Background: KIF17 is targeted to microtubule plus-ends by EB1 and promotes microtubule stabilization in epithelial cells.
Results: KIF17 motor and tail domains have direct and distinct effects on microtubule polymerization.
Conclusion: The KIF17 motor domain is sufficient to regulate microtubules, but catalytic activity is modulated by EB1 and the KIF17 tail.
Significance: KIF17 can function as a direct regulator of microtubule dynamics and stability.
Keywords: Cytoskeleton, Epithelial Cell, Kinesin, Microtubules, Protein Domains
Abstract
KIF17 is a kinesin-2 family motor that interacts with EB1 at microtubule (MT) plus-ends and contributes to MT stabilization in epithelial cells. The mechanism by which KIF17 affects MTs and how its activity is regulated are not yet known. Here, we show that EB1 and the KIF17 autoinhibitory tail domain (KIF17-Tail) interacted competitively with the KIF17 catalytic motor domain (K370). Both EB1 and KIF17-Tail decreased the K0.5MT of K370, with opposing effects on MT-stimulated ATPase activity. Importantly, K370 had independent effects on MT dynamic instability, resulting in formation of long MTs without affecting polymerization rate or total polymer mass. K370 also inhibited MT depolymerization induced by dilution in vitro and by nocodazole in cells, suggesting that it acts by protecting MT plus-ends. Interestingly, KIF17-Tail bound MTs and tubulin dimers, delaying initial MT polymerization in vitro and MT regrowth in cells. However, neither EB1 nor KIF17-Tail affected K370-mediated MT polymerization or stabilization significantly in vitro, and EB1 was dispensable for MT stabilization by K370 in cells. Thus, although EB1 and KIF17-Tail may coordinate KIF17 catalytic activity, our data reveal a novel and direct role for KIF17 in regulating MT dynamics.
Introduction
Kinesin family motors are microtubule (MT)2-stimulated ATPases that affect cell morphology and function by diverse mechanisms. Although best known for their role in protein transport (1), many kinesins also influence cellular organization by modulating MT dynamics. Regulation of MT dynamics by kinesins can be mediated by direct effects of motors on MTs, usually at MT ends, or indirectly by transporting cargoes to MT ends that affect MT dynamics (2–4). During mitosis, kinesins regulate the assembly and length of the mitotic spindle, kinetochore-MT attachments, and attachment of astral MTs to the cortex (for review, see Refs. 5–7). In interphase, kinesins also regulate the length, stability, and organization of cytoplasmic and ciliary MT arrays, with downstream effects on processes including cell migration and polarization (8–17).
Kinesins use varying mechanisms to reach MT ends. Kinesins lacking translocase activity (e.g. kinesin-13) can employ one-dimensional diffusion along the MT lattice to reach plus-ends, whereas those with translocase activity (e.g. kinesin-4, -7, -8, and -14) walk to the MT end (3). Accumulation and retention at these sites can occur through preferential association with GTP-tubulin; recognition of subtle differences in the structure of MTs at ends versus along the lattice; and in some cases, associations with MT plus-end tip-tracking machinery, such as EB1 (end-binding protein-1). EB1-mediated MT plus-end targeting can further affect kinesin activity locally by changing MT-binding affinity and catalytic activity (18–21). Once at MT ends, kinesins also have different ways of regulating dynamics, and some motors exhibit context-dependent effects on features of dynamic instability (3). Kinesin-8, -13, and -14 families act primarily as depolymerases but have also been shown to stabilize MTs by dampening dynamics at MT ends (9, 22–30). Kinesin-4 and -7 motors dampen dynamic instability or promote assembly but may also induce structural changes to the MT lattice that result in MT destabilization (31, 32). Kinesins without known direct effects on MTs, e.g. kinesin-1 and -2, can use their transport activity to deliver tubulin, MT effectors, and regulatory proteins to MT ends (13–15, 33).
We showed previously that the kinesin-2 motor KIF17 contributes to MT stabilization and polarization of epithelial cells (13). KIF17 interacts with EB1 and adenomatous polyposis coli, components of the MT capture and stabilization pathway (34), and localizes with them on a subset of MT plus-ends. In addition, EB1 is required for MT plus-end localization of endogenous KIF17; however, a constitutively active, extended conformation mutant of KIF17 localized to MT plus-ends and stabilized MTs independent of EB1. This suggests that EB1 may function as a KIF17 activator at MT ends by relieving head-tail autoinhibitory conformation. It is also possible that EB1 recruits KIF17 to MTs or prevents its dissociation from MTs upon reaching the plus-end, as described for Tea2 (kinesin-7) and KIF18B (kinesin-8) (18, 20). Consistent with these ideas, we show here that EB1 and the KIF17 C-terminal tail (KIF17-Tail) regulate the MT-binding and catalytic activities of the KIF17 motor domain. However, the KIF17 catalytic motor domain has direct effects on MT dynamics in vitro, promoting persistent MT elongation, and is sufficient to stabilize MTs in vitro and in cells. Because EB1 enhances binding of the KIF17 motor domain to MTs in vitro and recruits endogenous KIF17 to MTs in cells, we speculate that EB1 may serve to retain KIF17 at MT plus-ends, where it can exert effects on MT polymerization and stabilization locally. KIF17-Tail, which interacts with tubulin and MTs and has independent effects on MT polymerization, may also tether the motor to MTs and perhaps transport tubulin as a cargo for incorporation into MT growing ends in cells.
EXPERIMENTAL PROCEDURES
Constructs
KIF17 motor domains (K370, amino acids 1–370; and K490, amino acids 1–490) were amplified by PCR from human A549 or Caco-2 cells and cloned into Gateway expression vectors (Invitrogen) as recommended by the manufacturer. The 5′-primer used for all KIF17 motor domain constructs was GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC CTC CGA GGC GGT GAA; the 3′-primers for K370 and K490 were GGG ACC ACT TTG TAC AAG AAA GCT GGG TCC TAG CTC ATC TGT GTC AGG ATG GC and GGG ACC ACT TTG TAC AAG AAA GCT GGG TCC TAA AAA GCA GGC GGG TAC TCA GC. KIF17 constructs encoding the motor domain alone (K339, amino acids 1–339) and KIF17-Tail (amino acids 849–1029) and full-length EB1 constructs were described previously (13). K370R288A/R294A was generated by PCR from full-length KIF17R288A/R294A using the 5′- and 3′-primers described above for K370. The Drosophila melanogaster kinesin-1 heavy chain (DmKHC) motor domain was a generous gift from S. P. Gilbert (Rensselaer Polytechnic Institute). K490 (35) behaved similarly to K370 in all assays described (see Fig. 4) (data not shown).
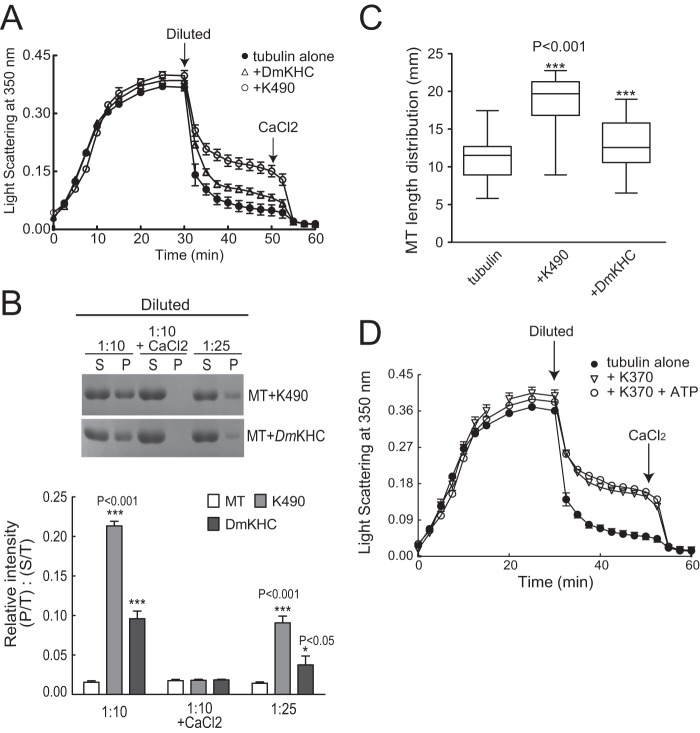
FIGURE 4.
