Background: The relations between the gut microbiota, food, and host play a crucial role in human health.
Results: Prevalent bacterial glycoside phosphorylases are able to break down dietary carbohydrates and the N-glycans lining the intestinal epithelium.
Conclusion: GH130 enzymes are new targets to study interactions between host and gut microbes.
Significance: Glycoside phosphorylases are key enzymes of host glycan catabolism by gut bacteria.
Keywords: Carbohydrate Metabolism, Enzyme Mechanisms, Glycoprotein, Intestinal Epithelium, Phosphorylase, GH130, N-Glycan, Human Gut
Abstract
To metabolize both dietary fiber constituent carbohydrates and host glycans lining the intestinal epithelium, gut bacteria produce a wide range of carbohydrate-active enzymes, of which glycoside hydrolases are the main components. In this study, we describe the ability of phosphorylases to participate in the breakdown of human N-glycans, from an analysis of the substrate specificity of UhgbMP, a mannoside phosphorylase of the GH130 protein family discovered by functional metagenomics. UhgbMP is found to phosphorolyze β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc and is also a highly efficient enzyme to catalyze the synthesis of this precious N-glycan core oligosaccharide by reverse phosphorolysis. Analysis of sequence conservation within family GH130, mapped on a three-dimensional model of UhgbMP and supported by site-directed mutagenesis results, revealed two GH130 subfamilies and allowed the identification of key residues responsible for catalysis and substrate specificity. The analysis of the genomic context of 65 known GH130 sequences belonging to human gut bacteria indicates that the enzymes of the GH130_1 subfamily would be involved in mannan catabolism, whereas the enzymes belonging to the GH130_2 subfamily would rather work in synergy with glycoside hydrolases of the GH92 and GH18 families in the breakdown of N-glycans. The use of GH130 inhibitors as therapeutic agents or functional foods could thus be considered as an innovative strategy to inhibit N-glycan degradation, with the ultimate goal of protecting, or restoring, the epithelial barrier.
Introduction
The human gut microbiota is a dense and complex ecosystem, which plays a crucial role in maintaining human health. The many meta-omic studies carried out in the last few years have shown that certain metabolic diseases, such as obesity, and some inflammatory diseases, such as inflammatory bowel diseases (IBD)2 (1–4), are associated with structural and functional imbalances of the microbiota (5). The catabolism of complex carbohydrates by gut bacteria plays a key role in the microbial colonization and equilibrium of the digestive tract (6, 7). Indeed, the plant polysaccharides (cellulose, hemicelluloses, pectin, and resistant starch) that compose dietary fiber supply the main source of carbon for gut bacteria, in the form of monosaccharides produced by polysaccharide breakdown, such as glucose, xylose, arabinose, uronic acids, and to a lesser extent, galactose, mannose, and rhamnose (8). In particular, mannose is found in plant cell walls, in mannan and glucomannan, the backbones that consist of a homopolymer of β1,4-linked d-mannopyranosyl residues and a heterogeneous sequence of β1,4-linked d-glucopyranosyl and d-mannopyranosyl units (9). There is, however, a further source of complex carbohydrates in the gut, namely the heavily O- and N-glycosylated glycoproteins that line the intestinal epithelium to form a protective barrier against pathogens and chemical and mechanical aggression (10). Although host O-glycans are rich in N-acetylhexosamines (GlcNAc, GalNAc, and Neu5Ac), galactose and fucose, many mature N-glycans contain eight α- and β-linked d-mannopyranosyl residues linked to chitobiose (Fig. 1) (11). Human glycan-microbial interactions in the gastrointestinal tract play a crucial role in determining the outcome of relations of both commensals and pathogens with the host. Indeed, alterations in the structure and/or quantity of host glycans due to biosynthetic defects or microbial degradation alter their barrier function and are thought to be involved in the initiation and the maintenance of mucosal inflammation in IBDs and in the development of intestinal cancer (12).
