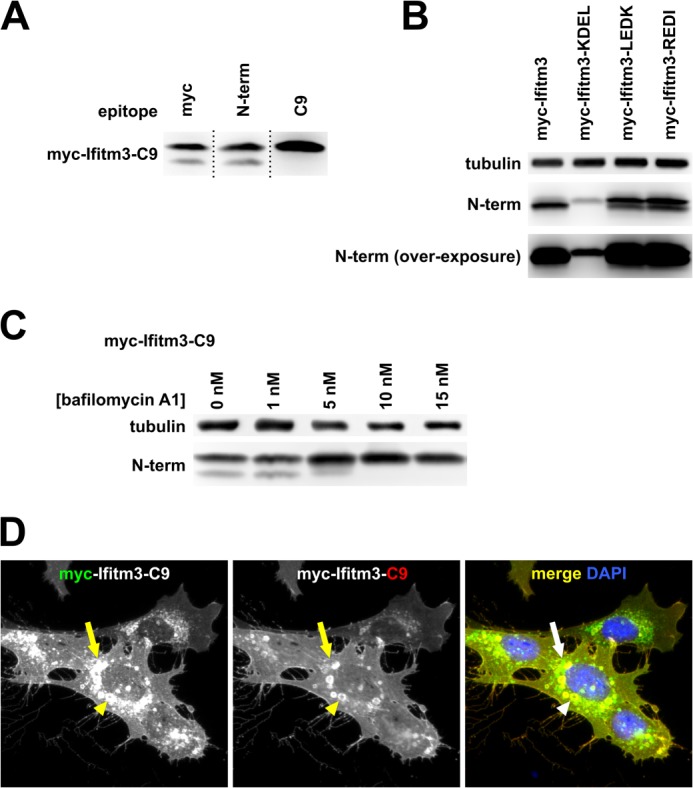
Ifitm3 C termini are degraded in lysosomes. A, lysates from MEFs expressing an N- and C-terminally tagged Ifitm3 (myc-Ifitm3-C9) were examined by Western blot with staining for the indicated epitopes. Dotted lines indicate the sites of reassembly of the membrane after staining. B, Western blot for Myc-Ifitm3 and Myc-Ifitm3-LEDK and -REDI is shown at two exposures to highlight the double band pattern for Ifitm3-LEDK and REDI (upper) and confirm the lack of a second band for Ifitm3-KDEL (overexposed, lower). C, bafilomycin A1 treatment inhibits degradation of the C-terminal tag as assessed by Western blot. D, MEFs expressing Myc-Ifitm3-C9 were co-stained for Myc (green) and C9 (red) epitopes. The plasma membrane is yellow/orange, indicating co-localization of the two epitopes (full-length Myc-Ifitm3-C9). Green cytoplasmic puncta (arrow indicates an example) show the sites of removal of the C9 tag. The C9 tag remained intact within the enlarged vacuoles that accompany IFITM protein overexpression (arrowhead).
We also noted that C-terminal LEDK- and REDI-tagged Ifitm3 from Fig. 5 showed a double band pattern on Western blot. The ER-sequestered KDEL mutant, however, showed no evidence of C-terminal cleavage, suggesting that the cleavage event occurred outside of the ER (Fig. 6B). We hypothesized that the Ifitm3 C terminus was exposed to the degradative interior of the lysosome and was removed by one or more lysosomal hydrolases. We treated cells with the vacuolar ATPase inhibitor bafilomycin A1 to inhibit lysosomal acid hydrolase activity. Bafilomycin A1 treatment efficiently prevented the degradation of the Ifitm3 C terminus (Fig. 6C). Fluorescence microscopy for the Myc-Ifitm3-C9 construct revealed that the C-terminal degradation occurred in punctate cytoplasmic structures consistent with lysosomes (although, interestingly, not in the enlarged, perinuclear endolysosomal compartments that accompany IFITM protein overexpression (4, 20) (Fig. 6D)).