Comparison of MT polymerization and stabilization by K490 and DmKHC. DmKHC contains the MT-binding domain, the neck-linker domain, and the first coiled-coil domain. To be internally consistent, we prepared a KIF17 construct encoding the motor, neck, and first coiled-coil domains (amino acids 1–490, K490). A, MT polymer (15 μm tubulin) analyzed by light scattering at A350 nm in the presence of K490 (0.15 μm) or DmKHC (0.15 μm) before and after dilution (1:1) and the subsequent addition of CaCl2 (5 mm). MTs polymerized with K490 (like K370) were resistant to dilution. B, sedimentation analysis of soluble and polymerized tubulin after 10- or 25-fold dilution or after 10-fold dilution with 5 mm CaCl2. Preformed MTs (30 μm) were incubated with K490 or DmKHC as indicated. Soluble tubulin (supernatant (S)) and polymerized tubulin (pellet (P)) were analyzed by SDS-PAGE. The graph below shows quantification of tubulin in pellets (P/T, where “T” is total (supernatant + pellet)) relative to the supernatant (S/T) at each time point. C, box-whisker plots showing the distribution of MT polymer lengths under the indicated conditions (n = 250 MTs measured per condition). Like K370, K490 promoted formation of long MTs (21.6 ± 0.86 μm). By comparison, the average length of MTs polymerized with DmKHC was 14.6 ± 1.2 μm. D, MT polymerization and stability (15 μm) analyzed by light scattering at A350 nm in the presence of K370 with or without 1 mm ATP before and after dilution (1:1) and when CaCl2 (5 mm) was added after dilution.
Protein Methods
N-terminal GST- and His-tagged protein constructs were transformed into Escherichia coli BL21(DE3) cells. His6- and GST-tagged versions of each protein behaved equivalently in all assays. Protein expression was induced with 0.5–1 mm isopropyl β-d-thiogalactopyranoside for 4 h prior to harvesting cells. Recombinant protein was purified on glutathione-Sepharose 4B (GE Healthcare) or nickel-nitrilotriacetic acid (Ni-NTA; Qiagen). Anion exchange with Q-Sepharose Fast Flow (Sigma) was performed to remove contaminants in preparations of KIF17 motor domains. Purified proteins were dialyzed and stored in 80 mm K-PIPES (pH 6.9) with 100 mm KCl, 2 mm MgCl2, 1 mm EGTA, 5% glycerol, and 0.5 mm DTT. Purified motor domains were stored at >1.5 mg/ml with 50 μm ATP. Protein concentrations were determined by the Bradford assay and are reported as monomer concentration unless stated otherwise. With the exception of EB1, proteins were prepared fresh for each set of experiments, as KIF17 protein fragments tended to aggregate and degrade upon freezing and thawing.
Immunoblotting was performed as described (13). The primary antibodies used were mouse anti-EB1 (clone 1A11, Abcam), mouse anti-β-actin (Sigma), and mouse anti-acetylated tubulin (clone 6-11B-1, Sigma). Proteins were detected using an Odyssey infrared imaging system (LI-COR Biosciences).
Hydrodynamic Analysis of Recombinant KIF17 Motor and Tail Constructs
7–25% sucrose gradients were formed by layering 7, 10, 15, 20, and 25% (w/v) sucrose (300 μl each) in 80 mm K-PIPES (pH 6.9) with 100 mm KCl, 2 mm MgCl2, 1 mm EGTA, 5% glycerol, and 0.5 mm DTT. Samples (100 μl) were loaded immediately and centrifuged at 55,000 rpm for 4 h in a Beckman TLS-55 swinging-bucket rotor. A mixture (5 μg each, final volume of 100 μl) of carbonic anhydrase (bovine erythrocytes), catalase (bovine liver), serum albumin (bovine), and ovalbumin (chicken egg) was used as a standard. 15 × 90-μl fractions were removed from the top for analysis. 20 μl of each fraction was boiled with an equal volume of 2× Laemmli buffer and resolved by 10% SDS-PAGE and Coomassie Blue staining.
Preparation of Tubulin/MTs
Tubulin was purified from bovine brain by two cycles of temperature-dependent assembly and disassembly, followed by ion exchange using DEAE-Sephadex A-50 (Sigma) as described (36, 37). Tubulin in BRB80 (80 mm K-PIPES (pH 6.8) with 1 mm EGTA and 1 mm MgCl2) was snap-frozen in single-use aliquots and stored at −80 °C. Before use, aliquots were thawed on ice and clarified by centrifugation at 90,000 rpm for 5 min at 4 °C in a Beckman TLA-100.4 rotor. Subtilisin (20 μg/ml) was used to remove the C termini of α- and β-tubulin as described (38). The concentration of tubulin dimer was determined by spectrophotometry using an extinction coefficient of 115,000 m−1 cm−1 at A280 nm.
For ATPase assays, tubulin (25 μm) was incubated in BRB80 with 1 mm DTT and 1 mm GMP-CPP (Jena Bioscience) on ice for 5 min before clarification. Clarified supernatant was then polymerized at 37 °C for 60 min. MTs were recovered in pellet fractions after centrifugation at 90,000 rpm for 5 min at 37 °C and resuspended in 0.8 volume (of original starting material) of prewarmed BRB80 with 1 mm DTT and 25 μm Taxol. For MT co-sedimentation assays, Taxol was omitted from all buffers. For MT polymerization and depolymerization assays, BRB80 contained only 1 mm GTP and 1 mm DTT.
MT-stimulated ATPase Activity
Steady-state ATPase activity was determined by measuring phosphate release at 25 °C with the PiColorLockTM reagent (Innova Biosciences). K370 (100 nm) alone or with KIF17-Tail (100 nm) and EB1 (200 nm) was incubated with MTs in BRB80 (pH 6.9) containing 75 mm KCl and 1 mm DTT. Phosphate release was plotted against MT concentration, and data were fit to a Michaelis-Menten equation using GraphPad Prism to determine Kcat and K0.5MT. No ATP hydrolysis was detected when MTs, EB1, or KIF17-Tail was assayed in the absence of K370. Data were derived from three to five independent experiments performed in triplicate.
Microtubule Co-sedimentation Assay
K370 (1 μm), KIF17-Tail (1 μm), and EB1 (1 μm) were mixed individually or together prior to incubation with 1.5 μm GMP-CPP-MTs in BRB80 containing 75 mm KCl. Samples were equilibrated at room temperature for 20 min and then centrifuged at 70,000 rpm for 20 min at 25 °C. Pellets were resuspended in BRB80 containing 75 mm KCl and 5 mm CaCl2 on ice (in a volume equal to the supernatant) for 10 min with vigorous pipetting. Pellets and supernatants were analyzed by 12% SDS-PAGE and detected with Coomassie Blue. The protein in each fraction was quantified by densitometry using NIH ImageJ. The Kd values of K370 and KIF17-Tail dimers for MTs were determined assuming a single binding site per tubulin dimer, but values did not change significantly when two binding sites per dimer were considered (data not shown). Data were fit to a bimolecular binding equation using GraphPad Prism under nonlinear regression conditions: Y = (Bmax × X)/(Kd + X), where Y is the fraction of bound protein, and X is the concentration of free MTs. Representative gels are shown in the figures. Binding data (Kd) were determined from three experiments.
