Background: Higher order actin filament structures are involved in many cellular processes.
Results: Arabidopsis CROLIN1 contains a predicted actin-cross-linking domain and shows F-actin binding, cross-linking, and stabilizing activities in vitro.
Conclusion: CROLIN1 functions as an actin-binding and cross-linking protein.
Significance: CROLIN1 is a previously undiscovered plant actin-cross-linking protein.
Keywords: Actin, Arabidopsis, Cell Polarity, Plant, Plant Biochemistry, CROLIN1, Actin-cross-linking Protein, Novel Actin-binding Protein
Abstract
Higher order actin filament structures are necessary for cytoplasmic streaming, organelle movement, and other physiological processes. However, the mechanism by which the higher order cytoskeleton is formed in plants remains unknown. In this study, we identified a novel actin-cross-linking protein family (named CROLIN) that is well conserved only in the plant kingdom. There are six isovariants of CROLIN in the Arabidopsis genome, with CROLIN1 specifically expressed in pollen. In vitro biochemical analyses showed that CROLIN1 is a novel actin-cross-linking protein with binding and stabilizing activities. Remarkably, CROLIN1 can cross-link actin bundles into actin networks. CROLIN1 loss of function induces pollen germination and pollen tube growth hypersensitive to latrunculin B. All of these results demonstrate that CROLIN1 may play an important role in stabilizing and remodeling actin filaments by binding to and cross-linking actin filaments.
Introduction
The actin cytoskeleton in eukaryotic cells is a highly organized and dynamic structure that plays a central role in the cell and is involved in numerous cellular processes, including cytoplasmic streaming, intracellular transport, cell growth, and organelle positioning (1). A regulatory system that contains actin filaments (F-actin) and actin-binding proteins (ABPs)3 is required for these processes, and the diverse actin cytoskeleton is directly controlled by different ABPs.
Actin bundles are central components of a variety of specialized cellular structures, including stress fibers, microvilli, and growth cones, in animal cells (2), and the main features of the plant cytoskeleton are conserved between plants and animals. For example, the amino acid sequence of actin shows 80–90% similarity between Arabidopsis and humans (3). Therefore, cytoskeleton-regulating factors, ABPs, in plants may be relatively well conserved and function in a fashion similar to those in animal cells. ABPs with bundling and cross-linking activities are responsible for generating and maintaining higher order actin structures. Recently, four conserved classes of actin-bundling factors have been identified in Arabidopsis; their biochemical functions are well documented and demonstrated that both bundling and cross-linking contribute to the formation of actin cables and/or bundles. For example, Arabidopsis VILLIN1, VILLIN4, and VILLIN5 all have been demonstrated to bundle F-actin in vitro (4–7). Tobacco NtWLIM1 and NtWLIM2 (8, 9), all Arabidopsis AtLIMs (10), and Lilium LlLIM1 (11) are recognized as actin-bundling proteins. In addition, fimbrins (both Fimbrin1 and Fimbrin5) bundle or cross-link F-actin in vitro (12, 13), and certain formins, such as rice OsFH5 (14, 15) and Arabidopsis AtFH1, AtFH4, AtFH8, and AtFH14 (16–19), also bundle actin in vitro.
Although several ABPs regulating the formation of actin bundles in plants have been identified (20), most conserved actin-bundling and actin-cross-linking proteins in animals, including facsin, α-actinin, and espin, are absent in the Arabidopsis genome (21). Recent research has reported several novel actin-binding proteins that show bundling activity in plants. The protein SCAB1 contains a unique and previously unreported actin-binding domain that participates in the regulation of F-actin reorganization during stomatal closure (22). THRUMIN1, which contains a conserved C-terminal glutaredoxin-like domain and a putative cysteine-rich zinc-binding domain, bundles F-actin in vitro (23). V-ATPase B subunits in Arabidopsis show actin binding, bundling, and stabilizing activities in vitro (24), despite an absence of reports on actin-bundling functions for members of this protein family in animals or yeast. Interestingly, Arabidopsis actin depolymerization factor 9 (ADF9) facilitates F-actin bundling in vitro (25), and SB401, a pollen-specific protein from Solanum berthaultii, also exhibits bundling activity (26). These results imply that plants may have mechanisms for the formation and regulation of higher order actin structures that are distinct from the mechanisms in animals.
Pollen tube growth relies on a dynamic and precisely organized actin cytoskeleton (27). During this process, F-actin is maintained as distinct structures and performs specific physiological functions (20, 28, 29). Additionally, many actin-bundling/cross-linking factors that maintain normal pollen germination and pollen tube growth have been identified in Arabidopsis pollen. For example, a VLN5 loss-of-function mutant displays delayed pollen tube growth and results in F-actin in both pollen grains and pollen tubes that are sensitive to latrunculin B (Lat B) (7). A loss-of-function mutant of Arabidopsis FIMBRIN5 also results in delayed pollen germination and inhibited tube growth, with both pollen germination and tube growth being hypersensitive to Lat B (13).
In the present study, we identified a functionally unknown gene family (named CROLIN) in the Arabidopsis genome that contains 1–2 predicted actin-cross-linking domains that are highly conserved. Remarkably, CROLINs are only found in the plant kingdom. Here, we mainly focus on CROLIN1, which is specifically expressed in pollen. Our results reveal that CROLIN1 exhibits actin binding, cross-linking, and actin depolymerization-inhibiting activities in vitro. CROLIN1 loss of function induces pollen germination and pollen tube growth hypersensitive to Lat B. Therefore, we demonstrate that CROLIN1, a previously undiscovered plant actin-cross-linking protein, is involved in the formation and maintenance of highly ordered actin structures in Arabidopsis.
EXPERIMENTAL PROCEDURES
Full-length cDNA Cloning and Plasmid Construction
The cDNA coding sequences of Arabidopsis CROLIN1, CROLIN1-N(33–165), and CROLIN1-ΔN165 were amplified from Arabidopsis flowers. For Escherichia coli expression, CROLIN1, CROLIN1-N(33–165), and CROLIN1-ΔN165 were cloned into the pET30a vector or pGEX-4T vector, accordingly.
For the complementation of CROLIN1 in Arabidopsis pollen, CROLIN1 was introduced into a modified binary vector pCAMBIA1300 that contains the pollen-specific promoter Lat52. To analyze promoter activity, the promoter of CROLIN1 (603 bp upstream from the ATG codon) was amplified with specific primers and then inserted into pBI121, which contains the GUS (β-glucuronidase) gene, to generate the pBI121-ProCROLIN1-GUS plasmid (see Table 1 for the primers used).
TABLE 1.
Primers used in this study
The restriction enzyme sites are underlined. F, forward primer; R, reverse primer.
| Primer name | Sequence (5′–3′) |
|---|---|
| CROLIN1 F | GGATCC/TCTAGACTCCAAGGATTGGATTGAAGATCAT |
| CROLIN1 R | GAGCTC/CCATGGGGTACC TCAAGAAGTGCTTGCAGCATC |
| N(33–165) F | GAGCTCCTCCAAGGATTGGATTGAAGATCAT |
| N(33–165) R | GTCGACTCATGACCTATGAGGTATATCAT |
| ΔN165 F | GGATCCATGACCACACAGGATTGGGTTTTA |
| ΔN165 R | GAGCTCTCAAGAAGTGCTTGCAGCATC |
| CROLIN1p F | AAGCTTCTACATGTTTACTAAATTAGGGT |
| CROLIN1p R | TCTAGATCATGTGAAACAATTTCAGGTAC |
| EF1A F | ACCACGAGTCTCTTCTTGAGGCAC |
| EF1A R | TGGCAGGGTCATCCTTGGAG |
| CROLIN1real-time F | TGGATGAAACAGCAAAGGAAGA |
| CROLIN1real-time R | TGGGAGGAAGATGTAACCGAAG |
| EF4A F | TTGGCGGCACCCTTAGCTGGATCA |
| EF4A R | ATGCCCCAGGACATCGTGATTTCAT |
Protein Purification
The CROLIN1 recombinant protein without the N-terminal 32 amino acids was expressed in the E. coli BL21 (DE3) strain by induction with 1 mm isopropylthio-β-d-galactopyranoside overnight at 28 °C. Recombinant CROLIN1 fused to a glutathione S-transferase (GST) tag was affinity-purified using glutathione-Sepharose 4B resin (GE Healthcare) according to the manufacturer's instructions. His-CROLIN1, His-CROLIN1-N(33–165), and His-CROLIN1-ΔN165 were affinity-purified using nickel-Sepharose (GE Healthcare) according to the manufacturer's instructions. The purified protein was dialyzed with buffer A3 (10 mm Tris-HCl, 200 μm CaCl2, 0.5 mm DTT, and 0.2 mm ATP, pH 7.0).
Actin was isolated from rabbit skeletal muscle according to a previous method (30). Pyrene-actin was prepared by labeling actin with pyrene iodoacetamide at Cys-374, as described previously for kinetic analyses (31).
High and Low Speed Co-sedimentation Assays
A high speed co-sedimentation assay was used to determine the F-actin binding activity of GST-CROLIN1 following a published method (32).