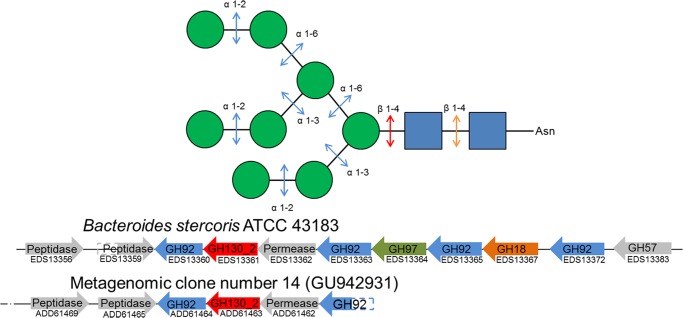
FIGURE 1.
Schematic representation of N-glycan processing by glycoside hydrolases and phosphorylases. Green spheres, mannosyl residues; blue squares, N-acetyl-d-glucosamine residues. The genomic cluster containing the GH130_2 encoding gene and other ones involved in N-glycan processing are shown for B. stercoris ATCC 43183 genome, and the UhgbMP encoding metagenomic clone (accession number GU94293). CAZyme encoding genes are colored according to the linkage they break down in the N-glycan structure, based on known biochemical data of the families to which they belong.
To breakdown these complex carbohydrates of either plant or human origin, gut bacteria produce a full repertoire of carbohydrate-active enzymes (CAZymes, listed in the CAZy database (13)), of which glycoside hydrolases (GHs) are the main constituents, as revealed by Gill et al. (14). In particular, the degradation of dietary mannans requires both endo-β-mannanases and β-mannosidases to release mannose (15). The hydrolysis of N-glycans by a broad consortium of endo- and exo-, α-, and β-glycosidases, in particular those produced by the prominent gut bacterium Bacteroides thetaiotaomicron (16–20), has also been described. Until August, 2013, only glycoside hydrolases (GHs) have been implicated in N-glycan breakdown.
However, other types of CAZymes participate in the breakdown of complex carbohydrates, particularly those of plant origin, by working in synergy with GHs as follows: carbohydrate esterases (CE), polysaccharide lyases (PLs), and glycoside phosphorylases (GPs). The presence of CEs and PLs is readily detectable in gut bacterial genomes, metagenomes, and metatranscriptomes, because they are sufficiently divergent from GHs as to be classified in their own CAZyme families. This is not the case for GPs, which are found both in the glycosyltransferase (GT) and glycoside hydrolase families, depending on the sequence and catalytic mechanism similarities shared with GT and GH archetypes, respectively. The prevalence of GPs and their role in the metabolism of carbohydrates is therefore difficult to evaluate on the basis of sequence data alone, as a given family of GHs (or of GTs) may include both GHs (or GTs) as well as GPs. GPs catalyze the breakdown of a glycosidic linkage from oligosaccharide or polysaccharide substrates with concomitant phosphate glycosylation to yield a glycosyl-phosphate product and a sugar chain of reduced length. These enzymes are also able to perform reverse phosphorolysis (in the so-called “synthetic reaction”) to form a glycosidic bond between the glycosyl unit originating from the glycosyl-phosphate, which acts as the sugar donor, and a carbohydrate acceptor (23). Retaining GPs, for which phosphorolysis occurs via overall retention of the substrate anomeric configuration, are found in CAZy families GT4, GT35, and GH13. Inverting GPs are classified in families GH65, GH94, GH112, and GH130. Inverting GH-related GPs and hydrolytic enzymes use a similar single displacement mechanism, differing in the requirement of GPs for a single catalytic residue (the proton donor), and inverting GHs for two catalytic residues. In GP-catalyzed reactions, the reaction begins with the direct nucleophilic attack by phosphate to the glycosidic bond with the aid of the catalytic residue, which donates a proton to the glycosidic oxygen atom and then proceeds through an oxocarbenium cation-like transition state. In GH reaction mechanisms, the nucleophilic attack of the C-1 of the glycoside is performed by a water molecule activated by the catalytic base. The natural structural and functional diversity of GPs thus appears to be highly restricted because of the following: (i) they are found