KIF17-Tail and EB1 Binding to K370
His-K370 (0.4 ml of 60 μm) was bound to 0.25 ml of Ni-NTA-Sepharose for 30 min at 25 °C. The resin was washed with 4 column volumes of 0.5 m NaCl and transferred to buffer containing 10 mm NaCl. In each experiment, 10–15% of loaded K370 bound to Ni-NTA-Sepharose. KIF17-Tail and EB1 (0.2 ml of 10 μm each) were loaded individually, together, or sequentially onto the column in 10 mm NaCl and saturated K370 on the beads, allowing analysis of competitive binding to K370. When loaded together, His-EB1 instead of GST-EB1 was used. Control experiments showed that His-EB1 did not bind to Ni-NTA-Sepharose preloaded with His-KIF1A-Tail (which does not interact with EB1). Unbound material was removed by washing with 3 column volumes of 10 mm NaCl. Bound KIF17-Tail and EB1 were then eluted from immobilized His-K370 with 0.75 ml of 25, 150, or 350 mm NaCl. Volumes of each fraction loaded on the gels were adjusted so that the protein concentration would be equivalent to that of the “preload” if 100% of protein was recovered in that fraction.
MT Polymerization and Stabilization Assay
Tubulin polymerization was measured by light scattering at 350 nm. Tubulin (15 μm) in BRB80 containing 1 mm GTP and 1 mm DTT was incubated at 37 °C with or without K370 (0.15 μm), KIF17-Tail (0.15 μm), and EB1 (0.25 μm) independently or together. Turbidity was measured at 30-s intervals for 30 min in a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA) at 37 °C. For analysis of depolymerization, samples were then diluted 1:1 with polymerization buffer, and turbidity was measured for another 22.5 min, followed by the addition of 5 mm CaCl2 to induce complete MT depolymerization. Similar results were obtained when K370, EB1, and KIF17-Tail were added after MT polymerization but before dilution (data not shown). For sedimentation analysis, 30 μm tubulin was polymerized for 20 min at 37 °C and divided into the appropriate number of tubes to compare the effects of K370, KIF17-Tail, and EB1 on MT stability after 1:10 and 1:25 dilutions in BRB80 containing 50 mm KCl with or without 5 mm CaCl2. After dilution, samples were incubated at room temperature for 15 min, followed by centrifugation at 70,000 rpm for 10 min at 37 °C in a Beckman TLA-100.4 rotor. Supernatants were removed, and pellets were resuspended (in a volume equal to the supernatant) in SDS sample buffer. Equal volumes of pellet and supernatant were loaded for SDS-PAGE.
For direct MT visualization, MTs were polymerized from a 10:1 mixture of unlabeled and rhodamine-labeled tubulin (Cytoskeleton, Inc.). After polymerization (or dilution), samples were fixed in prewarmed glutaraldehyde (0.2% in BRB80) and diluted 25-fold in BRB80. 5 μl of each sample was analyzed by fluorescence imaging on a Nikon Eclipse E400 microscope with a 60× 1.4 numerical aperture oil immersion objective. Images were acquired without binning using an ORCA-II-ER cooled CCD camera (Hamamatsu Photonics) controlled by MetaMorphTM (Molecular Devices).
Analysis of MT Regrowth after Nocodazole Washout
The effects of KIF17-Tail on MT regrowth after washing out nocodazole (NZ) were quantified by scoring cells as positive or negative based on the presence of (i) MTs regrowing from centrosomes 1 min after NZ washout and (ii) MTs with a uniform radial orientation 3 min after washout.
Cell Culture, Transfection, and Microinjection
Madin-Darby canine kidney (MDCK) cells were cultured in DMEM with 5% FBS and 20 mm HEPES (pH 7.2). Cells were seeded on sterilized coverslips and used at 80% confluence 2–3 days after plating. siRNA (0.5–3 μg) was transfected using Amaxa Nucleofector as recommended by the manufacturer. cDNAs (5–20 μg/ml) in 10 mm HEPES and 140 mm KCl (pH 7.4) were injected into cell nuclei using a Narishige micromanipulator mounted on a Nikon Eclipse TS100 microscope. Cells were incubated at 37 °C for at least 60 min to allow for expression of cDNAs and treated subsequently as indicated under “Results.”
Immunofluorescence
Cells were fixed in −20 °C methanol for 1–2 min. In MT regrowth experiments, cells were first washed twice with PEM buffer (100 mm PIPES, 2 mm EGTA, and 2 mm MgCl2 (pH 6.9)), and soluble tubulin was extracted by incubating cells at 37 °C for 45 s in prewarmed PEM buffer containing 0.1% Triton X-100, followed by fixation in −20 °C methanol. The primary antibodies used were mouse anti-acetylated tubulin (clone 6-11B-1) and rat anti-tyrosinated tubulin (clone YL1/2). Fluorescently conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. Fixed cell images were acquired on either a Nikon Eclipse E400 or Nikon TiE microscope using a 40× 1.0 numerical aperture Plan Apochromat oil immersion objective and an ORCA-II-ER or Neo sCMOS (Andor Technology) camera without binning. Images were scaled to highlight features of interest as indicated under “Results” and converted to 8-bit copies for figure assembly. Devices were controlled by MetaMorphTM or Nikon NIS-Elements.
RESULTS
KIF17 Activity Is Regulated by EB1 and KIF17-Tail
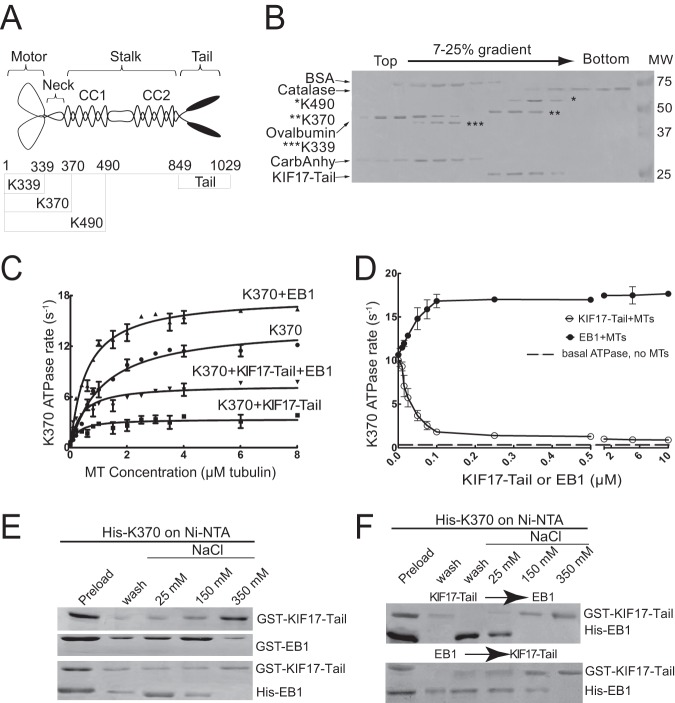
In cells, KIF17 localization on MTs was shown to be regulated by EB1 and KIF17-Tail (13, 35), both of which interact with the KIF17 N-terminal catalytic motor domain. To determine the significance of these interactions in KIF17 function, we analyzed the effects of purified EB1 and KIF17-Tail on the activity of the KIF17 motor domain (K370, encoding the MT-binding and neck-linker domains) (Fig. 1A). Hydrodynamic and native PAGE analyses (Fig. 1B) (data not shown) confirmed that purified K370, K490, and KIF17-Tail behaved as dimers, whereas K339, encoding the motor domain alone, behaved as a monomer. In addition, K339 had very low MT-stimulated ATPase activity at steady state (data not shown) and thus was not used further in this study.
FIGURE 1.