To determine the apparent equilibrium dissociation constant (Kd) values, various amounts of GST-CROLIN1 and His-CROLIN1-N(33–165) (0.5–3.5 μm) present in the pellet and supernatant were analyzed using ImageJ software (version 1.38). The Kd value for CROLIN1/CROLIN1-N(33–165) bound to F-actin was calculated by plotting the amount of bound CROLIN1 versus free CROLIN1/CROLIN1-N(33–165) and then fitting the data with a hyperbolic function using GraphPad Prism version 5.01 software (Synergy Software).
A high speed co-sedimentation assay was also employed to assess the F-actin-stabilizing activity of CROLIN1 with 2 μm ADF1 treatment. Preformed F-actin (3 μm) was incubated with 0, 0.5, 1, or 3 μm CROLIN1 at 20 °C for 1 h prior to treatment with 2 μm ADF1 for 1 h. The samples were then centrifuged at 100,000 × g for 1 h, and the resulting pellets and supernatants were analyzed by SDS-PAGE.
Low-speed co-sedimentation was then used to determine the actin bundling activity. Except for the rotational speed (13,500 × g), all of the steps were the same as in the high speed co-sedimentation assay.
Buffer A3 was used for the high and low speed co-sedimentation assays, whereas 1× F buffer (buffer A3 with the addition of 50 mm KCl, 2.5 mm MgCl2, and 0.25 mm ATP) was used for actin polymerization. All of the samples were separated on 12% SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue R250 (Sigma-Aldrich). ImageJ was used to analyze the percentages of actin or CROLIN1 in the pellets and supernatants.
Fluorescence Microscopy and Electron Microscopy Visualization of F-actin
To visualize F-actin using fluorescence microscopy, the samples were labeled with Alexa 488-phalloidin (Molecular Probes), as described in a previously published method (32). F-actin was observed using a Leica DFC420C fluorescence microscope equipped with a 5-megapixel CCD and Leica Application Suite software. For electron microscopy, F-actin was observed by negative staining. Prepolymerized F-actin at a concentration of 3 μm was incubated with 0, 0.75, and 2.25 μm His-CROLIN1 in a 200-μl reaction at 20 °C for 1 h. Negative staining was performed using saturated uranyl acetate, and the samples were observed using an FEI Tecnai G20 electron microscope.
Actin Depolymerization Assays
To test the effects of CROLIN1 on actin depolymerization, a depolymerization assay was performed, as described by a published method (32). The decrease in pyrene fluorescence intensity accompanying actin depolymerization was monitored for 500 s at room temperature using a FluoroMax®-4 spectrofluorometer (HORIBA Jobin Yvon).
Tag Removal
To remove the His tag, the purified His-CROLIN1 protein was incubated with thrombin (1:300) at 4 °C overnight. The sample was first purified using nickel-Sepharose (GE Healthcare), and then thrombin was removed using Streptavidin-agarose (Sigma-Aldrich).
CROLIN1 Dimer Assays
To assess the extent of CROLIN1 dimerization, cross-linking experiments were performed using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) (Sigma). EDC was added to the buffer containing 15 μm CROLIN1 to give a final concentration of 4 mm. The sample was incubated at room temperature for 1 h, and the reaction was stopped by the addition of SDS-PAGE loading buffer and boiling for 10 min; the sample was analyzed by SDS-PAGE.
Higher order structures were also analyzed by an acrylamide gel analysis, as described by Huang et al. (26). Native CROLIN1 protein and proteins of known molecular mass (ovalbumin, 44 kDa; albumin, 66 kDa; phosphatase b, 97 kDa; β-galactose, 116 kDa) were electrophoresed on a 10% native acrylamide gel.
RT-PCR Analysis
Total RNA was isolated from various tissues of WT plants using an RNA-extracting kit (Invitrogen). Total RNA (3 μg) from different tissues was used for reverse transcription with Moloney murine leukemia virus reverse transcriptase (Takara). To confirm the expression levels of CROLIN1 in different tissues, 1 μl of reaction product was used as a template for amplifying the cDNA fragments of CROLIN1; EF4A was used as an internal control. The PCR products were examined by 1% agarose gel electrophoresis.
Quantitative Real-time PCR Analysis
For real-time PCR, the flowers of WT plants, a T-DNA insertion mutant (SAIL_108507, obtained from the Arabidopsis Biological Resource Center), and a complemented line were used to obtain total RNA. We employed SsoFastTM EvaGreen® Supermix (Bio-Rad) and the iCycler iQ5TM multicolor real-time PCR detection system (Bio-Rad). EF1A was used as an internal control. The amplification was performed as follows: 95 °C for 35 s, 40 cycles at 95 °C for 10 s and 56 °C for 15 s, 72 °C for 20 s, and 78 cycles of 10 s at 56 °C (for determination of the melting curve). The analysis was performed in triplicate. The “comparative count” method was used to analyze the changes in the transcript levels, as described by Fan et al. (33). The primers used are shown in Table 1.
GUS Staining
At least 10 independent transgenic T3 lines were subjected to GUS staining. Staining for GUS activity was performed overnight, as described by Marrocco et al. (34), and images were obtained using a Leica DM4000 microscope equipped with Leica Application Suite software.
Pollen Germination and Pollen Tube Growth Measurement
According to a previously published method (7, 35), pollen was harvested from newly opened flowers and placed on a pollen germination medium. Various concentrations of Lat B were added to the germination medium to determine the effects of Lat B on pollen germination. To measure the pollen tube growth rate, the pollen grains were germinated for 2 h on a normal germination medium, and various concentrations of Lat B were added to the surface of the solid medium for an additional 3 h. The pollen grains and pollen tubes were observed using a Leica DFC420C microscope equipped with a ×10 objective.
Actin Staining of Pollen Grains and Tubes and Treatment with Lat B
To visualize the actin cytoskeleton in pollen tubes, the pollen tubes were germinated for 4 h and then treated with 0 and 50 nm Lat B for 30 min. The pollen tubes were then fixed with 1% polyoxymethylene and 0.025% glutaraldehyde (solubilized in 50 mm PIPES, pH 6.8) for 20 min, 2% polyoxymethylene and 0.05% glutaraldehyde for 20 min, and 4% polyoxymethylene and 0.1% glutaraldehyde for 20 min. The fixed pollen tubes were gently washed twice with 50 mm PIPES (pH 6.8) and then subjected to actin staining with 0.18 μm Alexa 488-phalloidin (Molecular Probes, Invitrogen). F-actin was subsequently visualized using a confocal laser-scanning microscope (Olympus DP72) equipped with a ×100 objective and a CCD camera. The fluorescent phalloidin was excited at a wavelength of 488 nm using an argon laser. The amount of F-actin was analyzed by measuring the pixel intensity (Int/mm2) of individual pollen tubes, and the images were subsequently processed and analyzed with ImageJ software by subtracting 50% of the background.
Statistical Analysis
Experiments that required an analysis of variance were analyzed using SPSS 17.0, followed by Dunnett's post hoc multiple comparisons. Other statistical analyses were performed using Student's t test. At least three independent experiments were performed.
Accession Numbers
The sequence data from this article can be found in the Arabidopsis Genome Initiative or the GenBankTM/EMBL databases under the following accession numbers: CROLIN1 (At3g28630, Q8GXC9); hisactophilin (ABR23227.1), and fascin (EAW87344.1). The accession numbers for the proteins used in this study are shown in supplemental Table 1.
RESULTS
Identification of the CROLIN Family of Novel Plant Actin-cross-linking Proteins in Arabidopsis
Utilizing a bioinformatic approach toward analyzing the Arabidopsis genome, we performed a BLAST search of the genes that contained the keyword “actin” in the TAIR database. The results included all of the genes that encode actin, most of the reported ABPs, and other putative actin-remodeling proteins. Among these proteins, we identified a functionally unknown gene family that contains a predicted actin-cross-linking domain (InterPro: IPR008999). According to an analysis by the Phyre Server, this domain typically forms a β-trefoil structure consisting of a triplet of β-hairpins packed against a six-stranded antiparallel β-barrel (36). Proteins with a structure similar to that of the CROLIN family include fascin (37), an animal protein that contains a tandem repeat with four copies of this domain, and hisactophilin (38, 39), a protein in Dictyostelium discoideum that contains only one domain. Both of these proteins are typical actin-bundling proteins, but their homologs have not been found in plants. Moreover, the actin-cross-linking domain of CROLIN1 has only 16% amino acid similarity with that of fascin (Fig. 1A) and hisactophilin (Fig. 1B). There are six members belonging to this family, which we named CROLIN1–CROLIN6 based on the actin-cross-linking domain (Fig. 1C). As shown in Fig. 1, D and E, these proteins contain 1–2 cross-linking domains and share 37–71% amino acid similarity with each other, and their actin-cross-linking domains are highly conserved (indicated by the black line). Remarkably, CROLIN1 has at least 87 homologs in the plant kingdom, as revealed by a BLAST search of the NCBI database (supplemental Table 1). CROLIN1 homologs have been identified in eudicots, monocots, ferns, and mosses but not in other non-plant species, such as animals and yeast. A phylogenetic tree of CROLINs in other species (supplemental Fig. 1) reveals that these proteins have high similarity. These data indicate that CROLINs may be novel plant actin-cross-linking proteins.