in only 7 of the 226 GH and GT families listed in the CAZy database (March 2013); (ii) approximately only 15 EC entries are currently assigned to GPs (24); (iii) their specificity toward glycosyl phosphates is limited to α- and β-d-glucopyranose 1-phosphate (25–29), which are the most prevalent substrates; α-d-galactopyranose 1-phosphate (30); N-acetyl-α-d-glucosamine 1-phosphate (31), and α-d-mannopyranose 1-phosphate (32). In July 2013, α-d-mannopyranose 1-phosphate specificity was described for just one enzyme produced by a human gut bacterium, the Bacteroides fragilis NCTC 9343 mannosylglucose phosphorylase (BfMP), which converts β-d-mannopyranosyl-1,4-d-glucopyranose and phosphate into α-d-mannopyranose 1-phosphate and d-glucose (32). This enzyme has been implicated in the catabolism of linear dietary mannans, assisted by a β-1,4-mannanase and a mannobiose 2-epimerase. During review of this study, Nihira et al. (33) reported the discovery of a metabolic pathway for N-glycans, which includes a β-d-mannosyl-N-acetyl-1,4-d-glucosamine phosphorylase named BT1033, produced by the human gut inhabitant B. thetaiotamicron VPI-5482. BfMP and BT1033 are two of the four enzymes to be characterized in the recently created GH130 family, which includes a total of 447 entries, from archaea, bacteria, and eukaryotes. The other characterized GH130 enzymes are RaMP1 and RaMP2 from the ruminal bacterium Ruminococcus albus NE1 (34). These enzymes have also been proposed to participate in mannan catabolism in the bovine rumen and are assisted by an endomannanase and an epimerase, via RaMP2- and RaMP1-catalyzed phosphorolysis of β-1,4-manno-oligosaccharides and 4-O-β-d-mannopyranosyl-d-glucopyranose, respectively (34). X-ray crystallographic studies show that GH130 enzymes share a 5-fold β-propeller fold. Currently, atomic coordinates data sets for four protein structures are available in the RCSB Protein Data Bank (35) as follows: BACOVA_03624 protein from Bacteroides ovatus ATCC 8483 (3QC2); BDI_3141 protein from Parabacteroides distasonis ATCC 8503 (3TAW); BT_4094 protein from Bacteroides thetaiotamicron VPI-5482 (3R67), and TM1225 protein from Thermotoga maritima MSB8 (1VKD). However, no function has yet been attributed to these four proteins, thus limiting the understanding of structure specificity relations for GH130 enzymes and the investigation of their catalytic mechanism.
Recently, the sequence of another GH130 enzyme, which we refer to as UhgbMP (unknown human gut bacterium mannoside phosphorylase, GenBankTM accession number ADD61463.1), was discovered by functional metagenomics of the human gut microbiota (36). The 36.6-kbp metagenomic DNA fragment containing the UhgbMP-encoding gene was taxonomically assigned to an as yet unidentified bacterium belonging to the genus Bacteroides. However, UhgbMP itself presents 99% protein sequence identity with the hypothetical protein BACSTE_03540 from Bacteroides stercoris ATCC 43183 (accession number EDS13361.1).
Here, we present an integrative approach, based on analyses of UhgbMP substrate specificity and of metagenomic and genomic data at the level of the entire human gut ecosystem, to reveal the role of UhgbMP and of 64 other GH130 enzymes produced by known gut bacteria in the breakdown of host and dietary mannose-containing glycans. In addition, we establish the molecular basis of GH130 enzyme catalysis, supported by the experimental results of rational engineering of UhgbMP and the analysis of its three-dimensional molecular model. Finally, we discuss the potential of this enzyme for the development of therapeutic agents and functional foods to inhibit N-glycan degradation, with the ultimate goal of protecting the epithelial barrier in the IBD context.
