Background: Ghrelin O-acyltransferase (GOAT) is a membrane protein that is responsible for octanoylating the metabolism-regulating peptide hormone ghrelin.
Results: We have used a combination of approaches to determine the topology of GOAT.
Conclusion: We have shown that GOAT has 11 transmembrane-spanning domains and one reentrant loop.
Significance: These findings serve as a reference for other membrane-bound O-acyltransferase family members.
Keywords: Endoplasmic Reticulum (ER), Lipids, Membrane Proteins, Metabolism, Peptide Hormones, Enzyme
Abstract
Ghrelin O-acyltransferase (GOAT) is a polytopic integral membrane protein required for activation of ghrelin, a secreted metabolism-regulating peptide hormone. Although GOAT is a potential therapeutic target for the treatment of obesity and diabetes and plays a key role in other physiologic processes, little is known about its structure or mechanism. GOAT is a member of the membrane-bound O-acyltransferase (MBOAT) family, a group of polytopic integral membrane proteins involved in lipid-biosynthetic and lipid-signaling reactions from prokaryotes to humans. Here we use phylogeny and a variety of bioinformatic tools to predict the topology of GOAT. Using selective permeabilization indirect immunofluorescence microscopy in combination with glycosylation shift immunoblotting, we demonstrate that GOAT contains 11 transmembrane helices and one reentrant loop. Development of the V5Glyc tag, a novel, small, and sensitive dual topology reporter, facilitated these experiments. The MBOAT family invariant residue His-338 is in the ER lumen, consistent with other family members, but conserved Asn-307 is cytosolic, making it unlikely that both are involved in catalysis. Photocross-linking of synthetic ghrelin analogs and inhibitors demonstrates binding to the C-terminal region of GOAT, consistent with a role of His-338 in the active site. This knowledge of GOAT architecture is important for a deeper understanding of the mechanism of GOAT and other MBOATs and could ultimately advance the discovery of selective inhibitors for these enzymes.
Introduction
GOAT6 is a polytopic integral membrane protein required for the octanoylation of ghrelin, a weight- and glucose-modulating peptide hormone produced primarily in the stomach, upper gastrointestinal tract, and brains of vertebrates (1–3). Octanoylation is required for activation of the ghrelin receptor, GHSR-1a, and is the only known example of protein octanoylation in higher organisms (4). Because dietary fats and those from local fatty acid metabolism can be directly conjugated to ghrelin (5–7), it can be viewed as a “fat sensor” from the brain to the body or within the brain itself, modulating energy balance. Supporting this possibility, inhibition of GOAT in mice prevented weight gain on a high fat diet and improved glucose control (8) and may represent an attractive target for the treatment of obesity and diabetes. Additionally, ghrelin has been implicated in a host of other physiologic processes, including gastric motility (9, 10); learning, memory, and reward behavior (11–13); cardiovascular function (14–16); and survival in extreme starvation (17–19).
GOAT is a member of the membrane-bound O-acyltransferase (MBOAT) superfamily (20), which is composed of many multispanning membrane proteins from prokaryotes and eukaryotes, including 11 in humans. MBOAT members have been shown to be critical for lipid biosynthesis, sterol acylation, and acyl modification of secreted proteins, including ghrelin, Hedgehog (HHAT), Wnt proteins (PORC), and yeast GPI-anchored proteins (GUP1) (21–26). MBOAT proteins contain a highly conserved asparagine (Asn-307 in GOAT) and an invariant histidine (His-338 in GOAT) separated by 30–45 residues; these residues have been proposed to be catalytic. There are two additional regions of conservation in the family, at the N- and C-terminal boundaries of loop 5 on GOAT, where it adjoins surrounding hydrophobic regions. The topology of a few MBOATs has been analyzed, and the invariant histidine has been shown to be lumenal in these cases (27, 28). Based on the modification of lumenal, secreted proteins and on lumenal location of the conserved histidine, the active site for the MBOAT family has been proposed to be conserved on the lumenal face (2, 27).
For several MBOATs, including GOAT, mutation of this conserved histidine eliminates acyl transfer activity (2, 23, 28–31). Detailed studies of catalysis by these purified enzymes is generally hindered by the challenge of solubilization in active conformations (32–34); notable exceptions to this are ACAT1, also known as SOAT1, a robust enzyme that transfers long chain fats from coenzyme A (CoA) to cholesterol and has been extensively characterized (reviewed in Ref. 35), and HHAT, which N-palmitoylates hedgehog proteins (36). In contrast to other MBOAT members, for HHAT, some catalytic activity was retained upon alanine mutation of the conserved histidine (37). Thus, the assignment of this histidine as catalytic is not yet definitive.
GOAT octanoylates serine-3 of proghrelin in the ER lumen after signal peptide cleavage. A range of fatty acids can be processed by GOAT, and the most likely acyl donors are acyl-CoAs (reviewed in Ref. 34). Because fatty acyl-CoAs are known to be cytosolic (e.g. those produced during fatty acid β-oxidation), the ER membrane represents a barrier that these acyl-CoAs must cross. In this regard, GOAT has been postulated to be a possible acyl-CoA transporter (2). Determination of the topology of GOAT is a critical step to defining its detailed structure and mechanisms. GOAT has been proposed to contain eight transmembrane helices (TMs) (2), but this has not been experimentally investigated.
In this study, we examine GOAT topology in detail, first computationally and then experimentally. Our data suggest that GOAT contains 11 TMs and segregate the invariant His-338 and conserved Asn-307 to opposite sides of the ER membrane. Photocross-linking binding studies map the binding of acyl ghrelin analogs to the C-terminal region of GOAT and demonstrate that purified GOAT can bind ligand as a monomer.
EXPERIMENTAL PROCEDURES
All reagents were purchased at the highest quality available from Sigma-Aldrich or Acros Organics unless otherwise indicated. Commercially available reagents were used without further purification.
Bioinformatics
GOAT sequences were identified using BLASTp. Annotation of chicken GOAT was corrected to include the first exon. Dog and green anole (Anolis carolinensis) GOAT sequences were identified by megaBLAST and tBLASTx searches from their draft genomes and annotated. Sequences were aligned using MUSCLE (38). Conserved domains and residues were identified using CDD (39), querying mouse GOAT against MBOAT pfam03062 (20). A battery of topology prediction servers were queried with GOAT sequences from various organisms, starting with mouse and human sequences. A multiple sequence alignment was submitted wherever possible. Older methods, such as TMHMM version 2.0 (40), TMpred (41), SOSUI (42), TopPred (43), and MEMSAT version 3 (44) gave very different results from species to species, even between the highly conserved human and mouse GOATs; the range was from 4 to 11 TMs. The consensus prediction servers TOPCONS (45) and ConPred II (46) both run a number of algorithms, including the above, in order to make better predictions. However, for GOAT, the results from both servers were similarly inconsistent between species and thus hard to interpret. HMMtop (47) and PolyPhobius (48) can use a multiple sequence alignment as input. Although this improved the results somewhat as compared with the aforementioned algorithms, these algorithms predict only seven and eight TMs, respectively. Both missed TMs predicted by MemBrain and MEMSAT-SVM that were subsequently validated experimentally. MemBrain and MEMSAT-SVM predictions were used as a basis for our study and are discussed under “Results.” For MemBrain, predictions for each organism were overlaid on the multiple-species alignment, and TM propensity plots were overlaid and aligned.
Cell Culture
HeLa cells were maintained in DMEM (Invitrogen) with 10% heat-inactivated FBS and penicillin/streptomycin. Transfections were done using Fugene 6 or Fugene HD (Promega) according to the manufacturer's instructions, with 2 μg of GOAT DNA and 6 μl of reagent per 35-mm dish or well of a 6-well plate. SF9 cells were maintained in ISFM medium, an affordable serum-free synthetic medium, made as reported in (49), with minor modifications.7
Cloning
For human cell expression, GOAT-3xFLAG was cloned into the CAG vector as described previously (8) with a modified multiple-cloning site. N- and C-terminal tags were placed immediately before the first or after the last residue of GOAT without any extraneous sequence by incorporating tag sequences into oligonucleotide primers. Internally V5-tagged GOAT clones were prepared using fusion PCR (50). Individual point mutants were made using a modified QuikChange protocol (Stratagene). Asn-free and Lys-free GOAT were made in one round of polymerization using MISO mutagenesis, as described in Fig. 2a of a previous study (51), as were V5Glyc clones. N-terminal deletion ΔN-H1 starts with a Met at residue 32. ΔN-H1-2, ΔN-H1-3, and ΔN-H1-3 constructs start with Met-56 and ATG codons substituted at positions 81 and 109, respectively, to define the first residue of the constructs. ΔC-H10-11 and ΔC-H11 are truncated after residues 357 and 399, respectively. N-IBV-1-GOAT and N-IBV-2-GOAT are GOAT-3xFLAG with an N-terminal fusion of the first 20 and 10 residues, respectively, of the M glycoprotein from infectious bronchitis virus (IBV; an avian coronavirus). Asn-3 and Asn-6 of this sequence are glycosylated in IBV-M, previously called the E1 protein (52). All clones were fully sequence-verified. Baculovirus constructs were made using the Bac2Bac system (Invitrogen) according to the manufacturer's instructions.
FIGURE 2.
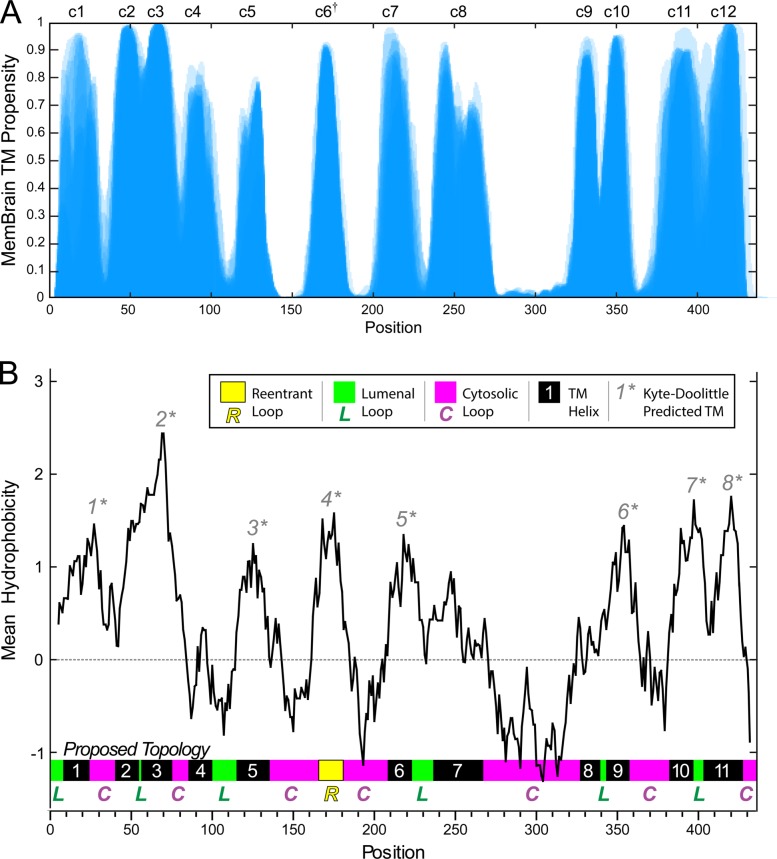
Comparison of new versus prior GOAT topology models. A, aligned overlay of MemBrain helical propensities by position for GOAT from 16 species. A darker color indicates a more consistent prediction. Candidate TMs are labeled c1–c12 at the top. †, candidate TM-6 was found to be non-TM. B, Kyte and Doolittle mean hydrophobicity profile for mouse GOAT, with an 18-residue scan window. *, TMs predicted by this method are shown above the hydrophobic peaks. Our proposed topology of GOAT is shown in the thick bar at the bottom of the graph, with TMs in black, lumenal loops in green, cytosolic loops in pink, and a possible reentrant loop in yellow.