FIGURE 1.
Multiple alignment and analysis of the amino acid sequences of the CROLIN family in Arabidopsis. A, predicted actin-cross-linking domain of CROLIN1 compared with four similar structures in fascin (EAW87344.1). The highest similarity is 16%. B, predicted actin-cross-linking domain of CROLIN1 compared with the functional domain of hisactophilin (ABR23227.1). The similarity is 16%. C, phylogenetic tree of Arabidopsis CROLINs. D, structures of CROLINs. The black curve represents a trans-membrane domain. The light and dark gray oblong regions represent the actin-cross-linking domains and other cDNA coding sequences, respectively. E, alignment of the amino acid sequences of Arabidopsis CROLINs. The predicted actin-cross-linking domain is indicated by a black line. The second cross-linking domains of CROLIN5 and CROLIN6 are at amino acids 230–360 and 244–378, respectively.
CROLIN1 Is Expressed Specifically in Pollen
According to microarray data, among the Arabidopsis CROLIN family members, CROLIN1 is predicted to be expressed only in pollen. Thus, tissue RT-PCR was performed for confirmation. As shown in Fig. 2A, the CROLIN1 transcript was preferentially detected in the flower and flower bud (lanes 4 and 5) but not in the root, stem, or leaf. This observation was further explored by measuring CROLIN1 promoter activity using a GUS (β-glucuronidase) reporter. There were strong GUS signals in the pollen tubes and pollen grains during all stages of flower development (Fig. 2, B–D), but CROLIN1 promoter activity was not detected in the vegetative organs or seedlings (Fig. 2, E–H). These results are consistent with the above RT-PCR assay, indicating that CROLIN1 may participate in pollen germination and pollen tube growth. To confirm this hypothesis, the biochemical properties and physiological functions of CROLIN1 were tested in vitro and in vivo, as described below.
FIGURE 2.
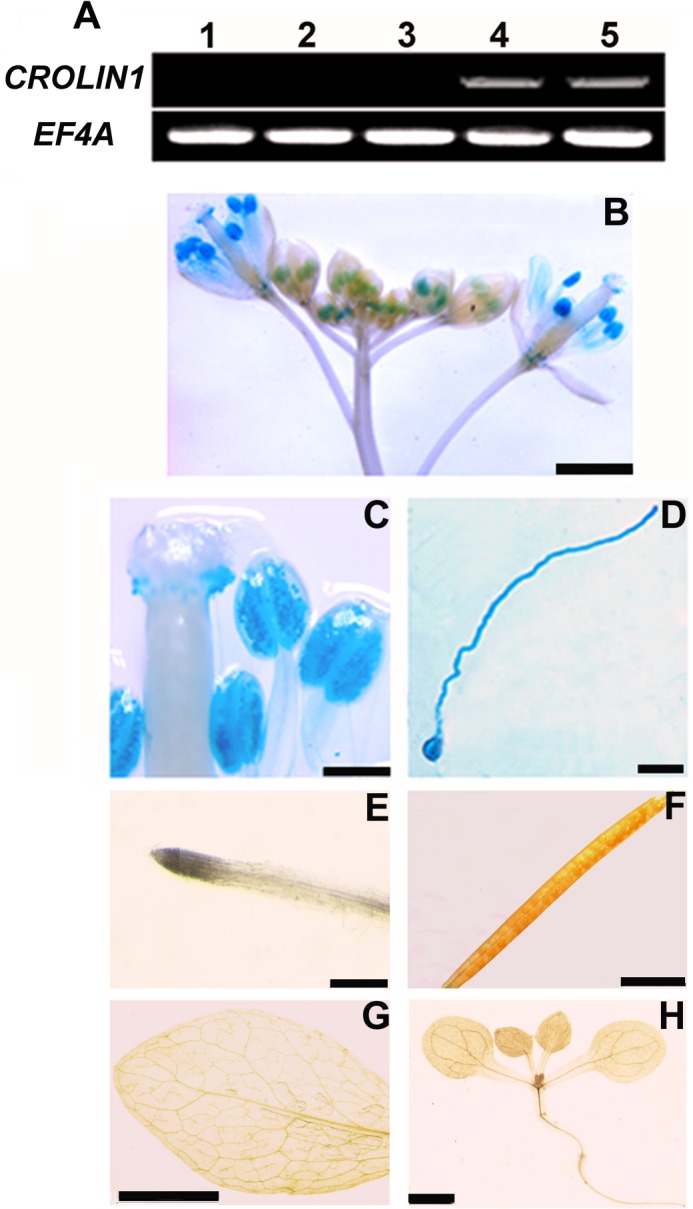
CROLIN1 is expressed specifically in pollen. A, the expression pattern of CROLIN1 was determined using RT-PCR. EF4A was used as a control. The samples are displayed as follows. Lanes 1–5, root, stem, leaf, flower, and flower bud, respectively. B–H, CROLIN1 promoter activity was detected using GUS as a reporter. B, inflorescence; C, stigma and anther; D, pollen grain and tube; E, root; F, silique; G, mature leaf; H, 10-day-old seedling. Bars in B–H, 1 mm, 200 μm, 50 μm, 2 mm, 2 mm, 5 mm, and 2 mm, respectively.
CROLIN1 Binds to F-actin in Vitro
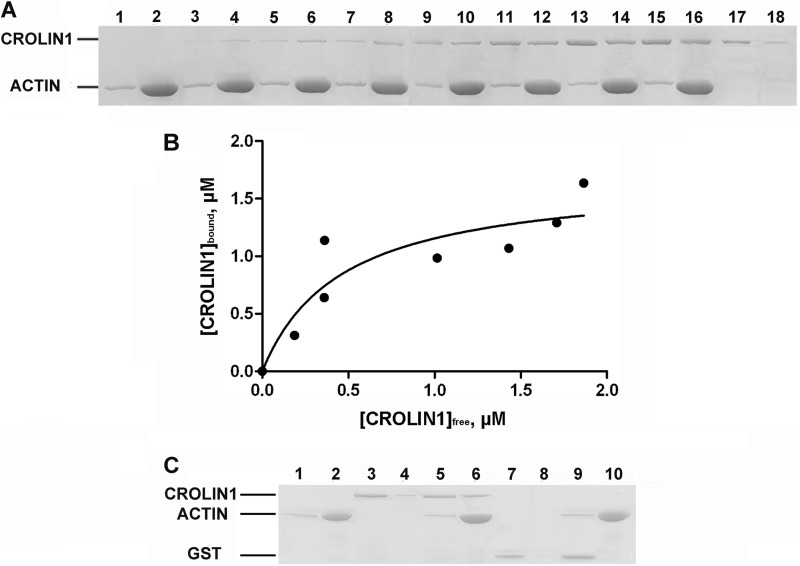
Because CROLIN1 contains a putative actin-cross-linking domain at the N terminus, indicating that it may interact with F-actin, the biochemical activities of CROLIN1 were investigated in vitro. However, due to the similar molecular masses of recombinant His-CROLIN1 and actin, the proteins could not be easily distinguished from each other on an SDS-polyacrylamide gel. Therefore, we used GST-CROLIN1 in high and low speed co-sedimentation assays. The ability of recombinant CROLIN1 to bind to F-actin was directly determined using a high speed co-sedimentation assay. GST-CROLIN1 was predominantly in the supernatant in the absence of the F-actin (Fig. 3A, lanes 17 and 18). However, a significant amount of GST-CROLIN1 sedimented with F-actin in a dose-dependent manner (Fig. 3A, lanes 3–16), demonstrating that CROLIN1 can bind to F-actin in vitro. The equilibrium dissociation constant (Kd) for this interaction was determined to be 0.324 ± 0.032 μm (Fig. 3B). GST alone was used as a negative control and demonstrated no actin binding activity (Fig. 3C).
FIGURE 3.
Recombinant A. thaliana CROLIN1 binds to F-actin in vitro. A, GST-CROLIN1 binds to F-actin in vitro. F-actin (3 μm) was incubated with various concentrations of CROLIN1. The samples in lanes 1–16 contain actin plus 0 (lanes 1 and 2), 0.5 (lanes 3 and 4), 1 (lanes 5 and 6), 1.5 (lanes 7 and 8), 2 (lanes 9 and 10), 2.5 (lanes 11 and 12), 3 (lanes 13 and 14), and 3.5 μm (lanes 15 and 16) CROLIN1; lanes 17 and 18 contain 2 μm CROLIN1 only. B, the equilibrium dissociation constant for CROLIN1 binding to F-actin was calculated. The x axis indicates the free concentration of CROLIN1, and the y axis indicates the concentration of CROLIN1 bound to F-actin. The Kd for this interaction was 0.324 ± 0.032 μm. C, GST does not affect the activity of CROLIN1 in F-actin binding. A mixture of 3 μm F-actin with 1.5 μm CROLIN1 or 1.5 μm GST was centrifuged at 100,000 × g for 1 h. The samples in lanes 1, 3, 5, 7, and 9 represent the supernatants of actin alone, CROLIN1 alone, actin plus CROLIN1, GST alone, and actin plus GST, respectively; the samples in lanes 2, 4, 6, 8, and 10 represent the respective pellets.