EXPERIMENTAL PROCEDURES
Recombinant UhgbMP Production and Purification
First, the UhgbMP encoding gene was PCR-amplified from the Escherichia coli metagenomic clone (GenBankTM accession number GU942931) using primers forward 5′-AGTATGAGTAGCAAAGTTATTATTCCTTGG-3′ and reverse 5′-TCAGATGATGCTTGTACGTTTGGTAAATTC-3′, by using the Expand Long Template PCR kit (Roche Applied Science).
To allow heterologous UhgbMP production in E. coli with His6 tag at the N-terminal extremity, the PCR product was purified and subsequently cloned into the pCR8/GW/TOPO entry vector (Invitrogen), and then into the pDEST17 destination vector (Invitrogen), according to the manufacturer's recommendations. E. coli BL21-AI cells (Invitrogen) harboring the UhgbMP-encoding plasmid were cultured at 20 °C for 24 h in ZYM-5052 autoinduction medium (37) supplemented with 100 μg/ml ampicillin, inoculated at A600 nm 0.1. Cells were harvested and resuspended in 20 mm Tris-HCl, pH 7.0, 300 mm NaCl, and lysed by sonication. Soluble lysate was applied to a TALON resin loaded with cobalt (GE Healthcare) equilibrated in 20 mm Tris-HCl, pH 7.0, 300 mm NaCl. After column washing with 8 volumes of the same buffer supplemented with 10 mm imidazole, the protein was eluted in 20 mm Tris-HCl, pH 7.0, 300 mm NaCl, 150 mm imidazole. Finally, the protein sample was desalted on a PD-10 column (GE Healthcare) and eluted in 20 mm Tris-HCl, pH 7.0, 0.1% Tween 80 (v/v). In these conditions, 84% of UhgbMP remained soluble after 8 days at 4 °C, thus allowing further functional characterization. The purity of the purified wild-type UhgbMP and mutants was evaluated higher than 95% by SDS-PAGE using Any kDTM Mini-PROTEAN® TGXTM Precast Gel (Bio-Rad) (supplemental Fig. 1). After migration, proteins were stained with the PageBlue Protein Staining Solution (Thermo Scientific) according to the manufacturer's recommendations. Protein concentrations were determined by spectrometry using a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) The NanoDrop® measurement error was 5%. The calculated extinction coefficient of the purified UhgbMP fused to an N-terminal His6 tag was 76 630 m−1·cm−1.
UhgbMP Mutagenesis
Site-directed mutagenesis was performed by using the pDEST17 plasmid harboring the wild-type UhgbMP encoding gene as PCR template and the primers 5′-ATGGCTGTGCCAATACCGTAACCG-3′ and 5′-TACGGTATTGGCACAGCCATAGTAT-3′ for D304N mutant, 5′-GTATGGCTACAACCCACGCGTGTGCT-3′ and 5′-ACGCGTGGGTTGTAGCCATACACCCA-3′ to obtain the D104N mutant, 5′-AACCGTACCAATGCATGGGCGATGT-3′ and 5′-ATGCATTGGTACGGTTCGCGC-3′ for the E273Q mutant, and finally 5′-TGGCGAGGACCCGCGCGT-3′ and 5′-TCCTCGCCATACACCCAGGTACCGA-3′ for the Y103E mutant. The PCR products, amplified with the Phusion® High Fidelity DNA polymerase (New England Biolabs), were purified and digested by DpnI (New England Biolabs) before E. coli TOP10 transformation. Protein production and purification were identical for wild-type UgbMP and its variants.
Enzyme Assays
All reactions were carried out at 37 °C (wild-type UhgbMP optimal temperature) in 20 mm Tris-HCl, pH 7.0 (wild-type UhgbMP optimal pH). Syntheses of manno-oligosaccharides from α-d-mannopyranose 1-phosphate were performed with 0.1 mg/ml purified UhgbMP during 24 h at 37 °C in 20 mm Tris-HCl, pH 7.0, with 10 mm of α-d-mannopyranose 1-phosphate (Sigma, reference M1755), and 10 mm of d-glucose, d-mannose, d-galactose, d-fructose, N-acetyl-d-glucosamine, β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose (Dextra, UK, reference MC0320), l-rhamnose, d-altrose, d-allose, d-fucose, l-fucose, d-mannitol, d-sorbitol, d-lyxose, xylitol, l-xylose, d-xylose, l-arabinose, or d-cellobiose.