KIF17-Tail and EB1 bind K370 directly and affect MT-stimulated ATPase activity. A, schematic diagram of KIF17 constructs used in this study. B, analysis of recombinant KIF17 motor domains (bands corresponding to each KIF17 motor domain are highlighted to their left by asterisks) and KIF17-Tail by sucrose gradient centrifugation. BSA (4.4 S), catalase (11.3 S), ovalbumin (3.6 S), and carbonic anhydrase (CarbAnhy; 3.2 S) were used as standards. C, ATPase activity of K370 (0.1 μm) with varied MT concentrations (0–8 μm) in the absence and presence of KIF17-Tail (0.1 μm), EB1 (0.2 μm), or both proteins. D, MT (2.5 μm)-stimulated ATPase activity of K370 (0.1 μm) with increasing concentrations of KIF17-Tail or EB1 (0–10 μm). E, binding of GST-KIF17-Tail (0.1 μm) and GST-EB1 (0.1 μm) incubated individually (upper panels) or together (lower panel; His-EB1 was used for simultaneous binding) with His-K370 and eluted at the indicated salt concentrations. Note that co-incubation of KIF17-Tail with EB1 resulted in elution of EB1 at lower salt concentrations, indicative of competitive binding. F, sequential incubation of GST-KIF17-Tail and His-EB1 with His-K370, followed by elution at the indicated salt concentrations. Note that EB1 was recovered almost entirely in the flow-through and 25 mm salt fractions when added after preincubation of KIF17-Tail with K370 (upper panel). Preincubation of EB1 with K370 had little effect on binding of KIF17-Tail to K370 (lower panel). Data are representative of three independent experiments. Error bars = S.E. (p < 0.01).
In phosphate release assays, K370 exhibited MT-dependent ATPase activity, with a Kcat of 11.72 ± 1.15 ATP s−1 (Fig. 1C and Table 1). The addition of purified EB1 reduced the K0.5MT of K370 by 52%, stimulating the rate of ATP hydrolysis. At saturating MT concentrations, EB1 increased Kcat by only 23% (Fig. 1C and Table 1). By contrast, KIF17-Tail inhibited K370 ATPase activity by 79% and decreased K0.5MT by 76% (Fig. 1C and Table 1). Increasing concentrations of KIF17-Tail suppressed ATP hydrolysis further, but K370 retained MT-stimulated ATPase activity that was 2–3 times above basal rates at even 100-fold excess (Fig. 1D). This and the decrease in K0.5MT suggests that KIF17-Tail may not completely displace K370 from MTs. In vitro binding analysis showed that EB1 and KIF17-Tail interacted competitively and directly with K370 and that KIF17-Tail bound K370 more tightly than EB1 (Fig. 1, E and F). Thus, when EB1 and KIF17-Tail were added to K370 simultaneously (Fig. 1C and Table 1) or sequentially (Fig. 1F), the inhibitory effects of KIF17-Tail on ATPase activity dominated.
TABLE 1.
MT-stimulated ATPase activities of K370 with or without EB1 or KIF17-Tail
Shown are the Kcat and K0.5MT values of 0.1 μm K370 alone or with 0.2 μm EB1 or 0.1 μm KIF17-Tail in steady-state ATPase activity assays. Data represent the means ± S.E. determined in three independent experiments (p < 0.01).
| Construct | Kcat | K0.5MT |
|---|---|---|
| s−1 | μm | |
| K370 | 11.72 ± 1.15 | 1.16 ± 0.4 |
| K370 + EB1 | 14.36 ± 0.8 | 0.56 ± 0.07 |
| K370 + KIF17-Tail | 3.03 ± 0.32 | 0.28 ± 0.09 |
| K370 + KIF17-Tail + EB1 | 5.91 ± 0.54 | 0.33 ± 0.1 |
EB1 and KIF17-Tail Enhance Association of K370 with MTs
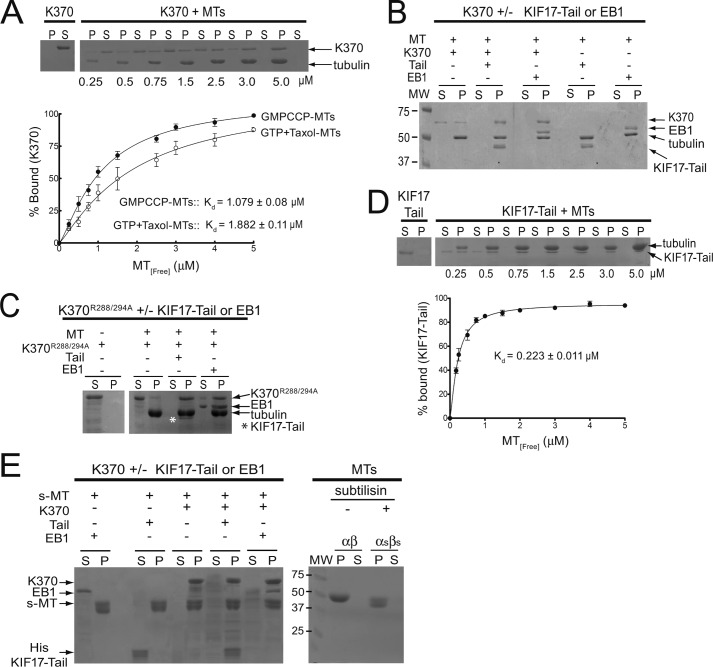
The effects of EB1 and KIF17-Tail on the K0.5MT of K370 suggest that both may influence binding of K370 to MTs. To test this, we incubated K370 with GMP-CPP-MTs in the absence or presence of EB1 or KIF17-Tail for 20 min and analyzed co-sedimentation with MTs after centrifugation. In the absence of MTs, we recovered K370 in the supernatant after centrifugation. The addition of MTs at increasing concentrations resulted in a shift of K370 to pellet fractions with complete MT co-sedimentation at ≥3 μm MTs and a dissociation constant (Kd) of 1.079 ± 0.08 μm (Fig. 2A). At 2 μm MTs, some K370 could still be detected in supernatants. However, the addition of EB1 or KIF17-Tail resulted in complete recovery of K370 in the MT pellet (Fig. 2B). Thus, EB1 and KIF17-Tail enhance K370 association with MTs. Interestingly, the Kd of K370 for MTs assembled with GTP and Taxol (1.882 ± 0.011 μm) (Fig. 2A, inset), where GTP-tubulin could be hydrolyzed to GDP-tubulin, was higher than that measured with GMP-CPP-MTs. This suggests that the KIF17 motor may bind preferentially to GTP-tubulin.
FIGURE 2.
Effects of KIF17-Tail and EB1 on K370 co-sedimentation with MTs. A, co-sedimentation of K370 (1 μm) with GMP-CPP-MTs (0–5 μm) or MTs assembled in the presence of GTP and Taxol. P, pellet; S, supernatant. B, co-sedimentation of K370 (1 μm) with MTs (1.5 μm) in the absence and presence of KIF17-Tail (1 μm) or EB1 (1 μm). C, co-sedimentation of mutant K370R288A/R294A (K370R288/294A; 1 μm) with MTs (3 μm) in the absence and presence of KIF17-Tail (1 μm; highlighted by asterisk) or EB1 (2 μm). D, co-sedimentation of KIF17-Tail (1 μm) with MTs (0–5 μm). E, left panel, co-sedimentation of K370 (1.5 μm) in the absence or presence of KIF17-Tail (1.5 μm) or EB1 (1.5 μm) with subtilisin-cleaved MTs (s-MT; 3.5 μm). Right panel, tubulin before (αβ) and after (αsβs) subtilisin treatment. Fractions of K370 or KIF17-Tail in pellets was graphed as a function of MT concentration and fit by nonlinear curve fitting to determine Kd. Data represent the average Kd determined in three experiments. Error bars = S.E. (p < 0.01).
To determine whether KIF17-Tail or EB1 recruits K370 to MTs, we used a MT-binding mutant (13) of the KIF17 motor domain, K370R288A/R294A, in MT co-sedimentation assays. K370R288A/R294A alone did not co-sediment with MTs and was detected almost entirely in supernatants, as expected. However, when EB1 or KIF17-Tail was added, K370R288A/R294A was recovered in MT pellets (Fig. 2C). Together with the data above, these results show that K370 can associate with MTs by both direct and indirect mechanisms.