CROLIN1 Bundles and Cross-links F-actin in Vitro
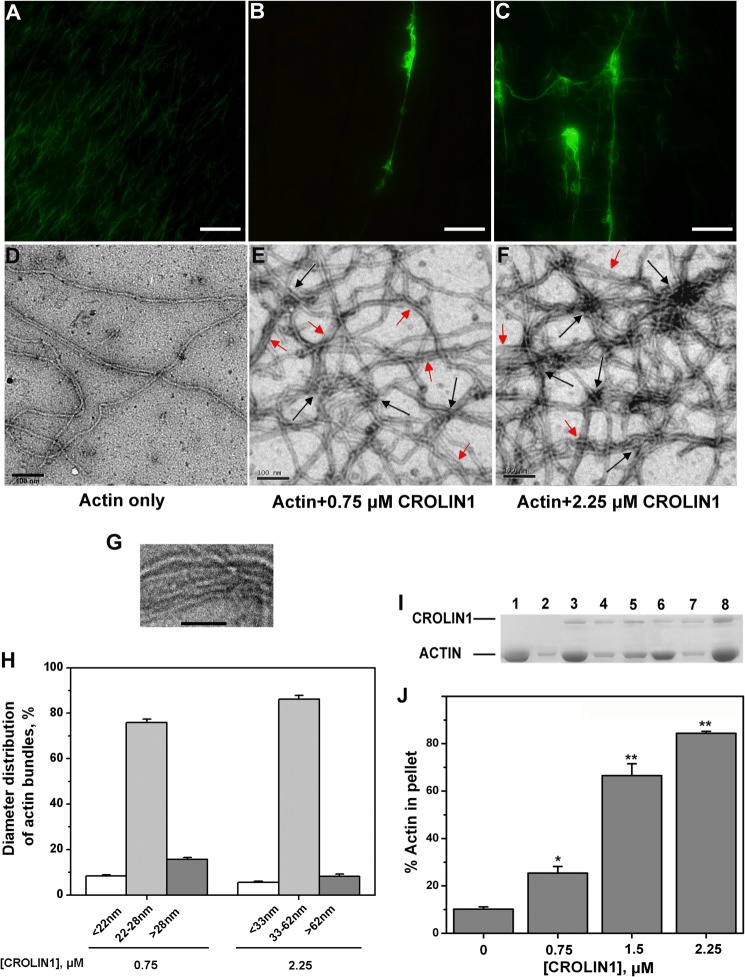
Fluorescence microscopy was used to directly visualize the formation of the higher order actin structures induced by CROLIN1. Prepolymerized F-actin was decorated with Alexa 488-phalloidin, and only individual F-actin was visualized in the absence of His-CROLIN1 (Fig. 4A). In contrast, long and thick actin bundles were observed when the F-actin was incubated with 0.75 μm His-CROLIN1 (Fig. 4B). When the concentration of CROLIN1 increased to 2.25 μm, a large amount of the cross-linking F-actin was observed (Fig. 4C). Subsequently, electron microscopy was employed to provide more detailed images. As shown in Fig. 4, D–F, F-actin was scattered throughout the visual field as single filaments in the absence of His-CROLIN1. However, when His-CROLIN1 was added to the reaction, the F-actin bundled together (indicated by red arrows) and also cross-linked and assembled into a large meshwork (indicated by black arrows). Based on the images of actin bundles shown in Fig. 4G, we measured the average distance between the F-actin of the actin bundles to be ∼4 nm.
FIGURE 4.
A. thaliana CROLIN1 bundles and cross-links F-actin in vitro. A–C, micrographs of F-actin stained with Alexa 488-phalloidin. F-actin (3 μm) was incubated with various concentrations of CROLIN1. Bars in A–C, 1 μm. A, individual F-actin in the absence of His-CROLIN1. B, F-actin bundles formed in the presence of 0.75 μm His-CROLIN1. C, larger F-actin bundles formed in the presence of 2.25 μm His-CROLIN1. D–F, electron micrographs of F-actin by negative staining. F-actin (3 μm) was incubated with various concentrations of CROLIN1. Bars in D–F, 100 nm. The red arrows indicate actin bundles, and the black arrows indicate the sites of cross-linking. D, individual F-actin in the absence of His-CROLIN1. E, F-actin bundles formed in the presence of 0.75 μm His-CROLIN1. F, thicker meshworks of F-actin bundles formed when 2.25 μm His-CROLIN1 was added. G, a representative image of actin bundles used to measure the average distance between the F-actin of the actin bundles. The average distance between the F-actin in the bundles was ∼4 nm. Bars, 50 nm. H, diameter distribution of actin bundles after incubation with various concentrations of CROLIN1. Error bars, S.E. (n ≥ 60). I, GST-CROLIN1 bundles F-actin in vitro. Lanes 1, 3, 5, and 7, supernatant of actin alone, actin plus 0.75 μm CROLIN1, actin plus 1.5 μm CROLIN1, and actin plus 2.25 μm CROLIN1, respectively. The samples in lanes 2, 4, 6, and 8 represent the respective pellets. J, quantification of the results from I. Error bars, S.E. (n = 3); *, p < 0.05; **, p < 0.01 (Student's t test).
We next measured the diameters of the actin bundles in the electron microscopy images. The diameter of a single F-actin was 5–8 nm, which is similar to that found in previous studies (40, 41). However, the diameter of most of the F-actin was 22–28 nm in the presence of 0.75 μm His-CROLIN1 and 33–62 nm in the presence of 2.25 μm His-CROLIN1 (Fig. 4H). These results demonstrate that CROLIN1 has actin-bundling and cross-linking activities.
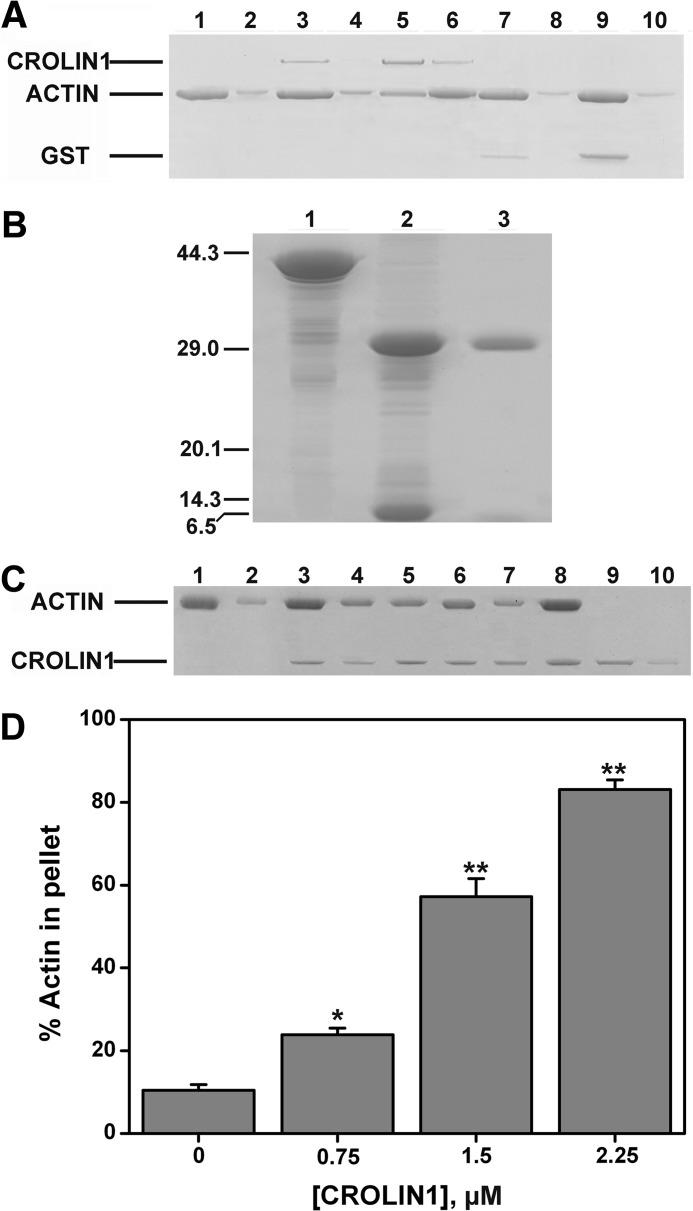
Based on these data, a low speed co-sedimentation assay was then performed to demonstrate that CROLIN1 could bundle F-actin. As shown in Fig. 4I, very little F-actin was sedimented at 13,500 × g in the absence of GST-CROLIN1 (lane 2). However, the F-actin sedimented in a concentration-dependent manner in the presence of GST-CROLIN1 (Fig. 4, I and J), indicating that CROLIN1 bundles F-actin in vitro. As a negative control, GST demonstrated no F-actin bundling activity in vitro, as shown in Fig. 5A. We wanted to verify that the actin bundling activity was due to CROLIN1 itself and not caused by a GST or His tag, which is known to form dimers and contain the high positive charge, respectively. Both of them may have the potential to affect the bundling activity of CROLIN1. Therefore, we utilized thrombin to remove the His tag. As shown in Fig. 5, B–D, CROLIN1 without the tag also displayed the same bundling activity as GST-CROLIN1. These data suggest that the GST and His tag do not affect the bundling activity of CROLIN1.