Assessment of the Activity of C-terminally Tagged Ifitm3
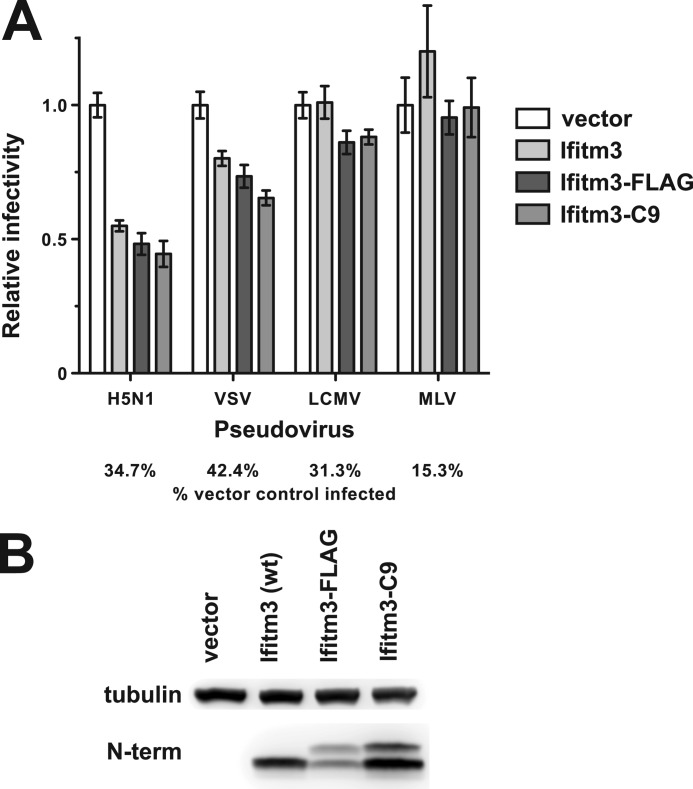
We also considered the possibility that the addition of a C-terminal epitope tag might, in and of itself, alter the topology of the Ifitm3. To exclude this possibility, we performed a viral entry assay using virus-like particles pseudotyped with the envelope proteins of a robustly Ifitm3-restricted virus (influenza A virus H5N1), a weakly restricted virus (vesicular stomatitis virus), and two Ifitm3-insensitive controls (lymphocytic choriomeningitis virus and MLV, Fig. 7). C-terminal tags (C9 and FLAG) interfered with neither Ifitm3 function nor expression levels, suggesting that these tagged constructs assume the same topology as the wild type protein.
FIGURE 7.
C-terminal tags do not impair the restriction activity or expression of Ifitm3. A, MEFs were transduced with a vector control or the indicated Ifitm3 constructs. Transduced MEFs were then infected with eGFP-encoding retroviruses pseudotyped with the envelope glycoproteins of four different viruses. Bars show the proportion of the percentage of infected (eGFP+) cells expressing each Ifitm3 construct relative to percentage of infected vector-transduced cells infected. Error bars show the median and range of triplicate wells. VSV, vesicular stomatitis virus; LCMV, lymphocytic choriomeningitis virus. B, expression levels of the constructs were determined by Western blot against the native Ifitm3 N terminus.
Analysis of the Isolated Intra/Transmembrane Domains of Ifitm3
We reasoned that if Ifitm3 were a type III transmembrane protein, both predicted transmembrane domains (TM1 and TM2) would function as signal anchor sequences. If it were a type II protein, however, only one of the two domains should be recognized as a signal anchor. To determine the functionality of these two domains, we added N-terminal FLAG and C-terminal C9 tags to the isolated TM1 and TM2 sequences, VVWSLFNTLFMNFCCLGFIAY and TLVLSILMVVITIVSVIIIVL, respectively, as annotated by UniProt (32). We transduced cells with these constructs and analyzed surface expression of the epitope tags by flow cytometry, excluding propidium iodide-positive cells as in Fig. 2. Cells effectively localized FLAG-TM2-C9 to the plasma membrane, albeit in both orientations (Fig. 8). FLAG-TM1-C9, however, could not be detected by flow cytometry or Western blot. Although the lack of expression of the isolated TM1 construct precludes any statements about its topology, TM2 clearly functions as a signal anchor, consistent with a transmembrane orientation of the second hydrophobic region of Ifitm3.
FIGURE 8.