Selective Permeabilization Immunofluorescence
200,000 HeLa cells were plated on glass coverslips, transfected 8–16 h after plating, and incubated for 40 h. For complete permeabilization, cells were fixed in 3% paraformaldehyde for 10 min, and fixative was then quenched using PBS containing 10 mm glycine and 0.2% sodium azide (PBS/gly). Cells were incubated for 3 min in 0.5% Triton X-100 in PBS/gly and then washed twice with PBS/gly. For selective permeabilization, dishes were placed on ice, washed with ice-cold KHM (20 mm HEPES, pH 7.4, 110 mm potassium acetate, 2 mm magnesium acetate), and kept on ice. The cells were permeabilized with 75 μg/ml digitonin (EMD Biosciences) in KHM for 10 min. Cells were washed twice with ice-cold KHM, moved to room temperature, and fixed as above.
Staining with primary and secondary antibodies was carried out for 20 min at room temperature by inverting coverslips onto Parafilm containing 45-μl drops of PBS/gly supplemented with 1% BSA and appropriate antibodies. Antibodies used were mouse anti-FLAG M2 (1:500; Sigma), rabbit anti-FLAG (1:1000; Sigma F7425), mouse anti-V5 (1:5000; Invitrogen), mouse anti-Myc 9E10 (1 μg/ml), rabbit anti-GFP (1:500; Molecular Probes), mouse anti-GFP (1:500; Roche Applied Science 11814460001), and rabbit anti-SSR1 (2 μg/ml; Sigma HPA011276; recognizes residues 230–286). Coverslips were washed and incubated analogously with secondary antibodies: Alexa Fluor® 488-conjugated anti-mouse IgG (1:1000) and Alexa Fluor 568-conjugated anti-rabbit IgG (1:1000). As controls, untransfected cells were stained identically, and primary antibodies were omitted. DNA was stained prior to imaging with Hoechst 33285 (0.1 μg/ml, 5 min), and coverslips were mounted using glycerol with 100 mm N-propyl gallate. Epifluorescent images were collected at room temperature using an Axioscop microscope with ×63 and ×40 objectives (Zeiss, Jena, Germany) equipped for epifluorescence using an ORCA-03G CCD camera (Hamamatsu, Japan) and iVision software (BioVision). For equal comparison of staining between permeabilizations, within each construct, for each antibody, an optimal exposure time was first determined, and then this fixed exposure time was used for both permeabilizations. Signal intensity was then assigned based on the optimal V5 exposure times as follows: ++++, <30-ms exposure; +++, 30–60-ms exposure; ++, 60–150-ms exposure; +, 150–500-ms exposure; −, not clearly above background. Image layouts were done in Adobe Photoshop CS6 and Illustrator CS6.
For co-localization of GOAT with CFP-KDEL, coverslips were treated as above except that fixation was in methanol for 20 min at −20 °C. Confocal images were acquired with a Cascade QuantEM 512SC camera (Photometrics) attached to a Zeiss AxioImager (×63 objective) with a Yokogawa spinning disk confocal scanner and Slidebook software (Intelligent Imaging Innovations).
Synthesis
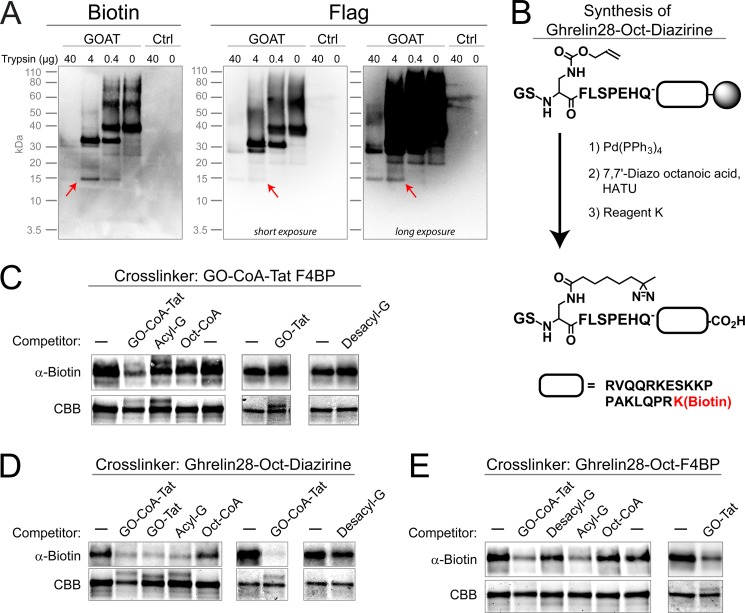
Peptide synthesis was performed using the Fmoc (N-(9-fluorenyl)methoxycarbonyl) strategy. GO-CoA-Tat, GO-Tat, and GO-CoA-Tat F4BP were prepared as described previously (8). In brief, amide-linked ghrelin analogs were prepared in which Ser-3 in ghrelin was replaced with Alloc (allyloxycarbonyl) protected 1,2-diaminopropionic acid, orthogonally deprotected using palladium catalysis, and then reacted with octanoic acid in the presence of HATU (1-(bis-(dimethylamino)methyliumyl)-1H-1,2,3-triazolo(4,5-b)pyridine-3-oxide hexafluorophosphate), reacted with octanoic anhydride, or reacted with derivatives thereof. For ghrelin-28-Oct-diazirine, 7,7′-diazooctanoic acid was used at this step. This was prepared by using the method described in the literature (53), and its structure and purity were confirmed by 1H NMR. Synthesized peptides were purified using a reversed-phase C-18 column, and their MALDI and ES mass spectrometry data were consistent with the calculated values. The final concentrations of the compounds in aqueous solution for assay were determined by amino acid analysis. Ghrelin sequences synthesized correspond to human ghrelin (GSSFLSPEHQRVQQRKESKKPPAKLQPKR).
GOAT Purification
Mouse GOAT with a C-terminal TEV-3xFLAG tag was expressed in SF9 cells at 3–6 × 106/ml in 1–6-liter aerated spinner flasks using a 72-h baculoviral infection with a multiplicity of infection of ∼10. Cells were harvested, and all subsequent steps carried out on ice or at 4 °C. Lysis was performed using a microfluidizer in 4 packed cell volumes of HBS + PI (50 mm HEPES, pH 7.0, 150 mm NaCl, 2 μg/ml aprotinin, 2.5 μg/ml leupeptin, 2 μg/ml pepstatin A, and 1 mm EDTA). Lysate was cleared for 10 min at 15,000 × g, and then microsomes were collected at 120,000 × g for 2–6 h, flash-frozen on liquid nitrogen, and stored at −80 °C. Microsomes were resuspended in 10 volumes of HBS + PI in a 40-ml Dounce homogenizer, solubilized for 1 h at 4 °C with 1% Fos-Choline 16 (FC-16; Anatrace), and cleared for 30 min at 100,000 × g. Supernatant was bound to pre-equilibrated FLAG-agarose in batch for 1–3 h and then transferred to a column, washed with HBS + PI + 0.05% FC-16 and with the same buffer containing 500 mm NaCl, and then washed with HBS without protease inhibitors and cleaved overnight with His6-TEV protease in 10 mm HEPES, pH 8.0, 100 mm NaCl, 0.005% FC-16, 2 mm DTT. Supernatant was collected, 20 mm imidazole was added, and then His6-TEV protease was removed using nickel affinity. The sample was then further purified by ion exchange chromatography (HiTrap SP FF, GE Healthcare), eluted in the same buffer containing 300 mm NaCl without imidazole, and concentrated to 2–10 mg/ml.
MALDI-TOF Mass Spectrometry
MALDI-TOF mass spectrometry of intact GOAT was performed by Tatiana Boronina in the Johns Hopkins Proteomics Core Facility using a Voyager DE-STR (Applied Biosystems). 3 μg of purified, tag-cleaved mouse GOAT was desalted using PLRP-S 300A/10–15-μm material (Polymer Labs/Agilent) packed in a pipette tip pre-equilibrated with 12% acetonitrile and 0.1% TFA; washed three times in the same buffer; and eluted with 90% acetonitrile, 0.1% TFA. 2 pmol of GOAT was spotted in a sinapinic acid matrix (Laser BioLabs) in either 50% ethanol, 0.1% TFA or 40% acetonitrile, 0.1% TFA. Calibration was with AB SCIEX C3 standards (5–17 kDa) in adjacent spots under identical conditions.
Analytical Ultracentrifugation
Sedimentation velocity analytical ultracentrifugation was done on a Beckman XL-I at 10 °C with an An-60Ti rotor at 40,000 rpm with a total run time of 12.5 h. Purified tag-cleaved GOAT samples in a buffer containing 10 mm HEPES, pH 7.0, 300 mm NaCl, and 0.0008% FC-16 were diluted to 0.8, 0.3, and 0.1 A280, corresponding to 6.8, 2.6, and 0.9 μm. Data shown (see below) are from the highest concentration; results from other cells were similar. Analysis was performed as described under “Results.”
Competition Photocross-linking
Each 200-μl reaction mixture in HBS + PI + 0.005% FC-16 contained 0.2–2.0 μm purified GOAT and 1 μm photocross-linking compound with or without 200 μm competitor. Competitors were preincubated for 1 h, and photocross-linkers were preincubated for 15 min at 4 °C. Reactions were cross-linked with a UV lamp with a 360-nm filter in a stirred quartz cuvette, water-cooled to 4 °C using a circulating bath. GO-CoA-Tat F4BP was exposed for 10 min, ghrelin-28-Oct-F4BP for 2 min, and ghrelin-28-Oct-diazirine for 15 min. Samples were then analyzed on SDS-PAGE, blotted for biotin (as above), and stained with colloidal Coomassie Brilliant Blue (Invitrogen) as a loading control.
Microsomal GOAT Octanoyltransferase Assay
SF9 microsomes containing GOAT-3xFLAG and control microsomes from empty vector SF9 cells were prepared as above except that lysis was performed using a Dounce homogenizer. Assays were carried out as described (2, 34) with modifications. 25 μg of SF9 microsomes (BCA) in 50 μl of HBS (pH 7.0) was incubated at 30 °C for 5 min with 10 μm C-terminally biotin-tagged human ghrelin (ghrelin-27-biotin), 60 μm palmitoyl-CoA, and 1 μm [3H]octanoyl-CoA (high specific activity; American Radiolabeled Chemicals). Reactions were quenched and solubilized by adding 1 ml of 2% SDS in TBS containing streptavidin beads and bound for >15 min. Beads were washed with 25 ml of TBS + 0.1% SDS on small columns and then scintillation-counted.
Microsomal Photocross-linking and Partial Proteolysis
Each1200-μl cross-linking reaction contained 5 μm GO-CoA-Tat F4BP and 5 mg of total microsome protein from either GOAT-3xFLAG containing microsomes or control microsomes, which were made using empty virus-infected SF9 cells. Reactions were carried out as described (8), solubilized by diluting to 1.5 mg/ml with 1% FC-16 for 30 min at 4 °C, and then cleared for 30 min at 100,000 × g. 0.4–40 μg sequencing grade modified trypsin (Promega) or buffer was added to each sample and incubated for 15 min at 37 °C. Protease was quenched with 20 μg/ml aprotinin and 20 μg/ml leupeptin, and samples were bound to pre-equilibrated anti-FLAG-agarose (Sigma) overnight. Beads were washed three times with HBS + PI with 0.1% FC-16 and eluted with 1 mg/ml 3xFLAG peptide (Sigma) in the same buffer and then separated with SDS-PAGE. Biotin detection was using streptavidin (Pierce) followed by polyclonal anti-streptavidin (Abcam).
RESULTS
Bioinformatic Predictions of GOAT Topology
To predict the membrane topology of GOAT, we started with the assumption that topology should be conserved across species. 16 GOAT sequences were identified and, in the cases of dog, green anole, and chicken, annotated. A battery of Web-based topology prediction servers were then tested, but most individual algorithms and consensus prediction servers gave very different results from species to species, even between the highly conserved human and mouse GOATs; the range was from 4 to 11 TMs (see “Experimental Procedures”).