FIGURE 5.
GST/His does not affect the bundling activity of CROLIN1. A, GST does not affect the F-actin bundling activity of CROLIN1. A mixture of 3 μm F-actin with 0.75 and 2.25 μm CROLIN1 or 0.75 and 2.25 μm GST was centrifuged at 13,500 × g for 1 h. The samples in lanes 1, 3, 5, 7, and 9 represent the supernatants of actin alone, actin plus 0.75 μm CROLIN1, actin plus 2.25 μm CROLIN1, actin plus 0.75 μm GST, and actin plus 2.25 μm GST, respectively. The samples in lanes 2, 4, 6, 8, and 10 represent the respective pellets. B, thrombin removed the His tag of the fusion protein. Lanes 1–3, His-CROLIN1, thrombin incubated with His-CROLIN1 at 4 °C overnight, and CROLIN1 without any tag, respectively. C, CROLIN1 without His tag bundles F-actin in vitro. F-actin (3 μm) was incubated with various concentrations of CROLIN1 and sedimented at 13,500 × g for 1 h. The supernatant and pellet samples were separated by SDS-PAGE. Lanes 1, 3, 5, 7, and 9, supernatants of actin alone, actin plus 0.75 μm CROLIN1, actin plus 1.5 μm CROLIN1, actin plus 2.25 μm CROLIN1, and 1.5 μm CROLIN1 alone, respectively. The samples in lanes 2, 4, 6, 8, and 10 represent the respective pellets. D, quantification of the results from C. Error bars, S.E. (n = 3); *, p < 0.05; **, p < 0.01 (Student's t test).
CROLIN1 Stabilizes F-actin in Vitro
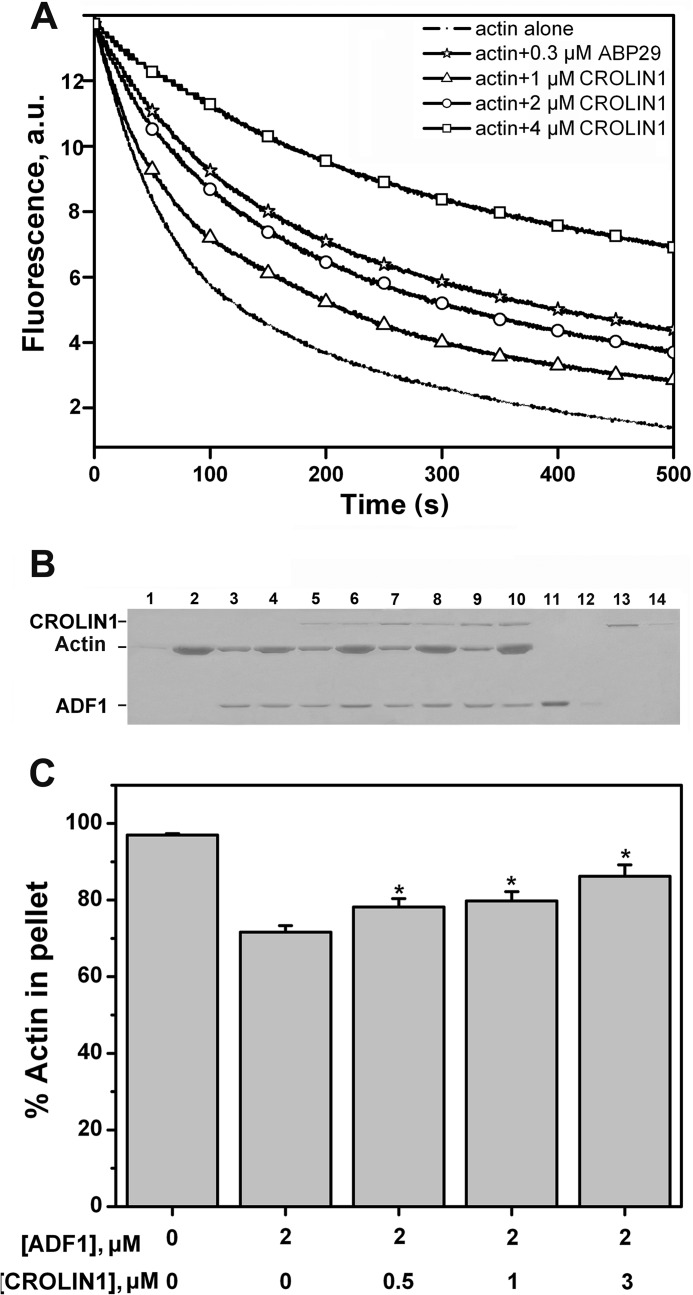
To determine whether CROLIN1 is able to stabilize F-actin in vitro, a dilution-mediated actin depolymerization assay was performed. As shown in Fig. 6A, CROLIN1 reduced the rate of depolymerization in a concentration-dependent manner. ABP29 was used as a positive control, as described previously (32).
FIGURE 6.
A. thaliana CROLIN1 stabilizes F-actin in vitro. A, CROLIN1 protects F-actin from dilution-mediated depolymerization. B, CROLIN1 relieves the effect caused by ADF1 in vitro. F-actin (3 μm) was incubated with different concentrations of CROLIN1 and ADF1. Lanes 1, 3, 5, 7, 9, 11, and 13, supernatants of actin alone, actin plus 2 μm ADF1, actin plus 2 μm ADF1 plus 0.5 μm CROLIN1, actin plus 2 μm ADF1 plus 1 μm CROLIN1, actin plus 2 μm ADF1 plus 3 μm CROLIN1, 2 μm ADF1 alone, and 1 μm CROLIN1 alone, respectively. Lanes 2, 4, 6, 8, 10, 12, and 14 represent the respective pellets. C, quantification of the experiment in B. Error bars, S.E. (n = 3); *, p < 0.05 (Student's t test). a.u., absorbance units.
Arabidopsis actin depolymerization factor 1 (ADF1), a well documented actin-depolymerizing and -severing protein (5, 42), was further employed to demonstrate that CROLIN1 could stabilize F-actin in vitro. Without GST-CROLIN1 and ADF1, the percentage of actin in the pellet was ∼92%, whereas the percentages of actin in the pellet varied to statistically significant degrees (*, p < 0.05) when ADF1 and different concentrations of CROLIN1 were added to the assay (Fig. 6, B and C). Taken together, these results indicate that CROLIN1 stabilizes F-actin in vitro.
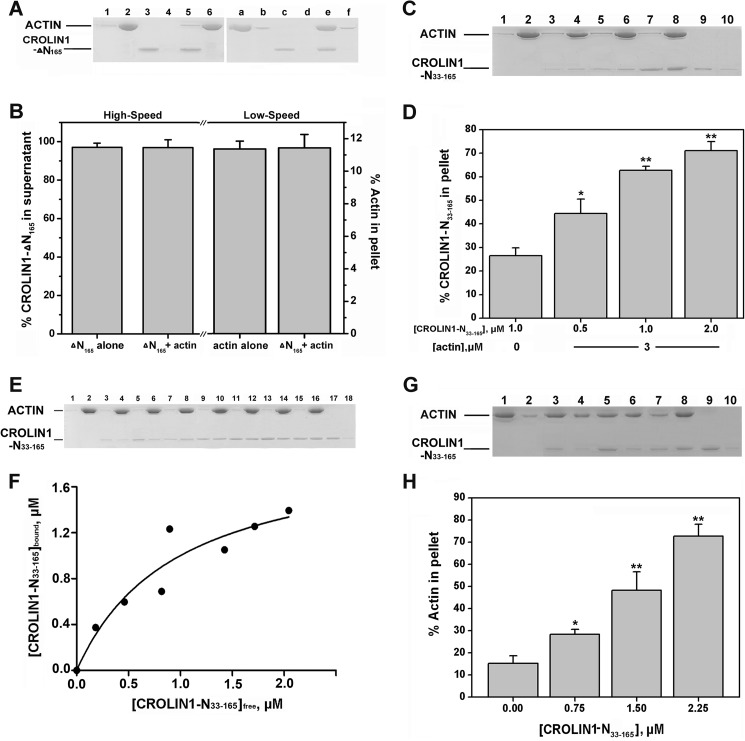
The Predicted Actin-cross-linking Domain Is Sufficient for the Bundling Activity of CROLIN1
We suspected that the ability of CROLIN1 to bind and bundle F-actin depended on its actin-cross-linking domain. To confirm this hypothesis, we deleted the predicted cross-linking domain and found that CROLIN1 lost both its binding and cross-linking activities (Fig. 7, A and B). Then we purified a His-tagged N-terminal region of CROLIN1 (amino acids 33–165) containing only the actin-cross-linking domain and performed high/low speed co-sedimentation assays. As shown in Fig. 7, C and D, this domain exhibited a concentration-dependent actin binding ability, with a calculated Kd value that did not differ from full-length CROLIN1 at 0.388 ± 0.048 μm (Fig. 7, E and F). Additionally, this domain showed concentration-dependent actin bundling activity in vitro (Fig. 7, G and H), which directly demonstrates that the actin binding and bundling activity of CROLIN1 depends on this domain.