Phosphorolysis kinetic parameters were determined with 0.01 mg/ml of purified enzyme by quantifying α-d-mannose 1-phosphate release rate from 0.5 to 10 mm inorganic phosphate (10 mm corresponding to the intracellular concentration of inorganic phosphate that was previously measured in bacteria (38)) and 1–10 mm of β-d-mannopyranosyl-1,4-d-glucose (Carbosynth, UK, reference OM04754), 0.4–4 mm β-1,4-d-mannan (Megazyme, Ireland, reference P-MANCB), 1–20 mm β-d-mannopyranosyl-1,4-d-mannose (Megazyme, Ireland, reference O-MBI), 0.1–1 mm pNP-β-d-mannopyranose, or 0.05–0.5 mm of β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose (Dextra, UK, reference MC0320). Reverse phosphorolysis kinetic parameters were determined with 0.01 mg/ml purified enzyme by quantifying α-d-mannopyranose 1-phosphate consumption rate from 0.5 to 10 mm α-d-mannopyranose 1-phosphate and 5–40 mm of d-mannose, d-glucose, d-galactose, d-fructose, or N-acetyl-d-glucosamine, or 0.1–40 mm of N,N′-diacetylchitobiose (Dextra, UK, reference C8002). The apparent kinetic parameters for phosphorolysis and reverse phosphorolysis at fixed initial concentrations of inorganic phosphate and α-d-mannose 1-phosphate, respectively, were determined by fitting the initial rates of α-d-mannose 1-phosphate release and consumption to the Michaelis-Menten equation. The kinetic parameters for phosphorolysis and synthesis of β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose and β-d-mannopyranosyl-1,4-d-mannose were determined by varying carbohydrates, α-d-mannose 1-phosphate or inorganic phosphate concentrations, and by fitting the initial rates of α-d-mannose 1-phosphate release and consumption to the sequential random Bi Bi mechanism equation (39). Nonlinear regression was performed with SigmaPlot enzyme kinetics module, version 1.3 (Systat Software, Inc., San Jose, CA). UhgbMP-specific activity toward β-1,4-d-manno-oligosaccharides, was determined with 0.1 mg/ml purified enzyme by quantifying α-d-mannopyranose 1-phosphate release rate from 10 mm inorganic phosphate and 10 mm β-d-manno-oligosaccharides of polymerization degree 2–6 (Megazyme, Ireland). The percentage of UhgbMP inhibition by exogenous carbohydrates or polyols was measured with 0.1 mg/ml purified enzyme by quantifying α-d-mannopyranose 1-phosphate consumption rate from 10 mm α-d-mannopyranose 1-phosphate as glycosyl donor, with and without 10 mm of l-rhamnose, d-altrose, d-allose, d-fucose, d-mannitol, d-sorbitol, d-lyxose, xylitol, l-xylose, d-xylose, l-arabinose, or d-cellobiose.
The percentage of reverse phosphorolysis activity of the D104N, D304N, and E273Q variants, compared with that of the wild-type enzyme, was determined with 0.1 mg/ml of purified proteins by quantifying α-d-mannopyranose 1-phosphate consumption rate from 10 mm α-d-mannopyranose 1-phosphate as glycosyl donor and 10 mm d-mannose as acceptor. α-d-Mannopyranose 1-phosphate was quantified by using high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Carbohydrates and α-d-mannopyranose 1-phosphate were separated on a 4 × 250 mm Dionex CarboPak PA100 column. A gradient of sodium acetate (from 0 to 150 mm in 15 min) and an isocratic step of 300 mm sodium acetate in 150 mm NaOH was applied at a 1 ml·min−1 flow rate. Detection was performed using a Dionex ED40 module with a gold working electrode and a Ag/AgCl pH reference. Finally, the hydrolytic or phosphorolytic behavior of the wild-type UhgbMP and its Y103E variant was assessed by using 1 mm pNP-β-d-mannopyranose, in the absence or presence, respectively, of 10 mm inorganic phosphate. The pNP release was monitored at A405 nm on a carry-100 UV-visible spectrophotometer (Agilent Technologies). Between three and five independent experiments were carried out to determine initial activity, kinetic constants, and percentage of inhibition by carbohydrates and polyols of wild-type UhgbMP and its mutants. For all reaction rate measurements, it was checked by HPAEC-PAD that less than 10% of substrate was consumed and that the amount of consumed or released α-d-mannopyranose 1-phosphate increased linearly with time.