KIF17 Motor and Tail Domains Bind Different Regions of the MT
KIF17-Tail also co-sedimented independently with MTs, with a Kd of 0.223 ± 0.011 μm (Fig. 2, B and D). Because it has higher affinity for MTs compared with K370, we tested if KIF17-Tail inhibits K370 activity by interfering with its binding to MTs. For this purpose, we polymerized MTs from subtilisin-treated tubulin. Subtilisin cleaves ∼10–20 amino acids from the C terminus of α- and β-tubulin, removing the E-hooks from both subunits, and was shown to inhibit binding of the kinesin-1 tail domain to MTs (39). In co-sedimentation assays with subtilisin-cleaved MTs, K370 was recovered in the subtilisin-cleaved MT pellet fraction (Fig. 2E), showing that the KIF17 motor domain interacts with MTs outside the extreme C terminus of tubulin. Conversely, KIF17-Tail and EB1 did not co-sediment with subtilisin-cleaved MTs and were instead recovered in supernatant fractions unless K370 was also present (Fig. 2E) (40, 41). Thus, KIF17 motor and tail domains bind different regions of the MT, consistent with the idea that intramolecular binding of the tail domain of KIF17 to its motor domain does not necessarily inhibit catalytic activity by displacing the motor from MTs.
K370 Promotes Polymerization of Fewer but Longer MTs in Vitro
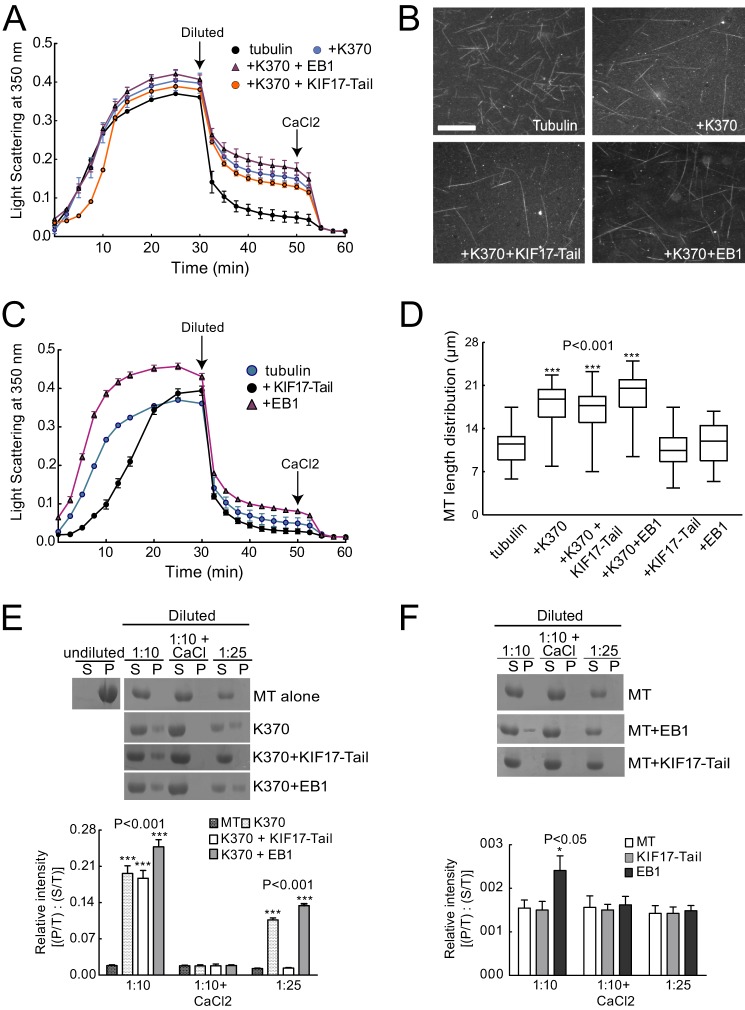
We showed previously that a constitutively active, conformational KIF17 mutant (KIF17G754E) dampens MT dynamics, reduces MT polymerization rates, and increases the level of stable MTs in epithelial cells (13). To determine how KIF17 affects MTs directly, we tested the effects of K370 on MT polymerization in vitro. K370 was incubated with unassembled tubulin on ice and then warmed to induce MT polymerization. Analysis of turbidity over time showed no significant difference in tubulin polymer mass in the absence or presence of K370 (Fig. 3A). 10-Fold higher concentrations of K370 (1.5 μm) also had no effect on total polymer mass or polymerization rate (data not shown). However, direct fluorescence visualization of MTs polymerized with K370 revealed that fewer MTs were formed, but that these MTs were 57% longer (17.8 ± 1.13 μm) than those polymerized without K370 (11.4 ± 1.04 μm) (Fig. 3, B and D). These findings suggest that K370 promotes tubulin addition to MT ends or alters rescue or catastrophe frequency. Either could result in elongation of existing MTs at the expense of MT number.
FIGURE 3.
K370 induces polymerization of longer but fewer MTs that are resistant to dilution-induced depolymerization. A, MT polymerization (15 μm tubulin) analyzed by light scattering at A350 nm with or without K370 (0.15 μm) in the absence and presence of KIF17-Tail (0.15 μm) or EB1 (0.25 μm). Arrows indicate when MTs were diluted (1:1) and when CaCl2 (5 mm) was added after dilution. Data represent three experiments performed in duplicate. B, direct fluorescence visualization of MTs polymerized for 30 min with or without K370 (0.15 μm) in the absence and presence of KIF17-Tail (0.15 μm) or EB1 (0.25 μm). Scale bar = 10 μm. C, MT polymerization (15 μm tubulin) analyzed by light scattering at A350 nm as described for A in the absence and presence of KIF17-Tail (0.15 μm) or EB1 (0.25 μm). D, box-whisker plots showing the distribution of MT polymer lengths under the indicated conditions (n = 250 MTs measured per condition). E, sedimentation analysis of soluble and polymerized tubulin after 10- or 25-fold dilution or after 10-fold dilution with 5 mm CaCl2. Preformed MTs (30 μm) were incubated with K370 alone or with KIF17-Tail or EB1. The graph shows densitometric quantification of tubulin in pellets (P/T, where “T” is total (supernatant + pellet)) relative to the supernatant (S/T) at each time point. F, sedimentation analysis of soluble and polymerized tubulin as described for E, but for EB1 or KIF17-Tail alone. Error bars = S.E. (p < 0.01). Note that tubulin detected in pellets after incubation with EB1 alone appeared to be protein aggregates by fluorescence microscopy (not shown).
The addition of EB1 with K370 induced a small but reproducible increase in MT polymer mass and MT length (19.4 ± 1.27 μm) (Fig. 3, A, B, and D). These changes could be due to independent effects of EB1 on MT polymerization in the absence of K370 (Fig. 3C), although EB1 alone did not significantly affect MT length (Fig. 3D). The addition of KIF17-Tail with K370 had no detectable effect on MT polymer mass or MT length and number (Fig. 3, A, B, and D). Interestingly, however, we did measure a reproducible decrease in the initial rate of tubulin polymerization when KIF17-Tail was present (Fig. 3A). This lag was more prominent when KIF17-Tail was incubated alone with tubulin (Fig. 3C) and reflects attenuated incorporation of tubulin into MTs as determined by sedimentation analysis and direct MT visualization (see Fig. 6). Together, these data indicate that KIF17 motor and tail domains can independently influence MT polymerization in vitro.
FIGURE 6.