FIGURE 7.
The cross-linking domain of CROLIN1 is necessary for its activity. A, without the cross-linking domain, CROLIN1 lost its actin binding and bundling/cross-linking activity. Left, a mixture of 3 μm F-actin with or without 1.5 μm His-CROLIN1-ΔN165 was centrifuged at 100,000 × g for 1 h. The samples in lanes 1, 3, and 5 represent the supernatants of actin alone, ΔN165 alone, and actin + ΔN165, respectively. Lanes 2, 4, and 6 represent the respective pellets. Right, a mixture of 3 μm F-actin with or without 1.5 μm CROLIN1-ΔN165 was centrifuged at 13,500 × g for 1 h. The samples in lanes a, c, and e represent the supernatants of actin alone, ΔN165 alone, and actin plus ΔN165, respectively. Lanes b, d, and f represent the pellets. B, quantification of the results from A. Calculations of the percentages of His-CROLIN1-ΔN165 in the supernatants and actin in the pellets are shown on the left and right, respectively. Error bars, S.E. (n = 3). C, the cross-linking domain of CROLIN1 exhibited actin binding activity. The samples loaded in lanes 1, 3, 5, 7, and 9 represent the supernatants of actin alone, actin plus 0.5 μm His-CROLIN1-N(33–165), actin plus 1 μm His-CROLIN1-N(33–165), actin plus 2 μm His-CROLIN1-N(33–165), and 1 μm His-CROLIN1-N(33–165) alone, respectively. Lanes 2, 4, 6, 8, and 10 represent the respective pellets. D, quantification of the results from C. Error bars, S.E. (n = 3); *, p < 0.05; **, p < 0.01 (Student's t test). E, CROLIN1-N(33–165) binds to F-actin in vitro. F-actin (3 μm) was incubated with various concentrations of CROLIN1-N(33–165). The samples in lanes 1–18 contain actin plus 0 (lanes 1 and 2), 0.5 (lanes 3 and 4), 1 (lanes 5 and 6), 1.5 (lanes 7 and 8), 2 (lanes 9 and 10), 2.5 (lanes 11 and 12), 3 (lanes 13 and 14), and 3.5 μm (lanes 15 and 16) N(33–165); lanes 17 and 18 contain 2 μm N(33–165) only. F, the equilibrium dissociation constant for N(33–165) binding to F-actin. The x axis indicates the free concentration of CROLIN1-N(33–165), and the y axis indicates the concentration of CROLIN1-N(33–165) bound to F-actin. The Kd for this interaction was 0.388 ± 0.048 μm. G, the cross-linking domain of CROLIN1 can bundle F-actin. The samples in lanes 1, 3, 5, 7, and 9 represent the supernatants of actin alone; actin plus 0.75, 1.5, and 2.25 μm His-CROLIN1-N(33–165); and 1.5 μm His-CROLIN1-N(33–165) alone, respectively. Lanes 2, 4, 6, 8, and 10 represent the respective pellets. H, quantification of the results from G. Error bars, S.E. (n = 3); *, p < 0.05; **, p < 0.01 (Student's t test).
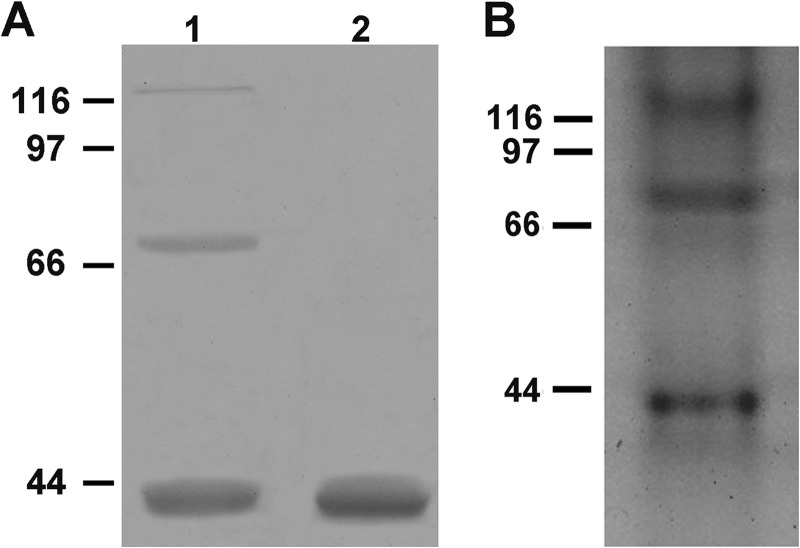
CROLIN1 Forms Oligomers
To investigate weather CROLIN1 formed oligomers, we conducted cross-linking experiments using EDC. After CROLIN1 was incubated with EDC, the mixture was separated on an SDS-polyacrylamide gel. Three bands were detected after incubation with EDC (Fig. 8A, lane 1), whereas only one band was present without EDC treatment (Fig. 8A, lane 2). To further confirm this observation, we next performed acrylamide gel analysis using native CROLIN1 and observed three protein bands on native gel at about 40, 80, and 120 kDa (Fig. 8B). These results demonstrate that CROLIN1 can form dimers and even trimers.
FIGURE 8.
CROLIN1 forms oligomers in vitro. CROLIN1 was analyzed after EDC treatment and by native acrylamide gel electrophoresis. A, CROLIN1 (15 μm) was incubated with 4 mm EDC for 1 h at room temperature and then separated on a 12% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. Three major bands can be observed on the gel (lane 1), whereas only one band was present without EDC (lane 2). The estimated molecular masses of the dimers and trimers were ∼80 and 120 kDa, respectively. B, native CROLIN1 protein was analyzed by 10% native acrylamide gel electrophoresis and stained with Coomassie Brilliant Blue. Three major bands were detected; the estimated molecular masses of the dimers and trimers were ∼80 and 120 kDa, respectively.
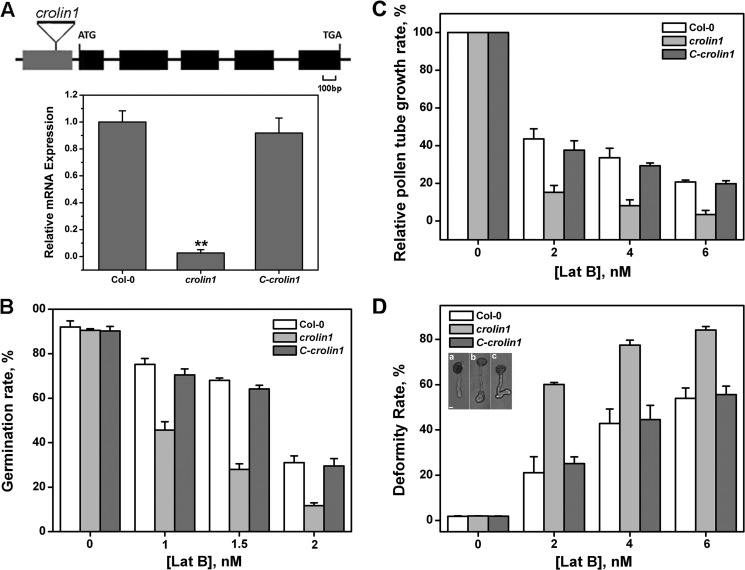
CROLIN1 Loss of Function Results in Pollen Hypersensitivity to Lat B, whereas Complementation of CROLIN1 Can Partially Recover Such Effects
To investigate the physiological function of CROLIN1 in vivo, a T-DNA insertion line (SALK_108507) named crolin1 and a complemented line were characterized. As shown in Fig. 9A, the expression of CROLIN1 was down-regulated in crolin1, and the complemented line displayed a transcript level similar to that of the WT plant.
FIGURE 9.