In contrast, two more recently developed programs, MemBrain (54) and MEMSAT-SVM (55), stood out in their consistency of results across species. MemBrain and MEMSAT-SVM incorporate newer machine-learning algorithms, phylogeny and PSI-BLAST strategies, and current knowledge about membrane protein structure, such as short and very long helices (<16 or >40 residues) and reentrant loops (also referred to as half-TMs or half-helices). Results from both programs were nearly identical, including predicted helical boundaries, except where described below. MemBrain-predicted TMs for each species are highlighted in Fig. 1, with a total of 12 candidate TMs identified in consensus. For clarity, we numbered TMs and loops in accordance with our final model. Corresponding TM propensities for each species are overlaid in Fig. 2A. Some species are predicted individually to be 11 or 13 TMs. Note that TM-1, -5, -8, and -9 were predicted to be either a TM or a reentrant loop, depending on species. TM-7 is predicted to be one long TM by MemBrain in 13 of 16 species, but in dogs, horses, and green anoles, it predicts this region as one TM and one reentrant loop.
FIGURE 1.
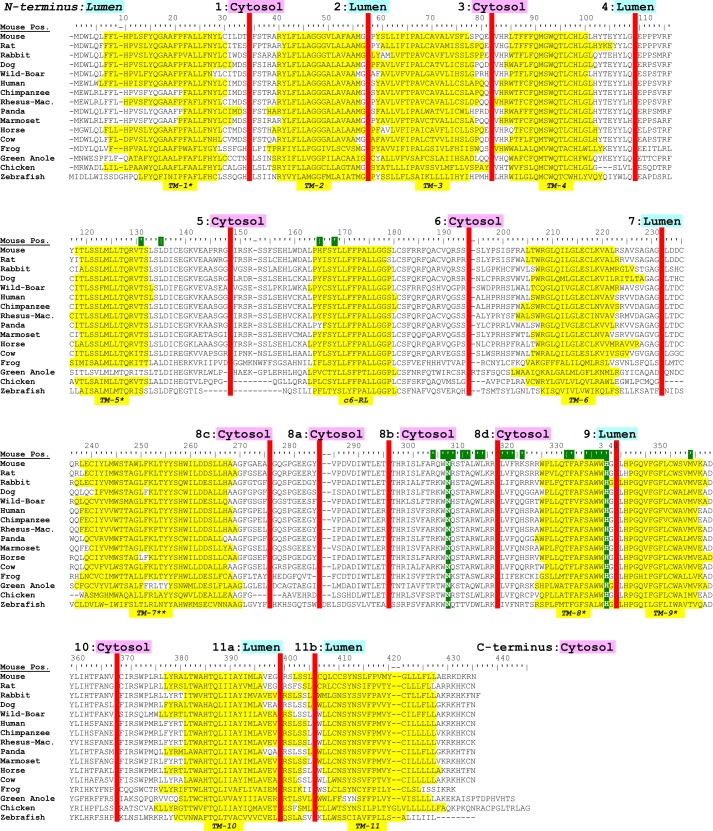
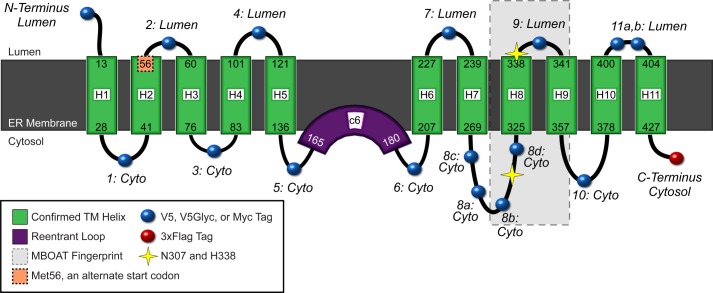
Bioinformatic analysis of GOAT sequences identifies 12 candidate TMs. Shown is GOAT multiple sequence alignment across 16 species with MemBrain-predicted candidate TMs highlighted in yellow; for clarity, TMs and loops are numbered in accordance with our final model. *, predicted to be either a full-length TM or a reentrant loop; **, TM-7 was predicted to be either one long TM or a reentrant loop plus one standard-length TM. c6-RL, candidate 6 reentrant loop; this region was found to be non-transmembrane, probably a reentrant loop. Highly conserved Asn-307 and invariant His-338 residues are highlighted in green; in every case, His-338 is predicted to be part of TM-9 (green + yellow highlight). Additional highly conserved MBOAT residues (NCBI conserved domain database pfam03062, 1.5 bits (39)) are highlighted in green in the top bar. Positions selected for epitope tag insertion are shown as red bars, and their localizations experimentally determined in this work are listed above. GOAT protein sequences were aligned using MUSCLE (38). Dog and green anole sequences were identified and annotated from their genomes; chicken annotation was corrected to include the first exon (which contains TM-1 and -2).
TM-8 and TM-9 are predicted to be two separate TMs by MemBrain in 14 of 16 species, with the 2–3-residue turn between the TMs containing MBOAT invariant His-338 at the border of TM-9. Our data (see below) suggest that this prediction is correct. In contrast, MemBrain predicts one long TM in this region for rabbits and zebrafish. MEMSAT-SVM predicts this region as one long TM in all cases. No signal peptide is predicted for GOAT by MemBrain, MEMSAT-SVM, PolyPhobius, or SignalP (56). MemBrain does not predict orientation for helices, but MEMSAT-SVM predicts that the GOAT N terminus is lumenal and that its C terminus is cytosolic.
We compared the new computational results with those obtained using a Kyte and Doolittle scanning hydropathy window (Fig. 2B). As reported previously (2), this method predicts eight TMs. In comparison, the consensus MemBrain results predict 12 TMs (Fig. 2, A and B (inset bar)).
Topology Determination Using Selective Permeabilization
To experimentally determine the topology of GOAT, we first verified its predicted localization to the ER by co-transfection with CFP-KDEL, an ER marker containing a strong, cleaved signal peptide and the KDEL retention sequence. Upon expression, GOAT-3xFLAG and CFP-KDEL colocalize (Fig. 3A).
FIGURE 3.
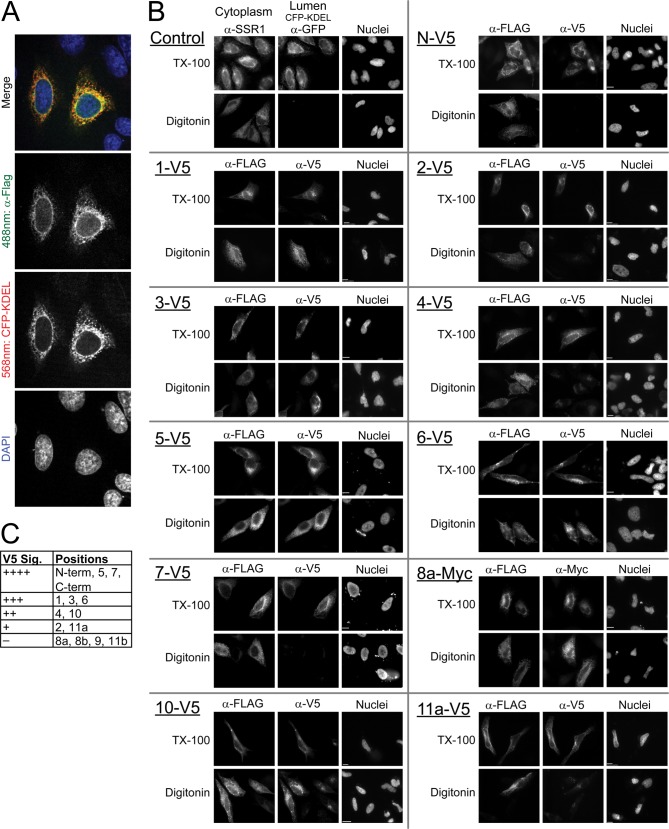
Mapping the topology of GOAT by selective permeabilization of the plasma membrane and indirect immunofluorescence. A, mouse GOAT bearing a C-terminal 3xFLAG tag is localized to the ER in HeLa cells, as seen by co-localization with co-transfected CFP-KDEL, an ER marker; images are a confocal projection. B, selective permeabilization experiments mapping the positions of internal V5 epitopes. Two dishes of HeLa cells were transfected with a GOAT cDNA bearing an internal V5 or Myc epitope tag; all constructs have a C-terminal 3xFLAG tag as an internal control. Selective permeabilization with digitonin revealed cytosolic but not lumenal epitopes, whereas full permeabilization with Triton X-100 (TX-100) revealed all accessible epitopes. Lumenal epitopes were thus visible with Triton X-100 only. As a control, two dishes were stained for proteins with known cytosolic and lumenal epitopes and permeabilized identically; in this case, we used endogenous SSR1 (cytosolic) and transfected CFP-KDEL (lumenal). Identical exposures and image normalization for both permeabilizations ensure fair side-by-side comparison. C, V5 signal strength in indirect immunofluorescence of V5-tagged constructs from Figs. 3B and 4D (see “Experimental Procedures”).
To test the topology predictions, we designed GOAT constructs containing N-terminal and C-terminal tags. In addition, GOAT constructs were generated with internal epitope tags inserted in the loops between candidate TMs, aligning to gaps where possible (Fig. 1, red bars). Because the boundary between TM-10 and -11 was not clearly defined across species, we created two positions, 11a and 11b. Position 8b was added later due to masking of V5 epitopes installed at 8a (see below), and positions 8c and 8d were added with V5Glyc tags (below). We next screened FLAG, 3xFLAG, and V5 tags for compatibility with proper translation of the GOAT N terminus, which contains a non-cleaved internal signal sequence (validated below). The V5 tag was selected for testing because its net charge is neutral. Staining of an N-V5 tag was indistinguishable from C-terminal 3xFLAG-tagged GOAT, but N-FLAG and N-3xFLAG tags resulted in poor expression and aggregated protein (not shown).
Mouse GOAT clones were then made with various internal and terminal epitope tags. The V5 tag was installed at the N and C termini and at all internal positions except for 8c and 8d (Fig. 1, red bars). To ensure that the internal tag did not perturb overall topology, “flipping” the C terminus to the opposite side of the membrane, all clones contain a constant C-terminal 3xFLAG tag (except when C-terminally V5- or 3xMyc-tagged, as noted). We then investigated the topology of these clones using indirect immunofluorescence with selective permeabilization of the plasma membrane. Parallel wells were transfected with constructs of interest and either fully permeabilized with Triton X-100 or selectively permeabilized at the plasma membrane with digitonin (Fig. 3B). To verify the success of the permeabilization, in each experiment, we employed control proteins of known localization in parallel: transfected lumenal CFP-KDEL and endogenous SSR1, an ER transmembrane protein with a cytosolic epitope. Both proteins were detected with Triton X-100 permeabilization, but only the cytosolic SSR1 epitope was detected with digitonin permeabilization.
The GOAT C terminus was consistently localized in the cytosol in all constructs, with the 3xFLAG tag readily detectable (Fig. 3B). The ability to detect the V5 tag varied from construct to construct (relative signal strength is shown in Fig. 3C), but overall, the technique was successful and allowed determination of topology at most positions. Notably, N and C termini were found to be on opposite sides of the membrane, requiring an odd number of TMs. Both positions 5 and 6 were found to be cytosolic, suggesting that the strongly predicted, hydrophobic c6 does not cross the membrane. We suggest that this segment represents a reentrant loop.
The C-terminal part of the protein was more challenging, with the V5 tag undetectable at position 8a, 8b, 9, or 11b. This was probably due to masking of the epitope rather than an expression problem, because the internally tagged proteins were expressed well, as revealed by C-terminal 3xFLAG staining (not shown). We therefore installed Myc tags in these positions as well as in positions 2, 4, 10, and 11a. The Myc tag was strongly detected at positions 8a and 8b, with clear cytosolic localization (Figs. 3B and 4D). Note that this localizes the conserved MBOAT fingerprint residue Asn-307 to the cytosol. Neither V5 nor Myc tag was visible at position 9 or 11b; the proteins were expressed, as seen by 3xFLAG staining, but these epitopes may have been masked in the context of the short loops. Myc tags at positions 2, 4, and 11a all gave results consistent with V5 tags at these positions but with a less robust signal/noise ratio. Myc signal from position 10 was not detectable.