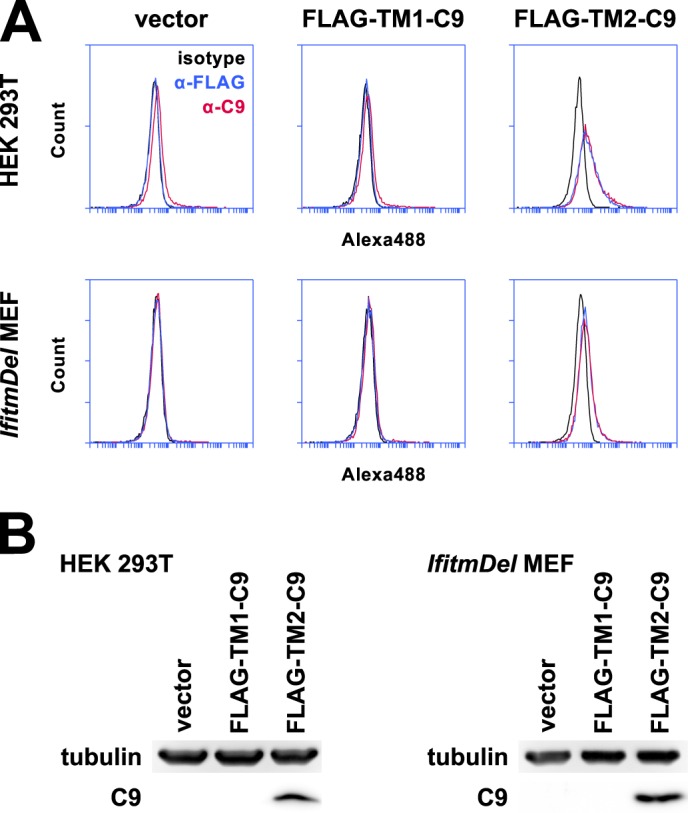
The second hydrophobic region of Ifitm3 acts as a signal anchor sequence. A, the first and second predicted intra/transmembrane domains were subcloned from Ifitm3 and expressed with the addition of N-terminal FLAG and C-terminal C9 tags (FLAG-TM1-C9 and FLAG-TM2-C9). 293T cells or MEFs expressing these constructs were stained by indirect immunofluorescence for both tags in addition to an isotype control antibody. FLAG-TM2-C9 but not FLAG-TM1-C9 could be detected at the surface of both cell types by flow cytometry. Permeabilized cells were excluded from analysis by PI gating as shown in Fig. 2. Alexa488, Alexa Fluor 488. B, FLAG-TM2-C9 but not FLAG-IM1-C9 could be detected by Western blot.
Analysis of a Forced Transmembrane Topology Mutant of Ifitm3
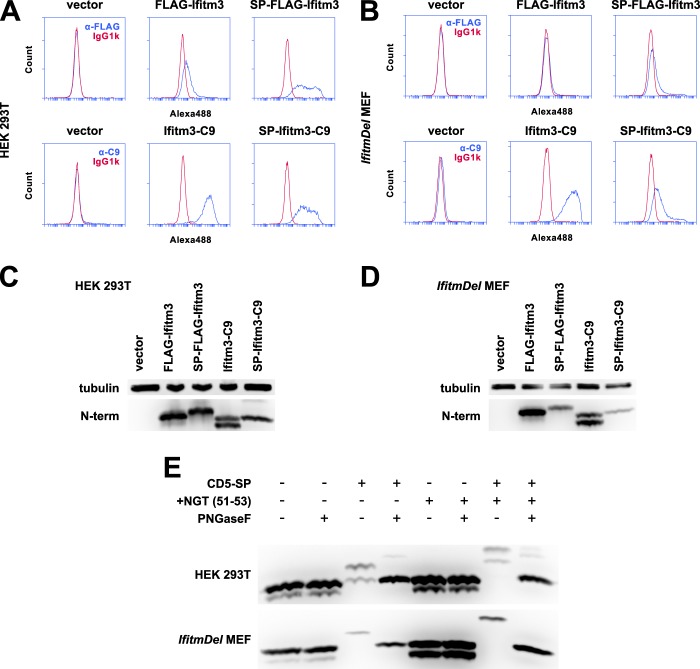
Although the dominant conformation of Ifitm3 appeared to be that consisting of an intracellular N terminus and extracellular C terminus, the presence of a minority population of Ifitm3 molecules with extracellular N termini led us to investigate this rare topology. To mimic the predicted type III topology, we introduced a signal peptide derived from CD5 (CD5SP) before the Ifitm3 coding sequence. Surface staining of CD5SP/FLAG-tagged Ifitm3 and CD5SP/C9-tagged Ifitm3 showed that the CD5 signal peptide effectively reoriented the N terminus toward the extracellular space but left the C terminus in its normal, extracellular orientation (Fig. 9).