The CROLIN1 loss-of-function mutation results in pollen germination and pollen tube growth hypersensitive to Lat B. A, genetic map of the Arabidopsis CROLIN1 gene. CROLIN1 contains five exons and four introns, which are indicated by filled boxes and lines, respectively. The position of the T-DNA insertion in mutant crolin1 (SAIK_108507) is indicated by triangles above the diagram. The insertion site is between bp 138 and 139, upstream from the ATG codon. The transcript levels in WT, crolin1, and the complemented line were determined by real-time PCR using EF1A as the internal control. Shown is the mean ± S.E. (error bars) of three biological replicates. **, p < 0.01 (Student's t test). B, germination of pollen grains from WT, crolin1, and the complemented line. The concentrations of Lat B used in this analysis were 0, 1, 1.5, and 2 nm. Error bars, S.E. (n ≥ 300). C, relative growth rate of the pollen tubes from WT, crolin1, and the complemented line. The concentrations of Lat B used in this analysis were 0, 2, 4, and 6 nm. Error bars, S.E. (n ≥ 150). D, pollen tube deformity rates of WT, crolin1, and the complemented line in the presence of 0, 2, 4, and 6 nm Lat B. Error bars, S.E. (n ≥ 150). The inset shows the deformity types. The main pollen tube deformities in the crolin1 mutant in the presence of Lat B are shown. a, normal pollen tube; b, enlargement; c, turns. Bars, 10 μm.
We next assessed the pollen germination rates and pollen tube growth rates in the crolin1 and complemented line and found no differences between the transgenic lines and WT plants under normal conditions (Fig. 9, B and C). However, when various concentrations of Lat B were applied, both the germination and tube growth rates were decreased to a greater extent in the crolin1 mutant compared with the WT plant (Fig. 9, B and C). These results indicate that CROLIN1 may be involved in stabilizing actin structures during pollen germination and pollen tube growth. In addition, we observed that the frequency of pollen tube deformities in crolin1 was clearly elevated (Fig. 9D). These deformities mainly appeared at the top of the pollen tube as enlargements and turns (Fig. 9D, insets a–c). The germination rate and growth rates were recovered in the corlin1 complemented lines (Fig. 9, B–D). These results illustrate that Lat52-CROLIN1 can complement the phenotype caused by CROLIN1 loss of function.
The Actin Cytoskeleton in Pollen Tubes Is Sensitive to Lat B in the crolin1 Mutants but Is Recovered in Complemented Plants
The above investigation indicated that CROLIN1 may be involved in stabilizing F-actin in pollen tubes. To investigate the effect of Lat B on F-actin in vivo, pollen tubes from WT, crolin1, and the complemented line were treated with 50 nm Lat B after germination in a standard medium for 4 h, and F-actin was then stained with Alexa 488-phalloidin. Without Lat B, the actin cytoskeleton was highly ordered in all of the pollen tubes, with no detectable differences among the plants (Fig. 10A, top). After treatment with Lat B, F-actin became notably shorter and less abundant in the WT pollen tubes (Fig. 10A, bottom). This effect is similar to that previously observed in maize (28), Papaver rhoeas (43), and Arabidopsis thaliana (7) pollen tubes. In contrast, F-actin almost completely disappeared in the pollen tubes of crolin1, whereas F-actin recovered to a level similar to that of the WT line in the pollen tubes of the complemented transgenic line. The relative intensity of F-actin was analyzed using ImageJ software (Fig. 10B). These results indicate that CROLIN1 stabilizes F-actin in vivo.
FIGURE 10.
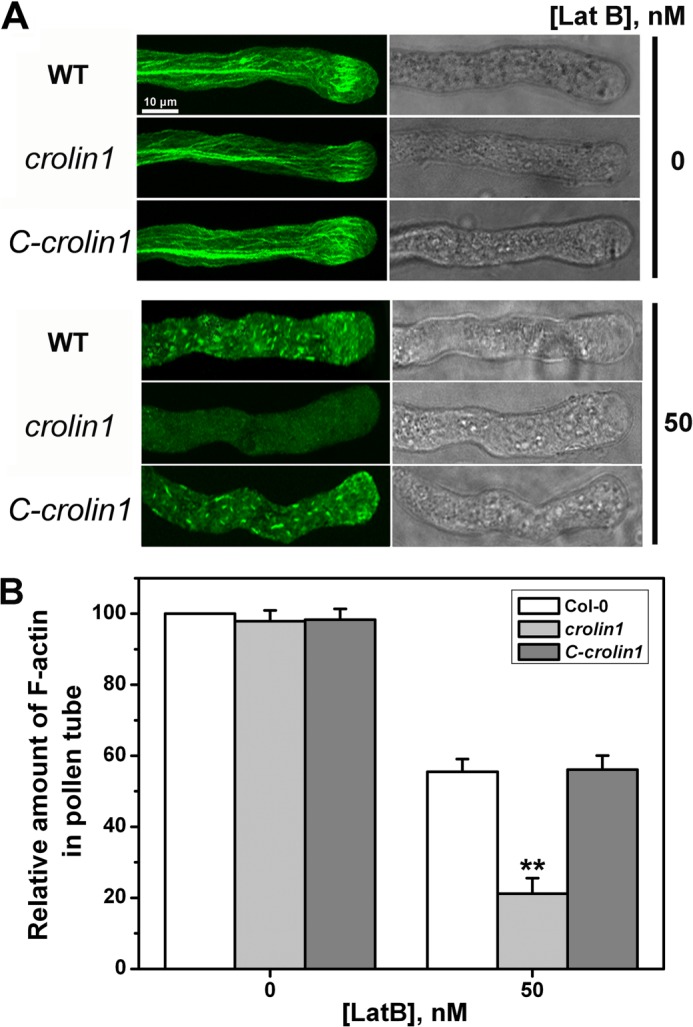
CROLIN1 down-regulation results in F-actin hypersensitive to Lat B, whereas CROLIN1 can partly recover this phenomenon. A, pollen tubes from WT, crolin1, and the complemented line with and without 50 nm Lat B treatment. Bar, 10 μm. B, quantification of the relative F-actin levels in pollen tubes using ImageJ software. The amount of F-actin in untreated WT pollen tubes was normalized to 100% as the control. Error bars, S.E. (n = 20). **, p < 0.01 (Student's t test).
DISCUSSION
Higher order actin structures play a crucial role in many physiological processes. For example, actin bundles in animals are important components of certain cellular structures, such as microvilli, stress fibers, filopodia, and growth cones (2). Over 23 classes of ABPs that are responsible for regulating the formation of actin structures have been discovered (2). In contrast to the situation in animals, actin bundles are distributed in virtually all plant tissues and cells, but many ABPs that exist in animal cells, such as fascin, α-actinin, and espin, are absent in plant genomes (21). Indeed, only a small number of actin-bundling proteins have been identified in plants to date. These proteins include the conserved ABP family proteins, such as villin, fimbrin, LIM, and formin (4–5, 8–13, 16, 18), plant-specific ABPs, such as SCAB1 (22), and plant ABPs with bundling activities, such as the V-ATPase B subunit and ADF9 (24, 25). However, the mechanism of higher order actin bundle formation remains unknown. In this study, we searched the Arabidopsis genome and identified a novel actin-binding protein family (the CROLIN family), which contains a predicted actin-cross-linking domain. Although similar structure exists in fascin and hisactophilin, the amino acid similarity between these proteins is very low. Interestingly, CROLIN homologs were identified in several plant proteins from monocots, eudicots, ferns, and mosses. Indeed, there are many CROLIN family members; for example, CROLIN has 26 isoforms in rice (Oryza sativa), 10 isoforms in maize (Zea mays), 18 isoforms in sorghum (Sorghum bicolor), 1 isoform in soybean (Glycine max), 8 isoforms in grape (Vitis vinifera), 9 isoforms in poplar (Populus trichocarpa), and 3 isoforms in castor (Ricinus communis) (supplemental Table 1). Our results demonstrate that the CROLIN family may be a novel group of ABPs that are expressed only in plants. In addition, a varying number of actin-cross-linking domains are present in the members of this family. There is only one such domain in Arabidopsis CROLIN1–CROLIN4, whereas there are two in CROLIN5 and -6. CROLIN2 also contains a predicted trans-membrane domain (Fig. 1D), suggesting that these proteins may have different regulatory mechanisms. Indeed, according to microarray data, Arabidopsis CROLINs may be specifically expressed in different tissues and during different developmental periods. This finding indicates that this gene family may have various distinct roles during plant development. Nonetheless, the physiological and biochemical functions of these genes require further investigation.
The members of the CROLIN family contain a predicted actin-cross-linking domain, suggesting that they may interact with actin. First, a high speed co-sedimentation assay was performed, and the results showed that CROLIN1 could bind to F-actin with high affinity (Fig. 3A). The Kd value was 0.324 μm (Fig. 3B), which is similar to that of FIM1 (0.71 μm) (12) and FIM5 (0.51 μm) (13). The results from low speed co-sedimentation, fluorescence microscopy, and electron microscopy assays further demonstrated that CROLIN1 was able to bundle F-actin into thick bundles, eventually cross-linking the bundles into networks. Because CROLIN1 contains only one actin-binding domain, we used cross-linking to examine the mechanism of CROLIN1-mediated F-actin bundling/cross-linking. Our experiments using cross-linked proteins and native gels demonstrated that CROLIN1 forms oligomers to bundle and cross-link F-actin. Moreover, the actin structure induced by CROLIN1 in vitro was similar to that of FIM1 and FIM5, which form higher order actin structures (12, 13). These results reveal that the mechanism of actin bundling by CROLIN1 is different from the mechanism of other ABPs, such as formin (16) and villin (7), which mainly form actin cables and bundles. CROLIN1 also protected F-actin from the effects of ADF1 (Fig. 6), suggesting that CROLIN1 stabilizes F-actin in vitro. Because the cross-linking domain alone is sufficient for actin binding and bundling/cross-linking in vitro (Fig. 7), the predicted cross-linking domain appears to be required for CROLIN1 activity. We conclude that CROLIN1 is a previously undiscovered plant actin-binding protein that functions in actin binding, bundling, and cross-linking.