FIGURE 4.
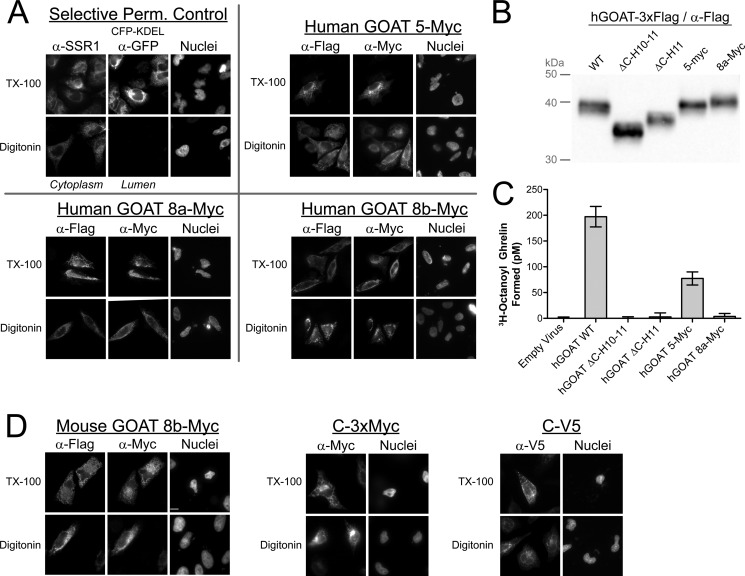
Selected human GOAT constructs recapitulate mouse GOAT results and one construct retains enzymatic activity. A, selective permeabilization of human GOAT constructs bearing the indicated internal Myc epitope tag with constant C-terminal 3xFLAG tags. B, anti-FLAG immunoblot of human GOAT constructs in SF9 cell microsomes. The ΔC-H10–11 construct lacks C-terminal helices 10 and 11, and ΔC-H11 lacks C-terminal helix 11. C, activity assay of constructs in b. [3H]Octanoyl-CoA (2,2′,3,3′) and C-terminally biotinylated human ghrelin were added to 25 μg of total microsome protein in a 50-μl reaction for 10 min at 30 °C, and then ghrelin was immobilized on streptavidin-linked polyacrylamide beads, washed, and scintillation-counted. Each bar represents an average of duplicates; error bars, S.D. D, selective permeabilization of mouse GOAT with three different C-terminal epitope tags and an additional internal Myc tag. The construct with a Myc tag at position 8b also has a C-terminal 3xFLAG tag.
To confirm the cytosolic localizations of positions 5 and 8, we made parallel constructs in human GOAT, repeated the selective permeabilizations (Fig. 4A), and expressed them in SF9 cells for activity assays (Fig. 4, B and C). We also made two C-terminal deletion mutants, ΔC-H11 and ΔC-H10–11, predicted to lack the final TM and final two TMs, respectively; all SF9 constructs have C-terminal TEV-3xFLAG-His tags. The 5-Myc construct showed GOAT activity within 3-fold of the untagged version, suggesting that the 5-Myc epitope tag was tolerated. In contrast, ΔC-H10–11 and ΔC-H11 as well as the 8a-Myc insertion showed no detectable GOAT enzyme activity, suggesting that these alterations might significantly perturb active site structure.
To reduce the possibility that the 3xFLAG tag caused artifactual cytosolic localization of the GOAT C terminus, we replaced the 3xFLAG tag in mouse GOAT with V5 or 3xMyc tags and repeated the selective permeabilization (Fig. 4D). These epitopes were all found to be cytosolic.
To summarize the results thus far, selective permeabilization with insertion of single epitope tags demonstrated that GOAT has an odd number of TMs, confirmed the existence of 9 of 12 predicted TMs, and suggested that one TM domain was a reentrant loop. However, using this method, we were unable to conclusively determine the topology around position 9 or the location of the conserved His-338.
The V5Glyc Tag, a Novel, Sensitive Dual Purpose Topology Reporter
To investigate the topology model for GOAT using a complementary approach and to probe the architecture around His-338, we developed the V5Glyc tag, a novel, small dual topology reporter (DTR) allowing sensitive readout of topology by both glycosylation gel shift Western blotting and selective permeabilization immunofluorescence microscopy. The design principles are diagrammed in Fig. 5. We wanted to leverage the fact that N-linked glycosylation only occurs in the ER lumen. However, glycosylation is context-sensitive, requiring both appropriate sequence context for recognition by oligosaccharyltransferase and a distance from the membrane of ∼12–14 amino acids (30–40 Å), corresponding to the distance of the oligosaccharyltransferase active site from the membrane (57). This is typically achieved by fusing a complete, folded protein (or domain) to the membrane protein of interest, often with a C-terminal truncation. For example, in yeast, the SUC2-HIS4C DTR, installed as a C-terminal fusion with truncation, has been used extensively to map the topology of a number of proteins, including other MBOATs. This DTR is reliable when used under particular conditions and has provided much insight, but it can be problematic when positioned too close to the membrane (27, 58–60). Additionally, this DTR is a very large modification of ∼130 kDa, much larger than the proteins of interest here, and it is inconvenient to clone in-frame without using yeast homologous recombination. Fusion of the SUC2-HIS4C to C termini (without truncation) has been shown to cause incorrect C-terminal localization in some cases (61, 62). Mapping internal sites by C-terminally truncating a membrane protein, as the tag is typically used, is likely to be of low reliability for a number of reasons. C-terminal truncations alone may result in major structural perturbations, resulting in the incorrect topology determination in the MBOATs ACAT1 and ACAT2 (35). Moreover, a very large protein insertion can further perturb the overall topology. In addition, the large size of the DTR combined with truncations of various lengths makes it challenging to verify that the fusion proteins are full-length.
FIGURE 5.
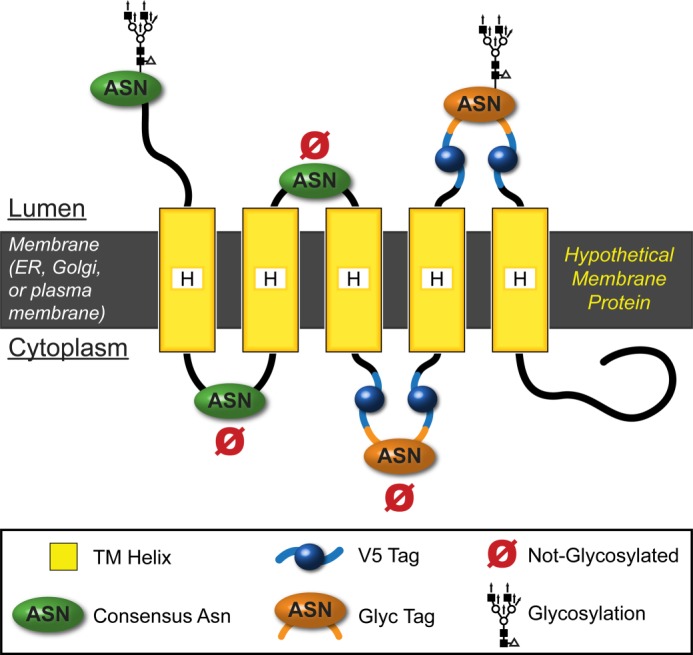
The V5Glyc tag is a novel dual purpose topology reporter. The V5Glyc tag is a neutral, hairpin-like tag containing an optimized 8-residue consensus for N-linked glycosylation, sandwiched between two V5 epitope tags. This tag was designed using the NetNGlyc server8 to provide a consistent context that can be glycosylated if located in the ER lumen. Here, we show a hypothetical membrane protein with asparagine residues in consensus sequences (i.e. NXS or NXT motifs) on both sides of the membrane. Only lumenal asparagines in the appropriate context and spaced far enough from the membrane can be N-glycosylated.
We hypothesized that a much smaller, flexible hairpin-like DTR consisting of two V5 epitope tags flanking a strong consensus glycosylation site could provide a consistent context for oligosaccharyltransferase, increase sensitivity for immunostaining and immunoblotting, and minimize epitope masking while minimizing protein perturbation by retaining a short sequence. It could also be readily installed in-frame in the context of a full-length protein of interest.
To create a strong consensus glycosylation site, we queried the NetNGlyc server, a neural network-based N-glycosylation prediction tool,8 with candidate synthetic V5Glyc sequences in the centers of lumenal loops 2 and 4 on GOAT. We started with NXS and NXT motifs and found that NXS was at best weakly predicted to be glycosylated (38–53% confidence), but NXT was stronger, with NVT optimal but still only with 62% confidence (7 of 9 “jury” agreeing). We iteratively tested a series of additional residues on either side, resulting in the sequence GYLNVTYV as a strong prediction (83 and 85% confidence when the sequence was installed in GOAT in positions 2 and 4, respectively, with 9 of 9 jury agreeing). The resulting V5Glyc tag is 36 amino acids long, and its complete sequence is GKPIPNPLLGLDST-GYLNVTYV-GKPIPNPLLGLDST (where underlining indicates the glycosylated Asn and hyphens separate the V5 sequences from the Glyc sequence).
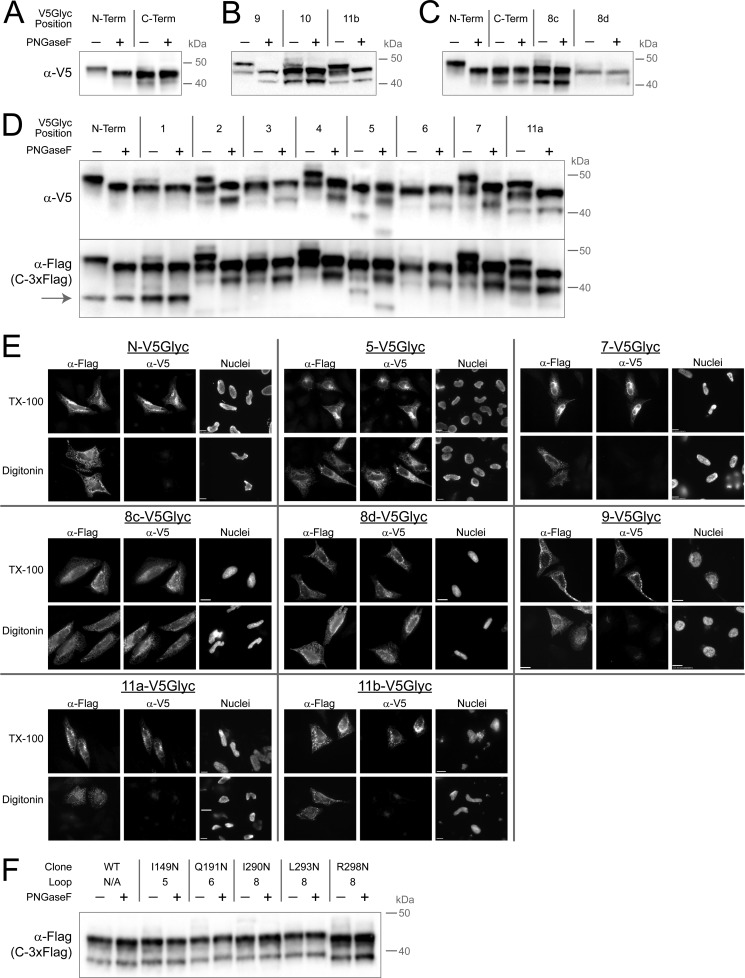
We then installed the V5Glyc tag at all previously referenced positions in mouse GOAT and used it to probe topology by gel shift immunoblotting (Fig. 6, A–D) and selective permeabilization immunofluorescence (selected positions shown in Fig. 6E). The N terminus of GOAT was lumenal with complete glycosylation, removable by peptide-N-glycosidase F, and its C terminus was cytosolic and thus not glycosylated (Fig. 6A). The technique also confirmed localizations of all internal positions, both for those that led to weaker expression (Fig. 6B) and those with strong expression (Fig. 6D). Position 9, immediately adjacent to the His-338 residue, was found to be lumenal by both peptide-N-glycosidase sensitivity and selective permeabilization. We show key positions in Fig. 6E; this approach was effective at all positions tested but two (positions 8a and 8b, where the construct did not produce full-length GOAT). Therefore, two new positions were tested in loop 8, 8c and 8d, and both were full-length and found to be cytosolic (Fig. 6, C and E). A summary of all topology data is presented in Table 1, and a full topology model taking into consideration all of the data is presented in Fig. 7.