FIGURE 9.
The addition of a signal peptide to the Ifitm3 N terminus mimics a type III topology. A, 293T cells expressing FLAG-Ifitm3, Ifitm3-C9, or the same constructs with the addition of an N-terminal CD5 signal peptide were stained for the appropriate epitopes and analyzed by flow cytometry. Alexa488, Alexa Fluor 488. B, MEFs were analyzed by flow cytometry as above. C, a Western blot for the native Ifitm3 N terminus shows the relative expression of each construct in 293T cells. Lysates were treated with peptide-N-glycosidase F to remove N-linked glycosylation of the N terminus. D, the relative expression of each construct in MEFs is shown by Western blot. E, the N terminus of Ifitm3 can be glycosylated but is not normally accessible to cellular glycosylation machinery. Cells were transfected (293T, top) or transduced (MEF, bottom) with constructs expressing C9-tagged Ifitm3 with or without a CD5 signal peptide (CD5-SP) and with or without introduction of an additional NGT motif (+NGT (51–53)). Lysates were treated with peptide-N-glycosidase F (PNGaseF) or mock-treated as indicated and analyzed by Western blot with antibodies against the native N terminus.
We next used the forced topology to investigate whether or not the Ifitm3 N terminus was exposed to the ER lumen. Yount et al. (27) previously noted a lack of N-linked glycosylation on an NHT motif on the wild type N terminus of IFITM3 (residues 2–4, conserved in mouse), but the addition of the CD5SP provided a positive control to determine whether or not this motif was, indeed, accessible to the cellular glycosylation machinery. We transfected (293T) or transduced (MEF) cells with constructs encoding Ifitm3-C9 with or without a CD5 signal peptide and with or without the introduction of an additional glycosylation motif (NGT inserted after residue Arg-50 of the N terminus). Glycosylation of Ifitm3 was inferred from the mobility shift on Western blot following peptide-N-glycosidase F digestion of the cell lysate (Fig. 9). In 293T cells, the wild type NHT motif of CD5SP-Ifitm3 was inefficiently glycosylated, but the NGT insertion was fully glycosylated. No corresponding peptide-N-glycosidase F-sensitive bands were noted for constructs lacking a signal peptide, consistent with the results of Yount et al. (27) indicating that the N terminus is not exposed to the ER lumen. A similar pattern was observed in Ifitm3-transduced MEFs.
DISCUSSION
The Ifitm3 C terminus is inserted into the ER lumen, extracellular at the plasma membrane, and degraded within the interior of the lysosome. The fact that little or no detectable Ifitm3-KDEL escapes the ER indicates that most, if not all, Ifitm3 molecules are oriented with ER-facing C termini. Similarly, the fact that plasma membrane staining of Ifitm3-C9 is equally bright on intact and permeabilized cells supports an entirely extracellular C terminus.
Although extracellular Ifitm3 N termini could be detected by flow cytometry, robust expression of this conformation was only detected in one of the two cell lines studied, and the intensity of staining was very faint when compared with the signal from staining of C-terminal epitopes. In contrast to the second transmembrane domain, the first predicted transmembrane domain fails to express in isolation, suggesting that it does not function as an effective signal anchor. Our results (summarized in Fig. 10) clearly indicate an extracellular C terminus and cytosolic N terminus. Thus, we conclude that Ifitm3 is a type II transmembrane protein.
FIGURE 10.
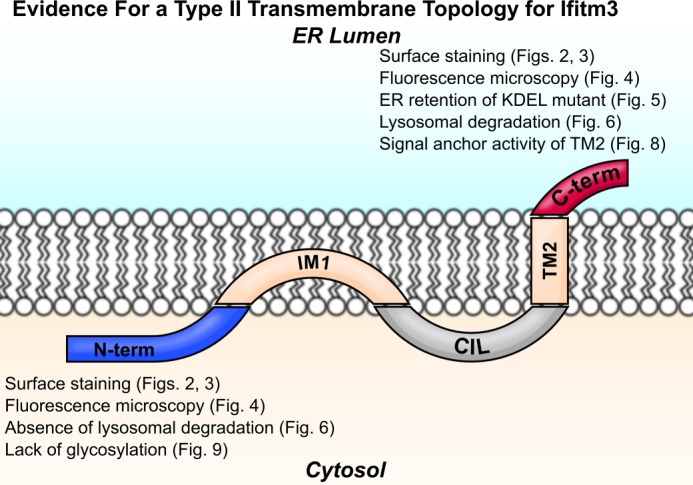
Ifitm3 is a type II transmembrane protein. Ifitm3 contains an N-terminal intramembrane domain (IM1) and C-terminal transmembrane domain (TM2) flanking the conserved intracellular loop (CIL) with an extracellular C terminus and intracellular N terminus. Lines of evidence from this study supporting a type II topology are listed next to the N and C termini of the molecule.