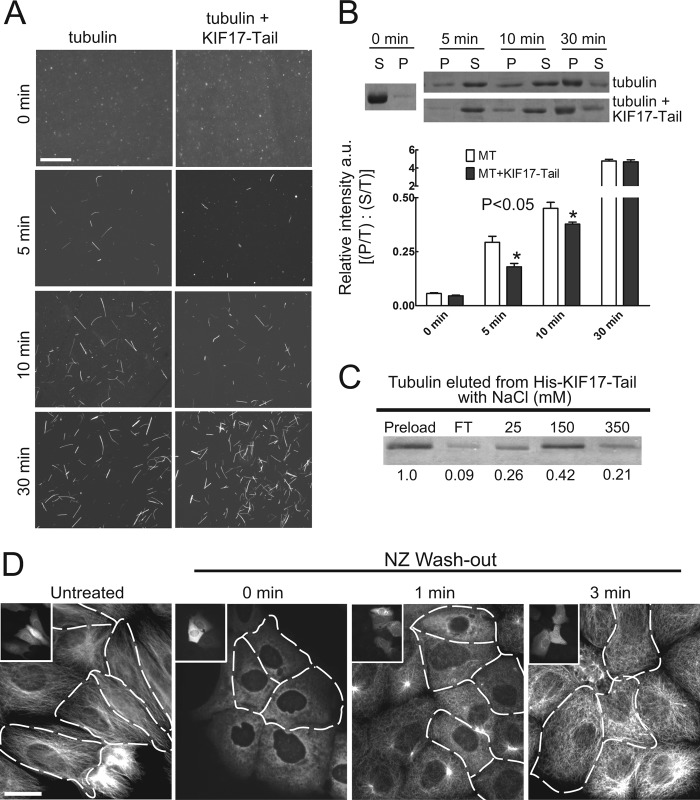
KIF17-Tail inhibits initial tubulin polymerization in vitro and in cells. A, MTs (15 μm) polymerized in the absence or presence of KIF17-Tail (0.15 μm) and processed at the indicated times for direct MT visualization as described in the legend to Fig. 3. Scale bar = 10 μm. B, Sedimentation analysis and quantification of soluble (S) and polymerized (P) tubulin (relative to total (T)) in the absence or presence of KIF17-Tail (0.15 μm) at each time point. C, binding of soluble tubulin heterodimers (no GTP) to His-KIF17-Tail. Numbers below each band show the proportion of total tubulin (normalized to total starting material = 1) eluted from KIF17-Tail at increasing salt concentrations. FT, flow-through fraction. D, MT regrowth in MDCK cells after NZ treatment and washout for the indicated times. Tubulin immunostaining showed that uninjected control cells contained newly growing centrosomal MT arrays 1 min after NZ washout, whereas cells expressing mCherry-KIF17-Tail (shown in insets and outlined in MT images) did not. After 3 min, cells expressing KIF17-Tail had MTs, but they were oriented randomly compared with MTs in controls. Scale bar = 20 μm.
K370 Is Sufficient for MT Stabilization in Vitro
To determine how changes in MT polymerization by KIF17 affect MT stability, we analyzed the effects of K370 on MT depolymerization in vitro by turbidity and sedimentation analysis. We polymerized tubulin in the absence or presence of K370 with and without EB1 or KIF17-Tail as described above. After polymerization, MTs were diluted 1:1, and turbidity was measured for an additional 20 min. Alternatively, MTs were diluted 1:10 and 1:25, below the critical concentration for tubulin assembly, and processed for sedimentation analysis. In the absence of K370, MT polymer mass decreased rapidly after dilution (indicated by a steep initial decrease in light scattering) (Fig. 3A), and tubulin was recovered only from supernatants after centrifugation (Fig. 3E). The inclusion of K370 significantly reduced the loss of polymer mass after dilution, and tubulin was recovered in both pellet and supernatant fractions after centrifugation (Fig. 3, A and E). By comparison, the kinesin-1 motor domain (42) was significantly less protective of MTs in these assays (Fig. 4, A–C), suggesting that MT stabilization activity is not necessarily a conserved kinesin function.
The addition of EB1 or KIF17-Tail to K370 had little effect on MT resistance to dilution in turbidity assays and had no effect on tubulin sedimentation profiles (Fig. 3, A and E). Thus, K370 is sufficient to protect MTs from dilution-induced depolymerization in vitro. However, the subsequent addition of CaCl2 (5 mm), which induces extreme curvature of MT ends and favors tubulin dissociation (43), resulted in complete MT depolymerization under all conditions tested (Fig. 3, A and E). MTs were not protected from dilution-induced depolymerization by EB1 or KIF17-Tail in the absence of K370 (Fig. 3, C and F).
Interestingly, K370 protected MTs from dilution-induced disassembly independent of ATP (Fig. 4D). This contrasts with our prior finding that a rigor mutant of full-length KIF17 (KIF17-FL) did not protect MTs from depolymerization by NZ in cells (13). Because expression of a monomeric KIF17 motor that lacks MT-stimulated ATPase activity (amino acids 1–339) can stabilize MTs,3 we speculate that this discrepancy reflects motility-independent differences in the association of K370 and the KIF17 holoenzyme with MT ends or the contribution of additional cytoplasmic factors to the regulation of KIF17 function in cells.
K370 Stabilizes MTs in Epithelial Cells
We next tested if KIF17 is sufficient to stabilize MTs in MDCK epithelial cells. We transiently expressed mCherry-tagged K370, KIF17-FL, or control empty vector by intranuclear injection of cDNAs, which are rapidly translated and visible by fluorescence microscopy 45–60 min after injection. Cells were fixed 3.5 h after injection and immunostained for dynamic (unmodified) and stable (post-translationally acetylated) MTs. We observed no significant difference in dynamic MT arrays labeled with anti-tyrosinated tubulin antibodies under any condition. However, cells expressing K370 or KIF17-FL were enriched in acetylated MTs compared with uninjected neighbors and cells expressing empty vector (Fig. 5A, dashed lines indicate injected cells). Expression of K370 or KIF17-FL also resulted in an increase in MTs that were resistant to treatment with the MT antagonist NZ (33 μm) for 45 min. In these assays, only 27% (n = 134) of control cells contained NZ-resistant MTs, whereas 71% (n = 183) and 70% (n = 135) of cells expressing K370 and KIF17-FL, respectively, contained NZ-resistant MTs.
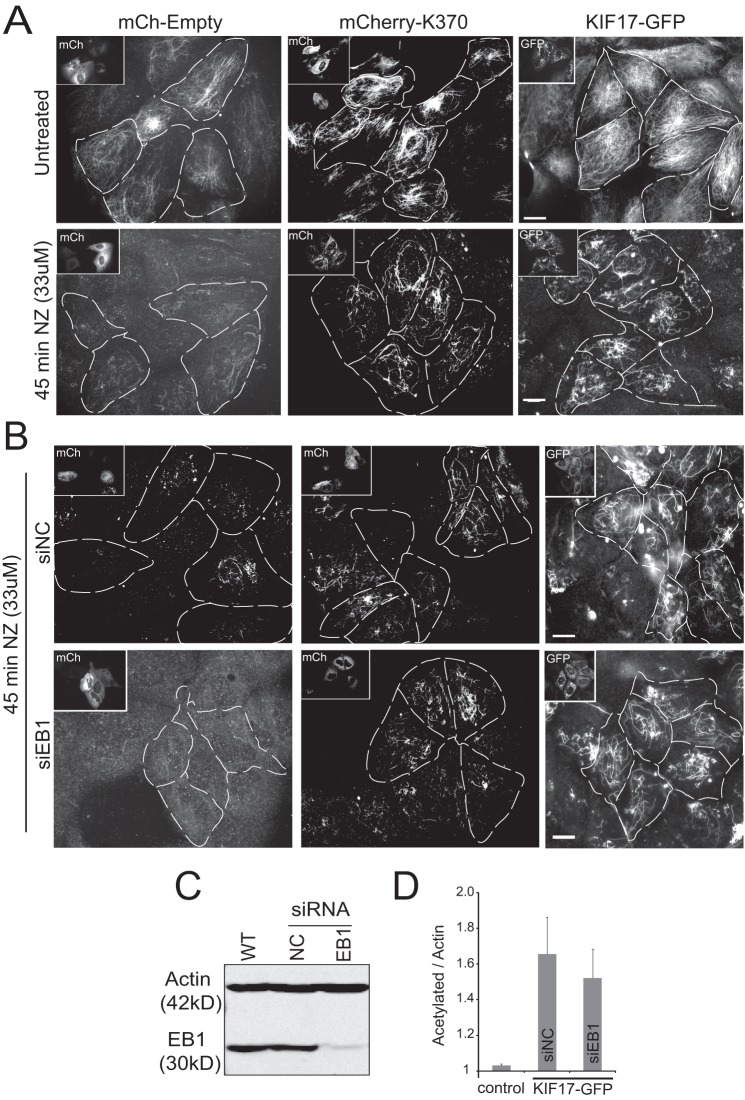
FIGURE 5.