Pollen tubes require a precisely regulated actin cytoskeleton to develop and maintain polarized growth (1). CROLIN1 is specifically expressed in pollen and stabilizes actin in vitro (Figs. 2 and 6); therefore, it may participate in the regulation of pollen tube growth by stabilizing F-actin in vivo. To confirm this hypothesis, we used a T-DNA insertion line and a complemented line to demonstrate the physiological function of CROLIN1. We also used Lat B to assess the germination, growth rate, and actin bundle stability. The results demonstrate that CROLIN1 loss of function resulted in pollen germination, pollen tube growth, and F-actin hypersensitive to Lat B treatment (Figs. 9 and 10). These results suggest that CROLIN1 may regulate pollen tube growth and pollen germination by stabilizing higher order actin structures.
Acknowledgments
We thank Dr. Jia Li for supplying the high speed centrifuge, Dr. Yi Wu for helping with electron microscopy, and Liping Guan and Wenliang He for technical assistance.
This work was supported by National Basic Research Program Grant 2014CB954200, National Natural Science Foundation of China Grants 31270326 and 30800079, and Fundamental Research Funds for the Central Universities Grant lzujbky-2013-94 (to Y. X.).

This article contains supplemental Table 1 and Fig. 1.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) Q8GXC9, ABR23227.1, and EAW87344.1.
- ABP
- actin-binding protein
- Lat B
- latrunculin B
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide.
REFERENCES
- 1. Staiger C. J. (2000) Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 257–288 [DOI] [PubMed] [Google Scholar]
- 2. Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009) Actin bundling in plants. Cell Motil. Cytoskeleton 66, 940–957 [DOI] [PubMed] [Google Scholar]
- 3. Meagher R. B., Williamson R. E. (1994) The plant cytoskeleton. in Arabidopsis (Meyerowitz E., Somerville C., eds) pp. 1049–1084, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 4. Huang S., Robinson R. C., Gao L. Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C. J. (2005) Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khurana P., Henty J. L., Huang S., Staiger A. M., Blanchoin L., Staiger C. J. (2010) Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22, 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y., Xiao Y., Du F., Cao L., Dong H., Ren H. (2011) Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol. 190, 667–682 [DOI] [PubMed] [Google Scholar]
- 7. Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C. J., Huang S. (2010) Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22, 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006) Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18, 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moes D., Gatti S., Hoffmann C., Dieterle M., Moreau F., Neumann K., Schumacher M., Diederich M., Grill E., Shen W. H., Steinmetz A., Thomas C. (2013) A LIM domain protein from tobacco involved in actin bundling and histone gene transcription. Mol. Plant 6, 483–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papuga J., Hoffmann C., Dieterle M., Moes D., Moreau F., Tholl S., Steinmetz A., Thomas C. (2010) Arabidopsis LIM proteins. A family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22, 3034–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang H. J., Wan A. R., Jauh G. Y. (2008) An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147, 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kovar D. R., Staiger C. J., Weaver E. A., McCurdy D. W. (2000) AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24, 625–636 [DOI] [PubMed] [Google Scholar]
- 13. Wu Y., Yan J., Zhang R., Qu X., Ren S., Chen N., Huang S. (2010) Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22, 3745–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., Zheng Y., Wang J., Zeng L., Li Q., Huang S., He Z. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23, 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., Yuan Z., Hu J., Ren H., Zhang D. (2011a) RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23, 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michelot A., Derivery E., Paterski-Boujemaa R., Guérin C., Huang S., Parcy F., Staiger C. J., Blanchoin L. (2006) A novel mechanism for the formation of actin-filament bundles by a non-processive formin. Curr. Biol. 16, 1924–1930 [DOI] [PubMed] [Google Scholar]
- 17. Deeks M. J., Fendrych M., Smertenko A., Bell K. S., Oparka K., Cvrcková F., Zársky V., Hussey P. J. (2010) The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule binding domain. J. Cell Sci. 123, 1209–1215 [DOI] [PubMed] [Google Scholar]
- 18. Li Y., Shen Y., Cai C., Zhong C., Zhu L., Yuan M., Ren H. (2010) The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22, 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue X. H., Guo C. Q., Du F., Lu Q. L., Zhang C. M., Ren H. Y. (2011) AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol. Plant 4, 264–278 [DOI] [PubMed] [Google Scholar]
- 20. Staiger C. J., Poulter N. S., Henty J. L., Franklin-Tong V. E., Blanchoin L. (2010) Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61, 1969–1986 [DOI] [PubMed] [Google Scholar]
- 21. Hussey P. J., Allwood E. G., Smertenko A. P. (2002) Actin-binding proteins in the Arabidopsis genome database. Properties of functionally distinct plant actin-depolymerizing factors/cofilins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y., Zhao S., Mao T., Qu X., Cao W., Zhang L., Zhang W., He L., Li S., Ren S., Zhao J., Zhu G., Huang S., Ye K., Yuan M., Guo Y. (2011) The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 23, 2314–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whippo C. W., Khurana P., Davis P. A., DeBlasio S. L., DeSloover D., Staiger C. J., Hangarter R. P. (2011) THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 21, 59–64 [DOI] [PubMed] [Google Scholar]
- 24. Ma B., Qian D., Nan Q., Tan C., An L., Xiang Y. (2012) Arabidopsis V-ATPase B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 287, 19008–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tholl S., Moreau F., Hoffmann C., Arumugam K., Dieterle M., Moes D., Neumann K., Steinmetz A., Thomas C. (2011) Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585, 1821–1827 [DOI] [PubMed] [Google Scholar]
- 26. Huang S., Jin L., Du J., Li H., Zhao Q., Ou G., Ao G., Yuan M. (2007) SB401, a pollen-specific protein from Solanum berthaultii, binds to and bundles microtubules and F-actin. Plant J. 51, 406–418 [DOI] [PubMed] [Google Scholar]
- 27. Hepler P. K., Vidali L., Cheung A. Y. (2001) Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17, 159–187 [DOI] [PubMed] [Google Scholar]
- 28. Gibbon B. C., Kovar D. R., Staiger C. J. (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11, 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren H., Xiang Y. (2007) The function of actin-binding proteins in pollen tube growth. Protoplasma 230, 171–182 [DOI] [PubMed] [Google Scholar]
- 30. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 31. Pollard T. D. (1984) Polymerization of ADP-actin. J. Cell Biol. 99, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P. J., Ren H. (2007) Actin binding protein 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19, 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan T. T., Zhai H. H., Shi W. W., Wang J., Jia H. L., Xiang Y., An L. Z. (2013) Overexpression of profilin 3 affects cell elongation and F-actin organization in Arabidopsis thaliana. Plant Cell Rep. 32, 149–160 [DOI] [PubMed] [Google Scholar]
- 34. Marrocco K., Lecureuil A., Nicolas P., Guerche P. (2003) The Arabidopsis SKP1-like genes present a spectrum of expression profiles. Plant Mol. Biol. 52, 715–727 [DOI] [PubMed] [Google Scholar]
- 35. Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009) Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21, 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protocols 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 37. Jawhari A. U., Buda A., Jenkins M., Shehzad K., Sarraf C., Noda M., Farthing M. J., Pignatelli M., Adams J. C. (2003) Fascin, an actin-bundling protein, modulates colonic epithelial cell invasiveness and differentiation in vitro. Am. J. Pathol. 162, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu C., Gaspar J. A., Wong H. J., Meiering E. M. (2002) Conserved and nonconserved features of the folding pathway of hisactophilin, a β-trefoil protein. Protein Sci. 11, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pintsch T., Zischka H., Schuster S. C. (2002) Hisactophilin is involved in osmoprotection in Dictyostelium. BMC Biochem. 3, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grazi E. (1997) Hypothesis what is the diameter of the actin filament? FEBS Lett. 405, 249–252 [DOI] [PubMed] [Google Scholar]
- 41. Ikawa T., Hoshino F., Watanabe O., Li Y., Pincus P., Safinya C. R. (2007) Molecular scale imaging of F-actin assemblies immobilized on a photopolymer surface. Phys. Rev. Lett. 98, 018101–018104 [DOI] [PubMed] [Google Scholar]
- 42. Carlier M.-F., Laurent V., Santolini J., Melki R., Didry D., Xia G.-X., Hong Y., Chua N.-H., Pantaloni D. (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover. Implication in actin-based motility. J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Snowman B. N., Kovar D. R., Shevchenko G., Franklin-Tong V. E., Staiger C. J. (2002) Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14, 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]