Since the discovery of the IFITM proteins, different investigators have provided conflicting evidence about IFITM protein topology. How have these discrepancies arisen? Our present work provides an explanation: rarely, Ifitm3 adopts an “alternate” topology with an extracellular N terminus. Whether this is a type III topology, as originally predicted, a type I topology resulting from recognition of the intramembrane domain as a signal anchor sequence, or a type I topology resulting from recognition of the transmembrane signal anchor in reverse orientation remains unclear. Nonetheless, it is possible for some investigators to identify IFITM protein N termini at the cell surface (26), whereas others isolate N-terminally phosphorylated (11) or ubiquitylated (27) molecules.
Our results are consistent with those of previous experiments that demonstrated ubiquitylation of Lys-24, phosphorylation of Tyr-20, and palmitoylation of Cys-71, Cys-72, and Cys-105 of human IFITM3 (11, 27) but at odds with the observation of palmitoylation of murine Ifitm1 on Cys-103 (28). Ifitm1 is unusual, however, in that its predicted extracellular C terminus consists of a single residue (Arg-104) and its TM2 may therefore be relatively poorly anchored.
Our observations raise additional questions. Because the alternate topology was not detected in MEFs, and Ifitm3 exhibits viral restriction activity in these cells, the presence of this alternate topology is probably not relevant to the restriction activity of Ifitm3. The alternate topology may, however, play some other role. For example, some of the earliest observations regarding IFITM1 revolved around the ability of IFITM1 antibodies to induce cell-cell adhesion (24, 25). The alternate topology might therefore preferentially associate with tetraspanins, integrins, or other cell surface proteins.
Cell type may also be an important factor in influencing IFITM protein topology. The early observations of IFITM1 antibody-induced intercellular adhesion were made in leukemic B- and T-cells (24, 25). In the present study, extracellular N termini were detected by flow cytometry when Ifitm3 was expressed in 293T cells but not when Ifitm3 was expressed in MEFs. Different cells may provide an environment conducive to the stability of one topology over another.
Another consideration is whether or not other members of the IFITM protein family “prefer” the alternate conformation over the type II. Because phosphorylation of Tyr-20 by Fyn kinase is necessary for localization of IFITM3 to endosomes (11), the cytoplasmic orientation of the IFITM3 N terminus is necessarily linked with its function. IFITM1, however, has no analogous tyrosine and may have evolved to preferentially adopt a different topology.
Whatever answers these questions might yield, the type II topology appears to be the predominant and immunologically active conformation of Ifitm3. This enhanced understanding of IFITM protein topology should further inform our search for Ifitm3 targets and cofactors.
This work was supported, in whole or in part, by National Institutes of Health Grants U54 AI057159 (Project 13) (to M. F.) and L40AI091009 (National Institutes of Health Loan Repayment Program) (to C. C. B.).