FIGURE 6.
V5Glyc tag allows mapping of GOAT topology using two different techniques. A–D, glycosylation gel shift assay for localization in transfected HeLa cells. Mouse GOAT constructs with internal or terminal V5Glyc tags and a constant C-terminal 3xFLAG tag were lysed 20 h after transfection and treated with peptide-N-glycosidase F (PNGaseF) or mock-treated in identical buffer and then subjected to anti-V5 immunoblotting. Only lumenal positions can be glycosylated, and peptide-N-glycosidase F treatment cleaves off the oligosaccharide. Constructs in B were less well expressed than those in D. The anti-FLAG blot in D shows an additional band present (gray arrow) not seen in the anti-V5 blot for the N-terminal construct and position 1; this band is shown below to represent a protein starting with Met-56. Note that positions 8a and 8b are not shown; no full-length GOAT could be detected from these constructs. In C, two additional positions were tested in loop 8 (8c and 8d), and both expressed well and confirmed cytosolic location. E, selective permeabilization of key V5Glyc constructs reports lumenal location for challenging positions 9 and 11 and cytosolic location of loop 8; other previously suggested positions are also confirmed. As above, GOAT bearing the indicated internal V5Glyc tag was used, and all constructs have a constant C-terminal 3xFLAG tag. The N terminus and positions 5 and 7 are shown because these were the most robust in our initial survey. F, NXS or NXT codons (not V5Glyc tags) were installed in all loops longer than 24 amino acids: loops 5, 6, and 8. Loop 8 is longer, so three constructs were generated. None of these epitopes could be glycosylated, consistent with cytosolic locations.
TABLE 1.
GOAT topology summary
L, lumenal; C, cytosolic. Plus signs indicate V5 or Myc signal level in indirect immunofluorescence. Alt glyc*, a variety of alternate glycosylation signal sequences were installed at the GOAT N terminus (Fig. 9B), and these were all lumenal. Endogenous Asn**, an endogenous Asn in a consensus NX(S/T/C) site was not glycosylated. NXS/T Mutant***, a mutation creating a consensus NX(S/T) glycosylation site was installed in the center of this loop but could not be glycosylated (Fig. 9B). Myc hGOAT, human GOAT was made with a Myc tag at this position, and results agree with the mouse GOAT result. ND, not done; GOAT with V5Glyc tag installed in positions 8a and 8b did not result in full-length protein, so positions 8c and 8d were tested with this tag. †, in a mass spectrometry experiment performed on full-length GOAT, we observed a peptide corresponding to ubiquitylation at residue Lys-141, suggesting cytosolic localization. Relative signal strength assignment shown with variable numbers of plus signs is described under “Experimental Procedures.”
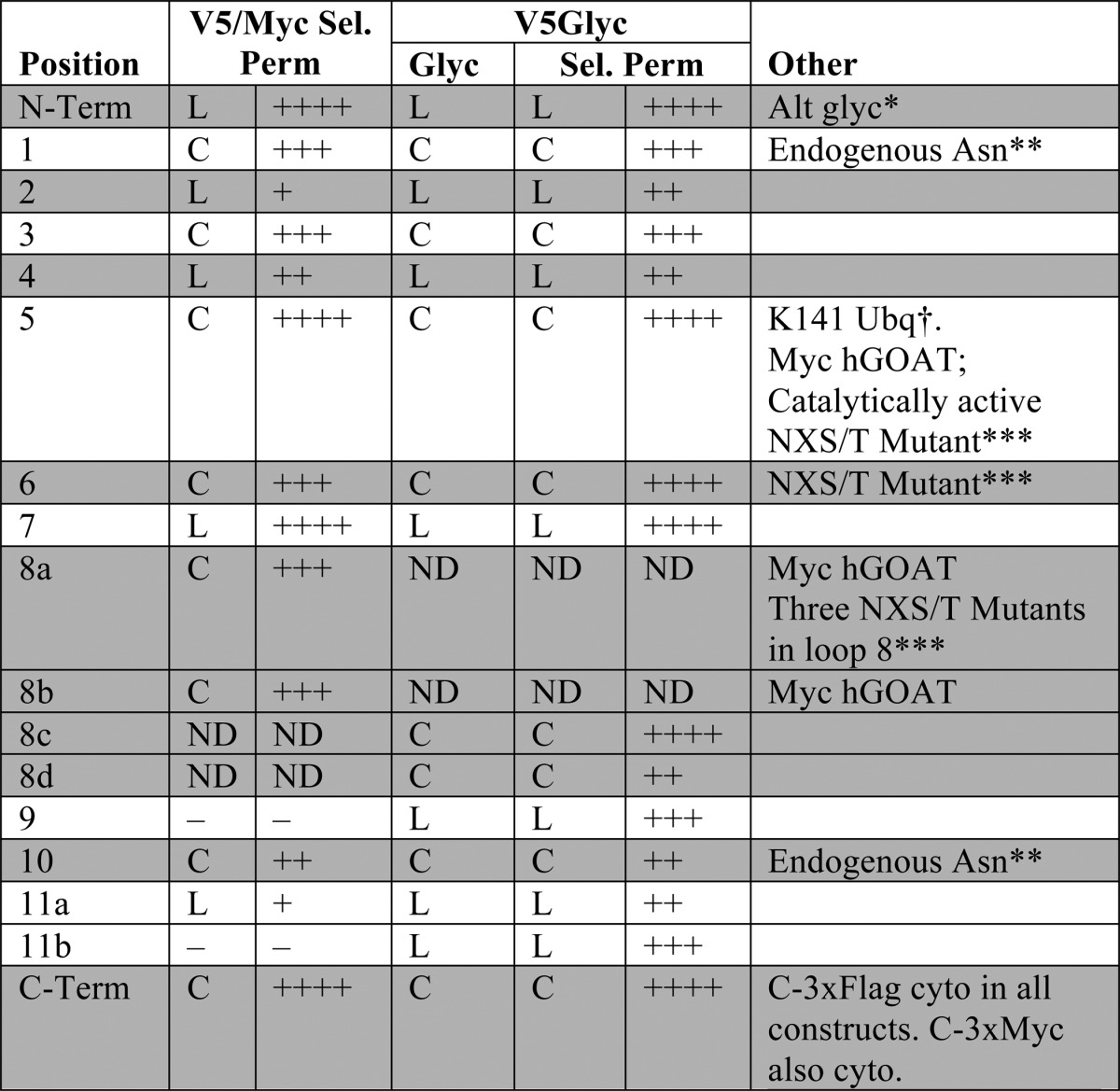
FIGURE 7.
GOAT topology model as an 11-TM protein with one reentrant loop. Confirmed TMs are shown in green, spanning the ER membrane (gray). The lumen is above, and cytosol is below. We suggest that the hydrophobic region of GOAT c6, predicted to be a TM domain, may be a reentrant loop, shown in purple. Relative loop length is shown by the size of the loops (black lines), but they are not precisely scaled. Variable epitope tag positions used are shown as blue spheres; the constant C-terminal 3xFLAG tag is a red sphere. The alternate translational start site (Met-56) is highlighted in orange. Positions of the two conserved MBOAT residues (Asn-307 and His-338 in mouse and human GOAT) are shown as yellow stars, and the most conserved region across all MBOATs is shown as a gray box (approximately from Arg-303 to Met-354 in GOAT, from pfam03062 (20)). The figure design is modified from the output of MEMSAT-SVM, with permission.
Met-56 Is an Alternate Start Codon in Mouse GOAT, Resulting in Two Species by SDS-PAGE
Interpretation of the gel shift blotting (Fig. 6, A–C) is somewhat complicated by the finding of at least two bands from mouse GOAT; the two predominant bands from HeLa cells have a nearly identical size difference as that induced by the gel shift from N-glycosylation. We noticed that in lumenal positions, the pattern of these bands generally shifts together upon peptide-N-glycosidase F treatment (e.g. position 9 (Fig. 6B) and positions 2, 4, and 7 (Fig. 6C)), suggesting that they were separate GOAT species that were both glycosylated.
We noticed that the lower band was absent from the blot for the modular V5Glyc tag but present at a smaller molecular weight in the C-terminal FLAG blot when the V5Glyc tag was installed at the N terminus and position 1 (Fig. 6D, gray arrow). This split suggested that the two bands represent GOAT proteins with different N termini (i.e. with two distinct translational start sites) and triggered us to map the lower band as GOAT initiating translation at Met-56 (see below).
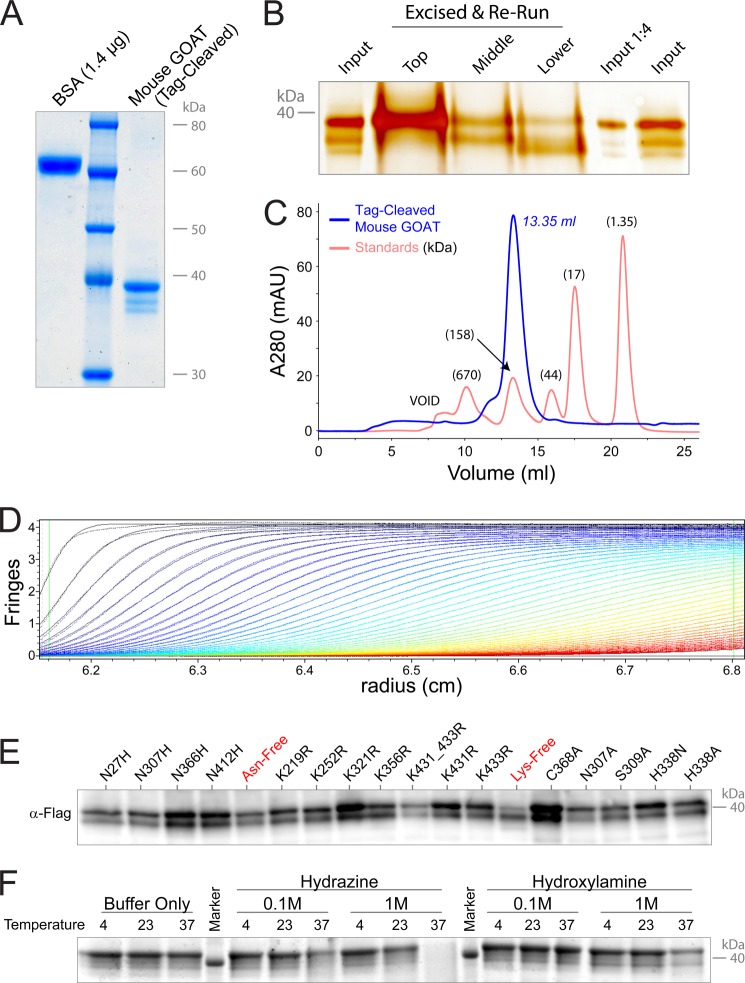
Mouse GOAT purified from SF9 cells (with C-terminal 3xFLAG tag cleaved) produces three distinct bands upon SDS-PAGE when the proteins are maximally separated and Coomassie-stained (Fig. 8A). The middle band is generally absent from immunoblots (Fig. 8E) and may represent a co-purified protein. Mobilities of the top and bottom bands when GOAT was produced in HeLa and SF9 cells were identical (not shown). We found that the two predominant bands did not change when all lysines or all asparagines were removed from GOAT by conversion to alanine residues (Fig. 8E), suggesting that they were not due to post-translational modification of these residues. Cys-368 was mutated as part of a potential NXC motif (see below), and S309A was mutated as a potential phosphopantetheinylation site, and these mutants too show both species. The banding pattern also did not change in response to hydroxylamine and hydrazine treatment of GOAT (Fig. 8F), suggesting that multiple banding did not arise from fatty acid esterification.