NMR Spectroscopy
Freeze-dried reaction media were exchanged twice with 99.9 atom % D2O and lyophilized. Deuterium oxide was used as the solvent, and sodium 2,2,3,3-tetradeuterio-3-trimethylsilylpropanoate was selected as the internal standard. 1H and 13C NMR spectra were recorded on a Bruker Advance 500-MHz spectrometer using a 5-mm z-gradient TBI probe at 298 K, an acquisition frequency of 500.13 MHz, and a spectral width of 8012.82 Hz. Spectra were acquired and processed using TopSpin 3.0 software. The various signals were assigned by comparison with signals obtained from α-d-mannopyranose 1-phosphate, β-d-mannopyranosyl-1,4-d-mannose (Megazyme, Irleland, reference O-MBI), β-d-mannopyranosyl-1,4-d-glucose (Carbosynth, UK, reference OM04754), or β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose (Dextra, UK, reference MC0320), used as standards.
Three-dimensional Molecular Modeling
The UhgbMP sequence was submitted to the I-TASSER server for automated protein structure and function prediction (40). The homologous T. maritima TM1225 structure (PDB accession code 1VKD) was used to provide spatial restraints. The three-dimensional model of UhgbMP predicted by I-TASSER was then further refined by energy minimization using the CFF91 force field implementation in the DISCOVER module of the InsightII software suite (Accelrys, San Diego). The CFF91 cross-terms, a harmonic bond potential, and a dielectric constant of 1.0 were specified in the energy function. An initial minimization was performed with positional restraints on the protein backbone using a steepest descent algorithm followed by conjugated gradient minimization until the maximum RMS energy gradient was less than 0.5 kcal mol−1 Å−1. The system was then fully relaxed without positional restraints. α-d-Mannopyranose 1-phosphate, β-d-mannoheptaose, and β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose were manually docked into the active site of UhgbMP. The ligand complexes were then optimized according to the minimization protocol described above with the ligand molecules free to move. Molecular graphics images were produced using PyMOL software (Schrödinger, LLC).
GH130 Multiple Sequence Alignment Analyses
The 369 public sequences of GH130 enzymes listed in the CAZy database in January, 2013, were aligned with MUSCLE version 3.7 (41). A distance matrix was generated from the multiple sequence alignment using the BLOSUM62 amino acid residue substitution matrix. The output result file was subjected to hierarchical clustering using Ward's method (39), and the resulting tree was visualized using DENDROSCOPE 3 (42). Two sequence clusters were clearly apparent, members of which were accordingly assigned to the GH130_1 or GH130_2 subfamilies.