- IFITM
- interferon-induced transmembrane
- TM
- transmembrane (domain)
- ER
- endoplasmic reticulum
- MEF
- murine embryonic fibroblast
- PI
- propidium iodide
- CD5
- cluster of differentiation 5
- CD5SP
- CD5 signal peptide
- IM
- intramembrane (domain)
- Sptlc1
- serine palmitoyl transferase 1
- eGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Zhang Z., Liu J., Li M., Yang H., Zhang C. (2012) Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7, e49265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Diamond M. S., Farzan M. (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., Rice C. M. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang I.-C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., Brass A. L., Ahmed A. A., Chi X., Dong L., Longobardi L. E., Boltz D., Kuhn J. H., Elledge S. J., Bavari S., Denison M. R., Choe H., Farzan M. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang D., Weidner J. M., Qing M., Pan X.-B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.-Y., Block T. M., Guo J.-T. (2010) Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weidner J. M., Jiang D., Pan X.-B., Chang J., Block T. M., Guo J.-T. (2010) Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brass A. L., Huang I.-C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anafu A. A., Bowen C. H., Chin C. R., Brass A. L., Holm G. H. (2013) Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 288, 17261–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mudhasani R., Tran J. P., Retterer C., Radoshitzky S. R., Kota K. P., Altamura L. A., Smith J. M., Packard B. Z., Kuhn J. H., Costantino J., Garrison A. R., Schmaljohn C. S., Huang I.-C., Farzan M., Bavari S. (2013) IFITM-2 and IFITM-3 but not IFITM-1 restrict rift valley fever virus. J. Virol. 87, 8451–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu J., Pan Q., Rong L., He W., Liu S.-L., Liang C. (2011) The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia R., Pan Q., Ding S., Rong L., Liu S.-L., Geng Y., Qiao W., Liang C. (2012) The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 86, 13697–13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li K., Markosyan R. M., Zheng Y.-M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J. C., Gratton E., Cohen F. S., Liu S.-L. (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y.-H., Zhao Y., Li N., Peng Y.-C., Giannoulatou E., Jin R.-H., Yan H.-P., Wu H., Liu J.-H., Liu N., Wang D.-Y., Shu Y.-L., Ho L.-P., Kellam P., McMichael A., Dong T. (2013) Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 4, 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanagata N., Li X., Morita H., Takemura T., Li J., Minowa T. (2011) Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J. Bone Miner. Metab. 29, 279–290 [DOI] [PubMed] [Google Scholar]
- 16. Cho T.-J., Lee K.-E., Lee S.-K., Song S. J., Kim K. J., Jeon D., Lee G., Kim H.-N., Lee H. R., Eom H.-H., Lee Z. H., Kim O.-H., Park W.-Y., Park S. S., Ikegawa S., Yoo W. J., Choi I. H., Kim J.-W. (2012) A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am. J. Hum. Genet. 91, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailey C. C., Huang I.-C., Kam C., Farzan M. (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 8, e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wakim L. M., Gupta N., Mintern J. D., Villadangos J. A. (2013) Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 14, 238–245 [DOI] [PubMed] [Google Scholar]
- 19. Lange U. C., Adams D. J., Lee C., Barton S., Schneider R., Bradley A., Surani M. A. (2008) Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 28, 4688–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feeley E. M., Sims J. S., John S. P., Chin C. R., Pertel T., Chen L.-M., Gaiha G. D., Ryan B. J., Donis R. O., Elledge S. J., Brass A. L. (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7, e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amini-Bavil-Olyaee S., Choi Y. J., Lee J. H., Shi M., Huang I.-C., Farzan M., Jung J. U. (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yount J. S., Moltedo B., Yang Y.-Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singer S. J. (1990) The structure and insertion of integral proteins in membranes. Annu. Rev. Cell Biol. 6, 247–296 [DOI] [PubMed] [Google Scholar]
- 24. Evans S. S., Lee D. B., Han T., Tomasi T. B., Evans R. L. (1990) Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood 76, 2583–2593 [PubMed] [Google Scholar]
- 25. Chen Y. X., Welte K., Gebhard D. H., Evans R. L. (1984) Induction of T cell aggregation by antibody to a 16kd human leukocyte surface antigen. J. Immunol. 133, 2496–2501 [PubMed] [Google Scholar]
- 26. Takahashi S., Doss C., Levy S., Levy R. (1990) TAPA-1, the target of an antiproliferative antibody, is associated on the cell surface with the Leu-13 antigen. J. Immunol. 145, 2207–2213 [PubMed] [Google Scholar]
- 27. Yount J. S., Karssemeijer R. A., Hang H. C. (2012) S-Palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hach J. C., McMichael T., Chesarino N. M., Yount J. S. (2013) Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 87, 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang I.-C., Li W., Sui J., Marasco W., Choe H., Farzan M. (2008) Influenza A virus neuraminidase limits viral superinfection. J. Virol. 82, 4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang B. L., Wong S. H., Low S. H., Hong W. (1992) Retention of a type II surface membrane protein in the endoplasmic reticulum by the Lys-Asp-Glu-Leu sequence. J. Biol. Chem. 267, 7072–7076 [PubMed] [Google Scholar]
- 32. UniProt Consortium (2013) Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41, D43–D47 [DOI] [PMC free article] [PubMed] [Google Scholar]