FIGURE 8.
Purification of GOAT as a monomer with at least two stable bands. A, colloidal Coomassie Brilliant Blue (CBB) staining of purified mouse GOAT after tag cleavage and ion exchange chromatography reveals three bands. Four identical lanes of mouse GOAT were used as input for electroelution. B, bands in A are stable species, not interconverting gel artifacts. Stained bands were excised, electroeluted, and concentrated and then rerun and silver-stained as compared with the original purified GOAT. C, size exclusion chromatography of cleaved GOAT and standard proteins. Sizes of proteins in the standard are shown in parentheses. The calculated molecular mass for monomeric GOAT in FC-16 micelles is 140.7 kDa. TEV-cleaved GOAT alone has a calculated molecular mass of 50.6 kDa. Empty FC-16 detergent micelles alone are ∼72.5 kDa and do not result in UV absorbance signal at the concentration used. D, interference optical monitoring data from analytical ultracentrifugation of GOAT were fit in SEDFIT to a C(s) model. Fringes over time are shown in progression from red to black; absorbance data (not shown) was similar. Approximately 90% of the signal in the sample fit to a single peak at 1.9 S, corresponding to an S (20 °C, water) of 5.4 and an approximate molecular mass of 110 kDa. E, anti-FLAG immunoblot of GOAT-3xFLAG in SF9 cells with the indicated mutations shows that multiple bands seen are not due to post-translational modification on asparagine or lysine or at residue Cys-368 or Ser-309. Asn-free and Lys-free GOATs include all of the Asn and Lys mutations listed in other constructs. F, treatment with hydrazine or hydroxylamine does not alter the banding pattern of purified GOAT. Mouse GOAT was incubated overnight at the indicated temperatures and concentrations with hydrazine and hydroxylamine (pH 8.0). A Coomassie Brilliant Blue-stained gel is shown; mouse GOAT used in this experiment was eluted from FLAG resin using 100 mm glycine, pH 3.5, and retains the C terminus of the 3xFLAG tag.
We were concerned that the two bands might represent a gel artifact because, like many other integral membrane proteins, GOAT aggregates when heated in SDS and therefore may not be fully denatured; it also runs at a smaller than predicted molecular weight, again common in membrane proteins due to hydrophobicity resulting in increased SDS binding (64). We therefore excised the three bands from the gel, electroeluted the proteins, and reran them in separate lanes (Fig. 8B). The three species ran true, within the limits of our ability to separate them using this technique, suggesting that they are distinctly covalent and non-interconverting structures.
We analyzed purified mouse GOAT in FC-16 detergent micelles by size exclusion chromatography. Under these conditions, the solution contains a mixture of detergent micelles containing GOAT and empty detergent micelles. The empty micelles are ∼72.4 kDa for FC-16, with an aggregation number of 178 detergent molecules/micelle (Anatrace). TEV-cleaved GOAT has a molecular mass of 50.6 kDa; as a monomer or dimer in micelles, the predicted mass would be ∼123 or ∼174 kDa, respectively. In size exclusion chromatography, GOAT ran as one major peak (Fig. 8C). As compared with standard (soluble) proteins, the size exclusion chromatography-calculated molecular mass for GOAT in its FC-16 micelle is 140.7 kDa. This size does not allow clear determination of whether purified GOAT is a monomer or dimer, although it is closer to a monomer position. Hydrophobic interactions between the GOAT protein and resin may result in non-ideal behavior. Note that the detergent does not result in detectable UV absorbance at these concentrations, as seen by the lack of a peak at ∼70 kDa corresponding to free detergent micelles.
To further investigate the quarternary structure of GOAT, we employed sedimentation velocity analytical ultracentrifugation. As shown in Fig. 8D, sedimentation velocity data using interference optical monitoring of GOAT were fit in SEDFIT to a C(s) model (65). Approximately 90% of the signal in the sample fit to a single peak at 1.9 S, which was normalized to 5.4 S when taking into account the temperature correction factor (20 °C, water). This sedimentation behavior corresponds to an approximate molecular mass of 110 kDa, consistent with monomeric GOAT in FC-16. Although the peak was slightly asymmetric, we were unable to resolve additional, smaller species. UV absorbance data (not shown) showed similar results. Fitting the data using the program dC/dT+, which employs alternative methods of fitting data to those used by SEDFIT, calculated a similar S value and identified one major species in the experiment (66).
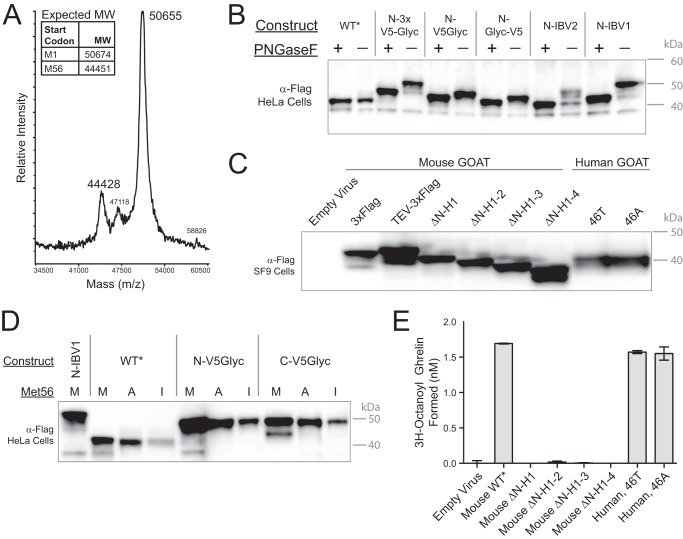
We next analyzed intact, purified GOAT by MALDI-TOF mass spectrometry (Fig. 9A). In addition to the major GOAT peak at the expected size, a minor species with a mass of 44,428 Da was found, consistent with a protein initiated at Met-56. Further support that this 44,428 Da peak corresponds to the lower band in SDS-PAGE can be found in its altered glycosylation pattern. The addition of various glycosylation sequences to the N terminus of GOAT (Figs. 6D and 9B), caused a size increase in the upper but not the lower band, and only the top band shifted further with glycosylation, suggesting that the faster migrating species initiates downstream of the first ATG.
FIGURE 9.
The lower GOAT band is a protein with N terminus Met-56. A, MALDI-TOF of intact, purified tag-cleaved mouse GOAT. Expected molecular weights (MW) with the indicated start codons are inset (estimated S.E. for these masses is ±50 Da). B, adding N-terminal sequence shifts the upper band but not the lower band in mouse GOAT. *, all constructs contain a C-terminal 3xFLAG tag. N-IBV1 and N-IBV2 are described under “Experimental Procedures”; 3xV5Glyc is V5-Glyc-V5-Glyc-V5; V5Glyc is V5-Glyc-V5; Glyc-V5 lacks the first V5 epitope of V5Glyc. C, anti-FLAG immunoblot of SF9 microsomes of full-length and helix-deleted constructs. ΔN-H1 construct lacks the first helix, ΔN-H1-2 lacks the first two helices, etc. The start codon for ΔN-H1-2 is Met-56; for ΔN-H1-3 and ΔN-H1-4, a Met was installed in place of residues 81 and 109, respectively, as the first residue of the construct. All deletion mutants have a C-terminal TEV-3xFLAG. Human GOAT proteins are labeled 46A and 46T because this position was expressed as either the conserved Ala or the recorded coding SNP Thr. 3-Fold more human GOAT-expressing microsomes were loaded than mouse GOAT-expressing microsomes, due to lower expression. D, mutation of Met-56 removes the lower band from mouse GOAT. Met-56 was mutated to alanine or isoleucine for the indicated constructs, and all were expressed in HeLa cells. Expression levels are reduced by 5–10-fold for mutant constructs (not shown); here, 4 μg of total protein was loaded for Met constructs, and 20 μg was loaded for Ala and Ile constructs. E, microsomal activity assay of TEV-3xFLAG tagged GOAT constructs in C. Each bar represents an average of duplicates; error bars, S.D.
Because Met-56 is predicted to be the last (lumenal) residue in TM-2, recovery of the lower band suggests that GOAT lacking its first two TMs might be stable. This may also be a physiologic product, and we note that Met-56 is the first ATG in the second exon of GOAT. We therefore made constructs for baculoviral expression from which the first, first two, first three, and first four helices had been removed (Fig. 9C). The size of the construct from which the first two helices have been deleted (ΔN-H1-2; start codon is Met-56) co-migrated with the lower band in the full-length construct, consistent with our hypothesis. However, none of the deletion mutants were active (Fig. 9E). We note that although human GOAT expressed at levels lower than mouse GOAT, it had similar activity. The two different human GOAT clones have Thr or Ala at position 46; we made both after noticing that the conserved Ala residue has also been reported as Thr (2). Indeed, a recorded human coding SNP specifies this polymorphism (rs7813902). Both clones performed identically in our assays.
To further confirm that Met-56 can act as an alternate start codon, we mutated this residue. Met-56 is conserved in all GOAT sequences evaluated except Green Anole, where it is Ile; we therefore made both M56A and M56I mutants in three different C-3xFLAG-tagged GOAT constructs and expressed them in HeLa cells (Fig. 9D). The lower band was no longer detectable in any of these mutants. We note that the expression of the mutant proteins was lower than that of WT, and we loaded 5-fold more total protein in those lanes. The N-IBV1 construct is shown to calibrate the lower band position.
Additional Data Supporting the Model
Mouse GOAT is not N-glycosylated (Figs. 8E and 9B). However, two Asn residues are predicted to be candidates for glycosylation by the NetNGlyc server; that neither can be glycosylated is consistent with their cytosolic localizations in our model. Asn-27 (79% and 9 of 9 jury, +++ rating) is at the cytosolic border of TM-1, and Asn-366 (62% and 9 of 9 jury, ++ rating) is in the middle of the cytosolic loop between TM-9 and -10. Conserved Asn-307 is in an NXS motif but is both cytosolic and not predicted to be glycosylated (48%, 7 of 9 jury, − rating).
Mouse GOAT has only three loops where central installation of NXS or NXT motifs by point mutations would also allow sufficient spacing from the membrane for potential glycosylation by oligosaccharyltransferase (12 residues on each side of the Asn, or 24 residues total). In loops 5 and 6, we installed sites on both sides of the reentrant helix (I149N and Q191N, NRS in both cases). These were predicted to be glycosylated by NetNGlyc (74 and 68%, respectively, both with 9 of 9 jury and ++ rating). Neither mutant GOAT could be glycosylated (Fig. 6F). We installed three more potential sites in loop 8, with mutants I290N, L293N, and R298N making NWT, NET, and NIS sequences (prediction 51, 58, and 67%, respectively, jury 6 of 9, 5 of 9, and 8 of 9, respectively, + rating). These positions were not glycosylated (Fig. 6F).
Loop 10 (20 residues) already contains Asn-366, which is centrally located and in a non-standard but putative NXC motif, so we made no changes here. In summary, the lack of glycosylation in any of these sites and mutants is consistent with our model.
Additionally, Lys-141 of GOAT made in SF9 cells was found to be ubiquitylated by electrospray mass spectrometry, with a trypsin site that was not cleaved and +114.04-Da shift corresponding to GG isopeptide linkage. This is consistent with cytosolic localization, because ubiquitylation can only occur on the cytosolic face of the ER membrane (67).