Position-dependent amino acid residue variation in multiple sequence alignment data were analyzed using the Shannon information entropy measure (HX), calculated using SEQUESTER software. The Shannon entropy (HX) at residue alignment position (X), corrected for the normalized frequency of residue type occurrence, is computed as shown in Equation 1,
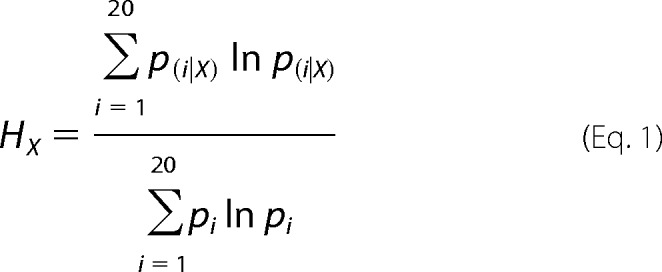 |
where pi‖X is the conditional probability of residue type i occurrence at alignment position X, and pi is the normalized probability of residue type i occurrence at any position. To minimize sampling bias, normalized residue type probability values were taken as those documented by Ranganathan and co-workers (43),3 garnered from sequence data for all natural proteins. Values of HX lie in the range 0–1; a zero value corresponds to a fully conserved residue position, and a value of unity represents a distribution in which each residue type has an equal chance of occurrence. SEQUESTER appends Shannon entropies at aligned residue positions in a chosen reference protein structure to an atomic coordinate data file for convenient three-dimensional visual display.
GH130 Genomic Context Analysis
The analysis of the habitat of the organisms displaying these 369 GH130 sequences, as referenced in the GOLD database, allowed us to sort out 28 public genomes of human gut bacteria that display GH130 sequences. The genomic context of these 63 sequences, as well as of the UhgbMP and the GenBankTM ADD61810 sequences belonging to the metagenomic sequences GU942931 and GU942945, respectively, was analyzed to identify the CAZy encoding genes that are present in the same multigenic cluster as a GH130_1 or a GH130_2 encoding gene. CAZy encoding genes were searched on the same DNA strand as the GH130 encoding gene, with an increment of 10 kbp maximum upstream or downstream the GH130 sequence. For each glycoside hydrolase (GH), polysaccharide lyase (PL), or CE family, the frequency of co-occurrence in a multigenic cluster with a GH130_1 or a GH130_2 sequence was calculated, pondered by the number of GH130_1 or GH130_2 sequences, and used as edge attributes in a Cytoscape representation. In total, 74 and 62 co-occurrences of GH, PL, or CE sequences with GH130_1 or GH130_2 sequences, respectively, were counted. Prediction of transmembrane topology and signal peptides was performed using PHOBIUS.
Analysis of GH130 Encoding Gene Prevalence in the Human Gut Microbiome
The 369 GH130 sequences referenced in the CAZy database in January, 2013, were searched by TBLASTN analysis (E-value = 0, identity ≥90%) in the catalog of prevalent genes (as defined by Qin et al. (3)) encoded on contigs of metagenomic sequences obtained from fecal samples of 162 individuals from the MetaHit cohort and of 139 individuals from the National Institutes of Health HMP cohort in the United States.
RESULTS
Uhgb_MP Substrate and Product Specificity
From the metagenomic sequence contained in the recombinant E. coli clone (accession number GU942931), we subcloned the UhgbMP encoding gene to produce a soluble protein tagged with a His6 tag at the N terminus, with a yield of 32 mg of purified protein per liter of culture. We first characterized the substrate specificity of UhgbMP for carbohydrate phosphorolysis. In contrast to RaMP1 and BfMP, but similar to RaMP2 and to BT1033, UhgbMP exhibits a relaxed specificity toward carbohydrate substrates (Table 1). UhgbMP is able to phosphorolyze β-d-mannopyranosyl-1,4-d-glucopyranose, β-1,4-linked d-manno-oligosaccharides, and mannan (β-d-Manp-1,4-(d-Manp)n, with n = 1–15), characterized by a notable increase in specific activity with the degree of polymerization (DP) (Fig. 2). UhgbMP is thus to date the only characterized GH130 enzyme that is able to break down mannan, a constituent of hemicellulose in grains and nuts (45). Interestingly, UhgbMP is also able to phosphorolyze β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose (β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc), a signature motif of human N-glycans. However, neither cellobiose (β-d-Glcp-1,4-d-Glcp) nor N,N′-diacetylchitobiose (β-d-GlcpNAc-1,4-d-GlcpNAc) could be phosphorolyzed, indicating that the UhgbMP subsite −1 is highly specific for mannosyl residues.