K370 and KIF17-FL are sufficient to protect MTs from nocodazole-induced depolymerization when expressed in MDCK cells. A, MDCK cells injected with cDNAs encoding mCherry (mCh)-K370 or mCherry-KIF17-FL. NZ (33 μm) was added 3.5 h after injection, and cells were incubated for an additional 45 min before fixation and immunostaining for tyrosinated and acetylated tubulin as markers of dynamic and stable MTs, respectively. Insets and dashed lines indicate cells expressing mCherry-K370 and mCherry-KIF17-FL. B, EB1-depleted MDCK cells injected with cDNAs encoding mCherry-K370 or mCherry-KIF17-FL after NZ treatment as described for A. Images show acetylated MTs remaining after 45 min of NZ (33 μm) treatment. Scale bars = 20 μm. siEB1, EB1 siRNA; siNC, negative control siRNA. C, immunoblot showing depletion of EB1 protein 48 h after siRNA transfection. D, quantification of acetylated tubulin by immunoblotting in control and EB1-depleted cells after transfection of KIF17-FL-GFP.
To determine whether EB1 is necessary for KIF17-mediated MT stabilization in cells, we depleted it with siRNA targeting EB1 as described (13). Despite ∼90% depletion of EB1 (Fig. 5C), cells expressing either K370 or KIF17-FL still had increased acetylated and NZ-resistant MTs compared with controls (Fig. 5, B and D). Although EB1 is required to localize endogenous KIF17 to MT plus-ends (13), these data show that K370 or KIF17-FL can stabilize MTs in the absence of EB1 when expressed exogenously in epithelia.
KIF17-Tail Delays Tubulin Polymerization in Vitro and in Cells
We showed above that KIF17-Tail attenuated initial rates of tubulin polymerization in vitro (Fig. 3C). Direct visualization of MTs at early time points during polymerization revealed that fewer MTs formed and a new, albeit small, population of very short MTs with an average length of ∼5 μm could be observed (Fig. 6A). In MT sedimentation assays, we measured a corresponding decrease in the proportion of tubulin in pellet fractions after centrifugation (Fig. 6B). Soluble tubulin bound reversibly to KIF17-Tail in vitro (Fig. 6C), suggesting that it may sequester or otherwise transiently inhibit incorporation of tubulin dimers into MTs.
To determine whether KIF17-Tail also affects MT polymerization in MDCK cells, we monitored MT regrowth after complete depolymerization with a combination of cold and NZ treatment (44), followed by NZ washout at 37 °C. Consistent with in vitro data, expression of KIF17-Tail resulted in a lag in initial regrowth of MTs (Fig. 6D, outlined cells). At 1 min after NZ washout, only 15% of cells expressing KIF17-Tail (n = 114) had regrown centrosomal MT asters compared with 96% of controls expressing empty vector (n = 87) (data not shown) or non-expressing neighboring cells. 3 min after NZ washout, 98% of control cells (n = 76) had dense centrosomal MT arrays. Although cells expressing KIF17-Tail contained numerous MTs at this time, only 32% (n = 84) contained centrosomal arrays; MTs in most cells were instead non-centrosomal and oriented randomly. Because tubulin could be eluted from KIF17-Tail with physiological salt concentrations, it is not yet clear if the tail domain of the holoenzyme would normally interact with soluble tubulin in cells. However, as the KIF17 ortholog in Caenorhabditis elegans may transport tubulin to promote cilial growth (33), it is possible that KIF17 acts in a similar manner to promote MT elongation in epithelial cells.
DISCUSSION
The data presented here reveal a novel role for KIF17 as a direct regulator of MT dynamics and stability. In vitro, the KIF17 catalytic motor domain is sufficient to induce persistent MT elongation, although it does not affect the rate or extent of tubulin polymerization. The corresponding reduction in MT number, increase in MT length, and 1.8-fold higher affinity of K370 for GMP-CPP-MTs suggest that KIF17 alters MT dynamic instability by acting at plus-ends, like kinesin-8, -13, and -14 (3). In support of this, we found that K370 protects MTs from end-mediated, dilution- or NZ-induced depolymerization in vitro and in cells. Furthermore, constitutively active KIF17 attenuates MT growth rate and dynamicity while increasing stable MTs in cells (13). Together, these data support a model wherein KIF17 could promote persistent MT growth and resistance to depolymerization by stabilizing structural features of the MT end that promote tubulin addition and suppress loss of the GTP cap. The ability of KIF17 to enhance persistent MT growth in cells may also involve a role for KIF17 as a MT polymerase, as KIF17-Tail binds both soluble tubulin and MTs. As such, it may act as a tubulin transporter, delivering subunits for incorporation at MT plus-ends as described for cilial elongation by the C. elegans ortholog OSM-3 (33). Alternatively, the MT-binding site in KIF17-Tail may serve to tether or enhance retention on MTs as described for kinesin-8 motors (22, 30). Additional studies are needed to determine whether KIF17-FL interacts with MTs and tubulin simultaneously and if this further enhances MT polymerization in vitro. It will also be important to identify the tubulin-binding site in KIF17-Tail to elucidate the significance of the interaction with tubulin and to appreciate fully the mechanism by which KIF17 regulates MT dynamics and stabilization in cells.
Although the KIF17 motor domain is sufficient to promote MT elongation and stability, we found that both EB1 and KIF17-Tail affected catalytic activity and binding of K370 to MTs. EB1 and KIF17-Tail each decreased the K0.5MT of K370 by ∼2-fold, but their effects on KIF17 function were strikingly different. Despite enhancing ATP hydrolysis at subsaturating MT concentrations, EB1 appears to act primarily in recruitment or retention of KIF17 on MTs. This is consistent with previous data showing that endogenous KIF17 is recruited to MTs by overexpressed EB1 in MDCK cells (13). Conversely, KIF17-Tail inhibits ATPase activity. Endogenous EB1 likely modulates KIF17 by promoting its accumulation at MT plus-ends. This could increase the probability of KIF17-mediated MT capture and stabilization in cells. EB1 could additionally affect KIF17 activity for MT stabilization by an allosteric mechanism, altering interactions with MTs, KIF17-Tail, and other KIF17-binding partners or cargos that contribute to MT regulation. Indeed, our data show that EB1 and KIF17-Tail interact competitively with K370. Further support for an allosteric effect of EB1 on KIF17 comes from analysis of KIF17 conformation in cells using FRET biosensors, which showed that EB1 recruits KIF17 to MTs and relieves autoinhibitory head-tail interactions in cells (46).
Unlike EB1, the decrease in K370 K0.5MT induced by KIF17-Tail was accompanied by inhibition of ATPase activity. However, K370 does hydrolyze some ATP in the presence of KIF17-Tail, and K370 and KIF17-Tail have different tubulin-binding sites and co-sediment simultaneously with MTs in vitro. From these data, we speculate that binding of KIF17-Tail to the catalytic motor domain could “stall” KIF17 without necessarily displacing it from MTs. This would allow KIF17 to reinitiate translocation on MTs rapidly when inhibition by the tail is relieved. Although consistent with the mechanism for regulation of kinesin-8 by its tail domain (30, 45), this contrasts with cellular assays showing that KIF17-Tail displaces exogenous KIF17-FL from MTs (35). We anticipate that future studies analyzing KIF17-FL in vitro will help determine unambiguously how the catalytic activity of KIF17 is regulated by the tail domain.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM087575 (to G. K.). This work was also supported by the Irma T. Hirschl Trust (to G. K.).
F. Jaulin and G. Kreitzer, unpublished data.
- MT
- microtubule
- DmKHC
- D. melanogaster kinesin-1 heavy chain
- Ni-NTA
- nickel-nitrilotriacetic acid
- GMP-CPP
- guanosine 5′-(α,β-methylene)triphosphate
- NZ
- nocodazole
- MDCK
- Madin-Darby canine kidney
- KIF17-FL
- full-length KIF17.