Characterization of the GOAT Active Site by Photocross-linking
We previously reported that both microsomal and solubilized, purified GOAT can be specifically photocross-linked by a biotinylated analog of the bisubstrate inhibitor of GO-CoA-Tat containing UV-activatable benzoylphenylalanine in place of Phe-4 (GO-CoA-Tat F4BP) (8). Note that solubilized GOAT is not active (32, 34); however, both with and without presolubilization, this cross-linking could be competed by the addition of excess, unlabeled GO-CoA-Tat, suggesting specificity of the reaction to the enzyme's active site and the maintenance of a physiologically relevant fold. Therefore, to further map the active site of GOAT, we performed partial proteolysis of cross-linked microsomal GOAT (Fig. 10A). This was done by first cross-linking and then solubilizing in FC-16, digesting with different amounts of trypsin, and then purifying C-terminal fragments using anti-FLAG. A 15-kDa fragment was detected in both anti-FLAG and biotin blotting, suggesting that at least some of the cross-linking occurs in approximately the C-terminal 15 kDa of GOAT (see “Discussion”). Attempts to characterize the exact locations of photocross-linking by mass spectrometry have so far been unsuccessful.
FIGURE 10.
Characterization of GOAT active site by photocross-linking. A, partial proteolysis of covalently photocross-linked mouse GOAT reveals a 15-kDa cross-linked fragment. GO-CoA-Tat F4BP (biotin-labeled) was photocross-linked to microsomal mouse GOAT-3xFLAG or to control microsomes (Ctrl; from empty-virus infected cells). Microsomes were then solubilized, digested with trypsin, and isolated using anti-FLAG beads (see “Experimental Procedures”). Samples were then immunoblotted using streptavidin or anti-FLAG. Note that although the biotin cannot be removed from GO-CoA-Tat F4BP during trypsin digest, the 3xFLAG tag contains multiple trypsin cleavage sites, and smaller tag-cleaved species may have been lost or may have fallen below the limit of detection. B, synthetic scheme to make ghrelin-28-Oct-diazirine, an acyl ghrelin product analog photocross-linker. As a complement to GO-CoA-Tat F4BP previously reported (8), we designed another set of photoaffinity labeling agents based on the fact that the amide analogs of the acyl ghrelin product have high affinity for GOAT (32). Photolabile diazirine was installed at the 7-position of the octanoic acid side chain, and biotinylated lysine was installed at the C terminus of ghrelin-28. Ghrelin-28-Oct-F4BP was made similarly, using unlabeled octanoic anhydride and replacing Phe-4 with benzoylphenylalanine. C–E, photocross-linking cross-competition binding assay. Each of three biotinylated photocross-linking compounds (GO-CoA-Tat F4BP (C), ghrelin-28-Oct-diazirine (D), and ghrelin-28-Oct-F4BP (E)) was cross-linked to purified, solubilized mouse GOAT in the presence of an excess of the indicated unlabeled competitors. Separate subpanels represent separate experiments. Coomassie Brilliant Blue (CBB) staining is used as a loading control.
We next characterized the binding of various analogs and inhibitors to GOAT using photocross-linking compounds and competitors. Based on the fact that ghrelin product analogs with amide lipid linkage have high affinity to GOAT (2), we synthesized two C-terminally biotinylated ghrelin product analogs with photocross-linking moieties in the lipid (ghrelin-28-Oct-diazirine; Fig. 10B) and on the adjacent residue in the peptide chain (ghrelin-28-Oct-F4BP). Purified GOAT in FC-16 was then cross-linked with these three analogs alone or in the presence of an excess of the following unlabeled competitors: GO-CoA-Tat, acyl ghrelin (amide form), desacyl ghrelin, octanoyl-CoA, and GO-Tat (amide-linked octanoyl ghrelin product analog containing 10 residues of ghrelin followed by Ahx linker and Tat sequence). Binding of the bisubstrate cross-linker GO-CoA-Tat F4BP was only efficiently competed by GO-CoA-Tat itself (Fig. 10C). This suggests that the bisubstrate inhibitor has a higher affinity for GOAT, perhaps by occupying a larger space in the binding site. On the other hand, the product analog photocross-linkers ghrelin-28-Oct-diazirine and ghrelin-28-Oct-F4BP behaved similarly to each other but distinctly from the bisubstrate (Fig. 10, D and E). Both product cross-linker analogs were competed by GO-CoA-Tat, suggesting that their binding to GOAT overlaps where the bisubstrate binds. Both product cross-linker analogs, which presumably attach to GOAT in distinct ways because of the location of their photoreactive groups, could also be competed by both GO-Tat and acyl ghrelin. These results suggest that the location of the photoreactive groups does not significantly affect analog binding. Octanoyl-CoA and desacyl ghrelin, which may bind more weakly to GOAT or in a non-overlapping way, did not compete or only minimally competed against both cross-linker product analog compounds.
DISCUSSION
These studies establish the location of the 11 transmembrane-spanning domains and one reentrant loop of GOAT. Using a combination of phylogeny, computational prediction, epitope mapping, and induced glycosylation, we obtained strong convergent evidence for this 11-transmembrane domain model. This new model contains three more transmembrane-spanning domains than previously predicted and also suggests that one domain predicted by the recently developed programs is instead a reentrant loop. This model also indicates that the topology of GOAT is quite structurally complex. Although our evidence strongly supports this overall 11-transmembrane model, we recognize that polytopic membrane proteins are complex, and dynamic alterations of topology are formally possible. We also appreciate that the loop insertions employed, although small in size and unable to flip the C terminus into the lumen, may perturb local structure.
Three other MBOATs have been fully mapped topologically: human ACAT1 and ACAT2 (official symbols SOAT1 and SOAT2, reviewed in Ref. 35) and yeast Gup1p (68). GOAT, ACAT1, and Gup1p have somewhat similar topologies, but ACAT2 is quite different than the other three. All four MBOATs have in common that the invariant histidine (His-338) is lumenal or buried in the ER membrane near the lumen; this was also shown for partially mapped yeast MBOATs Ale1p and Are1p (68). For GOAT, ACAT1, Gup1p, and Ale1p, the histidine is at the lumenal tip of two TMs separated by only a short 2–3-residue turn. Are2p is similar, except the C-terminal TM is probably instead a reentrant loop. The conserved asparagine (Asn-307 in GOAT) was also found to be cytosolic in all six cases.
The topology models of GOAT and Gup1p are the most similar, with 12 distinct hydrophobic regions separated by relatively short hydrophilic loops: N terminus lumenal, C terminus cytosolic, short terminal tails, a reentrant loop near the middle of the protein, and a relatively long cytosolic loop immediately N-terminal to the hydrophobic region containing the invariant His. However, for Gup1p, the N-terminal two regions were found to be reentrant loops, resulting in a model with nine TMs.
ACAT1 topology has been studied rigorously using a variety of methods. Overall, its topology is relatively similar to GOAT, with nine TMs, but ACAT1 has longer loops, two fewer helices, and opposite orientation of the N and C termini. In contrast, ACAT2 was shown to have only two transmembrane domains using selective permeabilization with two distinct epitope tags (29). These results are surprising, considering that ACAT2 has 10 regions predicted to be transmembrane or reentrant by MemBrain (data not shown), similar to the other MBOATs; some of the non-TM hydrophobic regions were modeled as reentrant loops. Also, with the exception of some subtle differences in predicted loop regions around the invariant histidine, the positions chosen should have efficiently interrogated these locations. It is possible, therefore, that ACAT2 has a distinct topology compared with the other MBOATs.
These studies have revealed that the GOAT C terminus can physically contact peptide and octanoyl-CoA substrates and that the bisubstrate analog encompasses both enzyme interaction surfaces. The lumenal location of His-338 suggests that it could well be in the active site of the GOAT enzyme, consistent with the hypothesis of a conserved lumenal active site for the MBOAT family. In contrast, the conserved extralumenal position of the conserved MBOAT residue Asn-307 indicates that it is unlikely to be directly involved in catalysis. However, its importance might be in substrate interactions, transport of substrates, or protein structural stability.
The finding of a FLAG-tagged ∼15-kDa fragment with GO-CoA-Tat F4BP cross-linking maps the ghrelin binding site to the C-terminal region of GOAT. This could be consistent with cross-linking to the regions around TM-8 and TM-9, depending on whether the cross-linked compound causes a gel shift of this smaller GOAT fragment. In any event, these results suggest that the GOAT C-terminal region is directly involved in substrate binding and/or catalysis. Additionally, this region contains three lysophospholipid acyltransferase motifs essential for activity in this subset of MBOATs (69).
The quaternary structure of GOAT has not been investigated previously; the only MBOAT for which the quaternary structure is known is ACAT1, which is a homotetramer both in cell culture and in vitro (63). We show by analytical ultracentrifugation that purified GOAT in detergent micelles is a monomer (Fig. 8D). Taken together with the specific binding of ghrelin photocross-linking analogs to GOAT under nearly identical conditions (Fig. 10, C–E), this suggests that GOAT can bind ghrelin and analogs as a monomer.
Additionally, the V5Glyc glycosylation strategy developed here may be of more general utility for the analysis of membrane proteins. In particular, the flanked sequence seems to be generally well tolerated for loop insertion and efficiently processed by oligosaccharyltransferase. It gives a robust signal on both immunofluorescence and immunoblotting, and this dual topology reporter is far smaller than others previously employed, suggesting that it is likely to be better tolerated for membrane protein applications.
Acknowledgments
We thank Hongbin Shen for helpful discussion on topology and for use of the MemBrain program. We thank Dan Leahy, Jennifer Kavran, Matthew Ward, and Thomas Cleveland for assistance with expression and purification. We thank Jun Liu and Shridhar Bhat for help with cross-linking experiments and helpful discussion. We thank Ann Hubbard and Ron Schnaar for assistance with solubilization and helpful discussion. We thank Geraldine Seydoux and Seth Zonies for confocal microscopy. We thank Jeremy Johnson for providing access to the draft anole genome, which greatly improved our multiple-species alignment. We thank Andreas Conzelmann for sharing information on the SUC2/His4C reporter.
This work was supported, in whole or in part, by National Institutes of Health Grants 8U54GM103520 and GM62437. This work was also supported by funds from the Pfeiffer Foundation.
T. E. Cleveland, 4th, M. D. Ward, M. S. Taylor, J. M. Kavran, and D. J. Leahy, manuscript in preparation.
R. Gupta, E. Jung, and S. Brunak, manuscript in preparation.
- GOAT
- ghrelin O-acyltransferase
- MBOAT
- membrane-bound O-acyltransferase
- ER
- endoplasmic reticulum
- TM
- transmembrane helix
- IBV
- infectious bronchitis virus
- FC-16
- Fos-Choline 16
- DTR
- dual topology reporter
- TEV
- tobacco etch virus.
REFERENCES
- 1. Gutierrez J. A., Solenberg P. J., Perkins D. R., Willency J. A., Knierman M. D., Jin Z., Witcher D. R., Luo S., Onyia J. E., Hale J. E. (2008) Ghrelin octanoylation mediated by an orphan lipid transferase. Proc. Natl. Acad. Sci. U.S.A. 105, 6320–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. (2008) Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 132, 387–396 [DOI] [PubMed] [Google Scholar]
- 3. Kojima M., Hosoda H., Date Y., Nakazato M., Matsuo H., Kangawa K. (1999) Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660 [DOI] [PubMed] [Google Scholar]
- 4. Ozawa A., Speaker R. B., 3rd, Lindberg I. (2009) Enzymatic characterization of a human acyltransferase activity. PLoS One 4, e5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishi Y., Hiejima H., Hosoda H., Kaiya H., Mori K., Fukue Y., Yanase T., Nawata H., Kangawa K., Kojima M. (2005) Ingested medium-chain fatty acids are directly utilized for the acyl modification of ghrelin. Endocrinology 146, 2255–2264 [DOI] [PubMed] [Google Scholar]
- 6. Kirchner H., Gutierrez J. A., Solenberg P. J., Pfluger P. T., Czyzyk T. A., Willency J. A., Schürmann A., Joost H. G., Jandacek R. J., Hale J. E., Heiman M. L., Tschöp M. H. (2009) GOAT links dietary lipids with the endocrine control of energy balance. Nat. Med. 15, 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. López M., Lage R., Saha A. K., Pérez-Tilve D., Vázquez M. J., Varela L., Sangiao-Alvarellos S., Tovar S., Raghay K., Rodríguez-Cuenca S., Deoliveira R. M., Castañeda T., Datta R., Dong J. Z., Culler M., Sleeman M. W., Alvarez C. V., Gallego R., Lelliott C. J., Carling D., Tschöp M. H., Diéguez C., Vidal-Puig A. (2008) Hypothalamic fatty acid metabolism mediates the orexigenic action of ghrelin. Cell metabolism 7, 389–399 [DOI] [PubMed] [Google Scholar]
- 8. Barnett B. P., Hwang Y., Taylor M. S., Kirchner H., Pfluger P. T., Bernard V., Lin Y. Y., Bowers E. M., Mukherjee C., Song W. J., Longo P. A., Leahy D. J., Hussain M. A., Tschop M. H., Boeke J. D., Cole P. A. (2010) Glucose and weight control in mice with a designed ghrelin O-acyltransferase inhibitor. Science 330, 1689–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trudel L., Tomasetto C., Rio M. C., Bouin M., Plourde V., Eberling P., Poitras P. (2002) Ghrelin/motilin-related peptide is a potent prokinetic to reverse gastric postoperative ileus in rat. Am. J. Physiol. Gastrointest. Liver Physiol. 282, G948–G952 [DOI] [PubMed] [Google Scholar]
- 10. Ejskjaer N., Wo J. M., Esfandyari T., Mazen Jamal M., Dimcevski G., Tarnow L., Malik R. A., Hellström P. M., Mondou E., Quinn J., Rousseau F., McCallum R. W. (2013) A phase 2a, randomized, double-blind 28-day study of TZP-102 a ghrelin receptor agonist for diabetic gastroparesis. Neurogastroenterol. Motil. 25, e140–e150 [DOI] [PubMed] [Google Scholar]
- 11. Carlini V. P., Monzón M. E., Varas M. M., Cragnolini A. B., Schiöth H. B., Scimonelli T. N., de Barioglio S. R. (2002) Ghrelin increases anxiety-like behavior and memory retention in rats. Biochem. Biophys. Res. Commun. 299, 739–743 [DOI] [PubMed] [Google Scholar]
- 12. Diano S., Farr S. A., Benoit S. C., McNay E. C., da Silva I., Horvath B., Gaskin F. S., Nonaka N., Jaeger L. B., Banks W. A., Morley J. E., Pinto S., Sherwin R. S., Xu L., Yamada K. A., Sleeman M. W., Tschöp M. H., Horvath T. L. (2006) Ghrelin controls hippocampal spine synapse density and memory performance. Nat. Neurosci. 9, 381–388 [DOI] [PubMed] [Google Scholar]
- 13. Egecioglu E., Jerlhag E., Salomé N., Skibicka K. P., Haage D., Bohlooly-Y M., Andersson D., Bjursell M., Perrissoud D., Engel J. A., Dickson S. L. (2010) Ghrelin increases intake of rewarding food in rodents. Addict. Biol. 15, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwenke D. O., Tokudome T., Kishimoto I., Horio T., Shirai M., Cragg P. A., Kangawa K. (2008) Early ghrelin treatment after myocardial infarction prevents an increase in cardiac sympathetic tone and reduces mortality. Endocrinology 149, 5172–5176 [DOI] [PubMed] [Google Scholar]
- 15. Baldanzi G., Filigheddu N., Cutrupi S., Catapano F., Bonissoni S., Fubini A., Malan D., Baj G., Granata R., Broglio F., Papotti M., Surico N., Bussolino F., Isgaard J., Deghenghi R., Sinigaglia F., Prat M., Muccioli G., Ghigo E., Graziani A. (2002) Ghrelin and des-acyl ghrelin inhibit cell death in cardiomyocytes and endothelial cells through ERK1/2 and PI 3-kinase/AKT. J. Cell Biol. 159, 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okumura H., Nagaya N., Enomoto M., Nakagawa E., Oya H., Kangawa K. (2002) Vasodilatory effect of ghrelin, an endogenous peptide from the stomach. J. Cardiovasc. Pharmacol. 39, 779–783 [DOI] [PubMed] [Google Scholar]
- 17. Goldstein J. L., Zhao T. J., Li R. L., Sherbet D. P., Liang G., Brown M. S. (2011) Surviving starvation. Essential role of the ghrelin-growth hormone axis. Cold Spring Harb. Symp. Quant. Biol. 76, 121–127 [DOI] [PubMed] [Google Scholar]
- 18. Li R. L., Sherbet D. P., Elsbernd B. L., Goldstein J. L., Brown M. S., Zhao T. J. (2012) Profound hypoglycemia in starved, ghrelin-deficient mice is caused by decreased gluconeogenesis and reversed by lactate or fatty acids. J. Biol. Chem. 287, 17942–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yi C. X., Heppner K. M., Kirchner H., Tong J., Bielohuby M., Gaylinn B. D., Müller T. D., Bartley E., Davis H. W., Zhao Y., Joseph A., Kruthaupt T., Ottaway N., Kabra D., Habegger K. M., Benoit S. C., Bidlingmaier M., Thorner M. O., Perez-Tilve D., Tschöp M. H., Pfluger P. T. (2012) The GOAT-ghrelin system is not essential for hypoglycemia prevention during prolonged calorie restriction. PLoS One 7, e32100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann K. (2000) A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 21. Chen M. H., Li Y. J., Kawakami T., Xu S. M., Chuang P. T. (2004) Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18, 641–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., Takada S. (2006) Monounsaturated fatty acid modification of Wnt protein. Its role in Wnt secretion. Dev. Cell 11, 791–801 [DOI] [PubMed] [Google Scholar]
- 23. Bosson R., Jaquenoud M., Conzelmann A. (2006) GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell 17, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petrova E., Rios-Esteves J., Ouerfelli O., Glickman J. F., Resh M. D. (2013) Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat. Chem. Biol. 9, 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hardy R. Y., Resh M. D. (2012) Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase. J. Biol. Chem. 287, 42881–42889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Resh M. D. (2012) Targeting protein lipidation in disease. Trends Mol. Med. 18, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pagac M., de la Mora H. V., Duperrex C., Roubaty C., Vionnet C., Conzelmann A. (2011) Topology of 1-acyl-sn-glycerol-3-phosphate acyltransferases SLC1 and ALE1 and related membrane-bound O-acyltransferases (MBOATs) of Saccharomyces cerevisiae. J. Biol. Chem. 286, 36438–36447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo Z. Y., Lin S., Heinen J. A., Chang C. C., Chang T. Y. (2005) The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. J. Biol. Chem. 280, 37814–37826 [DOI] [PubMed] [Google Scholar]
- 29. Lin S., Lu X., Chang C. C., Chang T. Y. (2003) Human acyl-coenzyme A:cholesterol acyltransferase expressed in Chinese hamster ovary cells. Membrane topology and active site location. Mol. Biol. Cell 14, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee H. C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. (2008) Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell 19, 1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McFie P. J., Stone S. L., Banman S. L., Stone S. J. (2010) Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the N terminus in dimer/tetramer formation. J. Biol. Chem. 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J., Zhao T. J., Goldstein J. L., Brown M. S. (2008) Inhibition of ghrelin O-acyltransferase (GOAT) by octanoylated pentapeptides. Proc. Natl. Acad. Sci. U.S.A. 105, 10750–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamaki H., Shimada A., Ito Y., Ohya M., Takase J., Miyashita M., Miyagawa H., Nozaki H., Nakayama R., Kumagai H. (2007) LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 34288–34298 [DOI] [PubMed] [Google Scholar]
- 34. Taylor M. S., Hwang Y., Hsiao P. Y., Boeke J. D., Cole P. A. (2012) Ghrelin O-acyltransferase assays and inhibition. Methods Enzymol. 514, 205–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang T. Y., Li B. L., Chang C. C., Urano Y. (2009) Acyl-coenzyme A:cholesterol acyltransferases. Am. J. Physiol. Endocrinol. Metab. 297, E1–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buglino J. A., Resh M. D. (2008) Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 283, 22076–22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buglino J. A., Resh M. D. (2010) Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PLoS One 5, e11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M. K., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Lu S., Marchler G. H., Song J. S., Thanki N., Yamashita R. A., Zhang D., Bryant S. H. (2013) CDD. Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 41. Hofmann K., Stoffel W. (1993) A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166 [Google Scholar]
- 42. Hirokawa T., Boon-Chieng S., Mitaku S. (1998) SOSUI. Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 43. Claros M. G., von Heijne G. (1994) TopPred II. An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686 [DOI] [PubMed] [Google Scholar]
- 44. Jones D. T. (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23, 538–544 [DOI] [PubMed] [Google Scholar]
- 45. Bernsel A., Viklund H., Hennerdal A., Elofsson A. (2009) TOPCONS. Consensus prediction of membrane protein topology. Nucleic Acids Res. 37, W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arai M., Mitsuke H., Ikeda M., Xia J. X., Kikuchi T., Satake M., Shimizu T. (2004) ConPred II. A consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32, W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tusnády G. E., Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 48. Käll L., Krogh A., Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction. The Phobius web server. Nucleic Acids Res. 35, W429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Inlow D., Shauger A., Maiorella B. (1989) Insect cell culture and baculovirus propagation in protein-free medium. J. Tissue Cult. Methods 12, 13–16 [Google Scholar]
- 50. Yon J., Fried M. (1989) Precise gene fusion by PCR. Nucleic Acids Res. 17, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mitchell L. A., Cai Y., Taylor M., Noronha A. M., Chuang J., Dai L., Boeke J. D. (2013) ACS Synth. Biol. 2, 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swift A. M., Machamer C. (1991) A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 115, 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Church R. F. R., Weiss M. J. (1970) Synthesis and properties of small functionalized diazirine molecules. Some observations on the reaction of a diaziridine with the iodine-iodide ion system. J. Org. Chem. 35, 2465–2471 [Google Scholar]
- 54. Shen H., Chou J. J. (2008) MemBrain. Improving the accuracy of predicting transmembrane helices. PLoS One 3, e2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nugent T., Jones D. T. (2012) Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinformatics 13, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 57. Nilsson I. M., von Heijne G. (1993) Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268, 5798–5801 [PubMed] [Google Scholar]
- 58. Kreft S. G., Wang L., Hochstrasser M. (2006) Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 281, 4646–4653 [DOI] [PubMed] [Google Scholar]
- 59. Sengstag C. (2000) Using SUC2-HIS4C reporter domain to study topology of membrane proteins in Saccharomyces cerevisiae. Methods Enzymol. 327, 175–190 [DOI] [PubMed] [Google Scholar]
- 60. Deshaies R. J., Schekman R. (1987) A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 105, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim H., Melén K., Osterberg M., von Heijne G. (2006) A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 11142–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim H., Melén K., von Heijne G. (2003) Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J. Biol. Chem. 278, 10208–10213 [DOI] [PubMed] [Google Scholar]
- 63. Yu C., Chen J., Lin S., Liu J., Chang C. C., Chang T. Y. (1999) Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. J. Biol. Chem. 274, 36139–36145 [DOI] [PubMed] [Google Scholar]
- 64. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Philo J. S. (2000) A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279, 151–163 [DOI] [PubMed] [Google Scholar]
- 67. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) The ubiquitylation machinery of the endoplasmic reticulum. Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 68. Pagac M., Vazquez H. M., Bochud A., Roubaty C., Knöpfli C., Vionnet C., Conzelmann A. (2012) Topology of the microsomal glycerol-3-phosphate acyltransferase Gpt2p/Gat1p of Saccharomyces cerevisiae. Mol. Microbiol. 86, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 69. Shindou H., Eto M., Morimoto R., Shimizu T. (2009) Identification of membrane O-acyltransferase family motifs. Biochem. Biophys. Res. Commun. 383, 320–325 [DOI] [PubMed] [Google Scholar]