REFERENCES
- 1. Hirokawa N., Noda Y., Tanaka Y., Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 2. Drummond D. R. (2011) Regulation of microtubule dynamics by kinesins. Semin. Cell Dev. Biol. 22, 927–934 [DOI] [PubMed] [Google Scholar]
- 3. Su X., Ohi R., Pellman D. (2012) Move in for the kill: motile microtubule regulators. Trends Cell Biol. 22, 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daire V., Poüs C. (2011) Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch. Biochem. Biophys. 510, 83–92 [DOI] [PubMed] [Google Scholar]
- 5. Ems-McClung S. C., Walczak C. E. (2010) Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 21, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gardner M. K., Odde D. J., Bloom K. (2008) Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 18, 307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanenbaum M. E., Medema R. H. (2010) Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell 19, 797–806 [DOI] [PubMed] [Google Scholar]
- 8. Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. (2005) Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235–245 [DOI] [PubMed] [Google Scholar]
- 9. Du Y., English C. A., Ohi R. (2010) The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 20, 374–380 [DOI] [PubMed] [Google Scholar]
- 10. Niwa S., Nakajima K., Miki H., Minato Y., Wang D., Hirokawa N. (2012) KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev. Cell 23, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi T., Tsang W. Y., Li J., Lane W., Dynlacht B. D. (2011) Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 145, 914–925 [DOI] [PubMed] [Google Scholar]
- 12. Kimura T., Watanabe H., Iwamatsu A., Kaibuchi K. (2005) Tubulin and CRMP-2 complex is transported via kinesin-1. J. Neurochem. 93, 1371–1382 [DOI] [PubMed] [Google Scholar]
- 13. Jaulin F., Kreitzer G. (2010) KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J. Cell Biol. 190, 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boehlke C., Kotsis F., Buchholz B., Powelske C., Eckardt K. U., Walz G., Nitschke R., Kuehn E. W. (2013) Kif3a guides microtubular dynamics, migration and lumen formation of MDCK cells. PLoS ONE 8, e62165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daire V., Giustiniani J., Leroy-Gori I., Quesnoit M., Drevensek S., Dimitrov A., Perez F., Poüs C. (2009) Kinesin-1 regulates microtubule dynamics via a c-Jun N-terminal kinase-dependent mechanism. J. Biol. Chem. 284, 31992–32001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Insinna C., Humby M., Sedmak T., Wolfrum U., Besharse J. C. (2009) Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev. Dyn. 238, 2211–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scholey J. M. (2012) Kinesin-2 motors transport IFT-particles, dyneins and tubulin subunits to the tips of Caenorhabditis elegans sensory cilia: relevance to vision research? Vision Res. 75, 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stout J. R., Yount A. L., Powers J. A., Leblanc C., Ems-McClung S. C., Walczak C. E. (2011) Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol. Biol. Cell 22, 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee T., Langford K. J., Askham J. M., Brüning-Richardson A., Morrison E. E. (2008) MCAK associates with EB1. Oncogene 27, 2494–2500 [DOI] [PubMed] [Google Scholar]
- 20. Browning H., Hackney D. D. (2005) The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J. Biol. Chem. 280, 12299–12304 [DOI] [PubMed] [Google Scholar]
- 21. Ten Hoopen R., Cepeda-García C., Fernández-Arruti R., Juanes M. A., Delgehyr N., Segal M. (2012) Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr. Biol. 22, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 22. Su X., Qiu W., Gupta M. L., Jr., Pereira-Leal J. B., Reck-Peterson S. L., Pellman D. (2011) Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell 43, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varga V., Leduc C., Bormuth V., Diez S., Howard J. (2009) Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183 [DOI] [PubMed] [Google Scholar]
- 24. Varga V., Helenius J., Tanaka K., Hyman A. A., Tanaka T. U., Howard J. (2006) Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8, 957–962 [DOI] [PubMed] [Google Scholar]
- 25. Gupta M. L., Jr., Carvalho P., Roof D. M., Pellman D. (2006) Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8, 913–923 [DOI] [PubMed] [Google Scholar]
- 26. Gardner M. K., Zanic M., Gell C., Bormuth V., Howard J. (2011) Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell 147, 1092–1103 [DOI] [PubMed] [Google Scholar]
- 27. Desai A., Verma S., Mitchison T. J., Walczak C. E. (1999) Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78 [DOI] [PubMed] [Google Scholar]
- 28. Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. (2003) The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell 11, 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sproul L. R., Anderson D. J., Mackey A. T., Saunders W. S., Gilbert S. P. (2005) Cik1 targets the minus-end kinesin depolymerase Kar3 to microtubule plus ends. Curr. Biol. 15, 1420–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stumpff J., Du Y., English C. A., Maliga Z., Wagenbach M., Asbury C. L., Wordeman L., Ohi R. (2011) A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell 43, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., Surrey T. (2004) A kinesin-like motor inhibits microtubule dynamic instability. Science 303, 1519–1522 [DOI] [PubMed] [Google Scholar]
- 32. Sardar H. S., Luczak V. G., Lopez M. M., Lister B. C., Gilbert S. P. (2010) Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr. Biol. 20, 1648–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hao L., Thein M., Brust-Mascher I., Civelekoglu-Scholey G., Lu Y., Acar S., Prevo B., Shaham S., Scholey J. M. (2011) Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat. Cell Biol. 13, 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gundersen G. G., Gomes E. R., Wen Y. (2004) Cortical control of microtubule stability and polarization. Curr. Opin. Cell Biol. 16, 106–112 [DOI] [PubMed] [Google Scholar]
- 35. Hammond J. W., Blasius T. L., Soppina V., Cai D., Verhey K. J. (2010) Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189, 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borisy G. G., Marcum J. M., Olmsted J. B., Murphy D. B., Johnson K. A. (1975) Purification of tubulin and associated high molecular weight proteins from porcine brain and characterization of microtubule assembly in vitro. Ann. N.Y. Acad. Sci. 253, 107–132 [DOI] [PubMed] [Google Scholar]
- 37. Murphy D. B., Borisy G. G. (1975) Association of high-molecular-weight proteins with microtubules and their role in microtubule assembly in vitro. Proc. Natl. Acad. Sci. U.S.A. 72, 2696–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodionov V. I., Gyoeva F. K., Kashina A. S., Kuznetsov S. A., Gelfand V. I. (1990) Microtubule-associated proteins and microtubule-based translocators have different binding sites on tubulin molecule. J. Biol. Chem. 265, 5702–5707 [PubMed] [Google Scholar]
- 39. Seeger M. A., Rice S. E. (2010) Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J. Biol. Chem. 285, 8155–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zanic M., Stear J. H., Hyman A. A., Howard J. (2009) EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS ONE 4, e7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu Z. C., Gupta K. K., Slabbekoorn A. R., Paulson B. A., Folker E. S., Goodson H. V. (2009) Interactions between EB1 and microtubules: dramatic effect of affinity tags and evidence for cooperative behavior. J. Biol. Chem. 284, 32651–32661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilbert S. P., Johnson K. A. (1993) Expression, purification, and characterization of the Drosophila kinesin motor domain produced in Escherichia coli. Biochemistry 32, 4677–4684 [DOI] [PubMed] [Google Scholar]
- 43. Weisenberg R. C., Deery W. J. (1981) The mechanism of calcium-induced microtubule disassembly. Biochem. Biophys. Res. Commun. 102, 924–931 [DOI] [PubMed] [Google Scholar]
- 44. Kreitzer G., Marmorstein A., Okamoto P., Vallee R., Rodriguez-Boulan E. (2000) Kinesin and dynamin are required for post-Golgi transport of a plasma-membrane protein. Nat. Cell Biol. 2, 125–127 [DOI] [PubMed] [Google Scholar]
- 45. Weaver L. N., Ems-McClung S. C., Stout J. R., LeBlanc C., Shaw S. L., Gardner M. K., Walczak C. E. (2011) Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr. Biol. 21, 1500–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Espenel C., Acharya B. R., Kreitzer G. (2013) A biosensor of local kinesin activity reveals roles of PKC and EB1 in KIF17 activation. J. Cell Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]