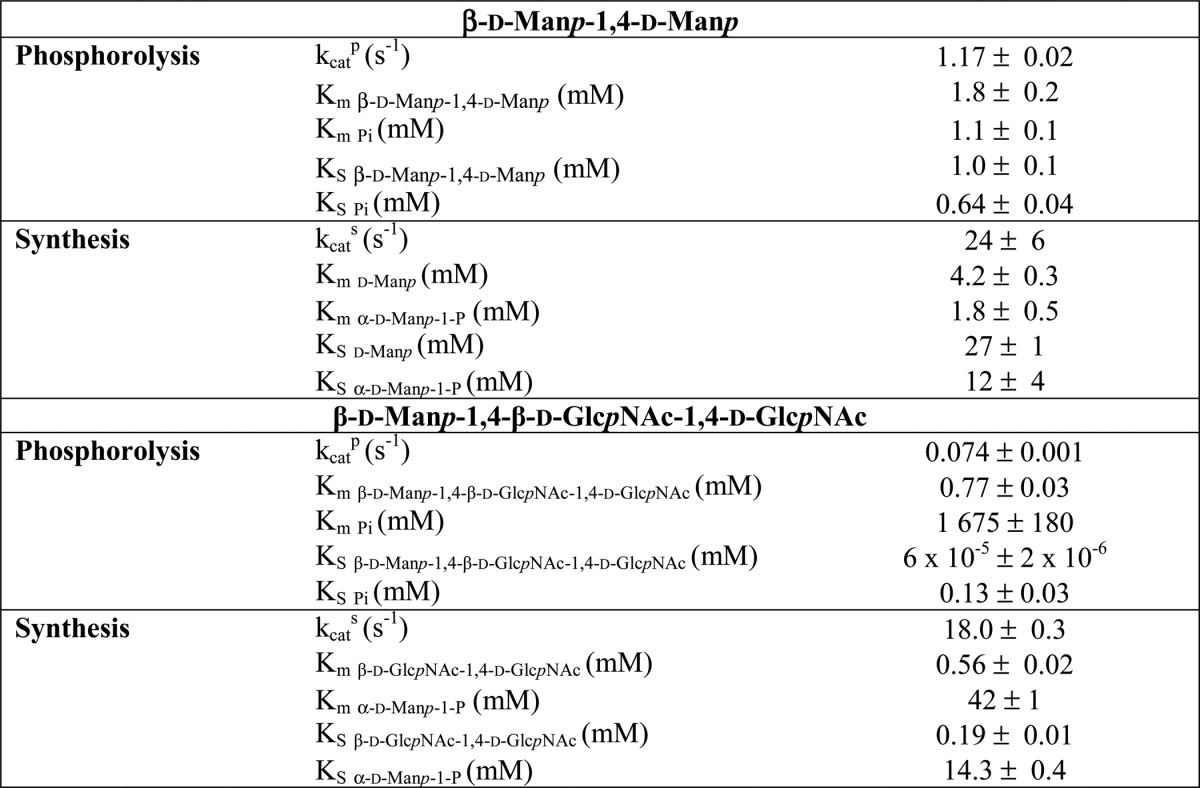
TABLE 1.
Apparent kinetic parameters for phosphorolysis and reverse phosphorolysis reactions catalyzed by UhgbMP, calculated from three to five independent experiments
Substrate concentrations were as follows: phosphorolysis, 10 mm inorganic phosphate and 1–20 mm β-d-mannopyranosyl-1,4-d-mannose; 1–10 mm β-d-mannopyranosyl-1,4-d-glucose, 0.4–4 mmβ-1,4-d-mannan, or 0.1–1mm pNP-β-d-mannopyranose, and 5 mm inorganic phosphate and 0.05–0.5 mm β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose; reverse phosphorolysis, 10 mm α-d-mannopyranose 1-phosphate and 5–40 mm d-glucose, d-mannose, d-galactose, d-fructose, or N-acetyl-d-glucosamine, and 5 mm α-d-mannopyranose 1-phosphate and 0.1–1 mm N,N′-diacetylchitobiose.
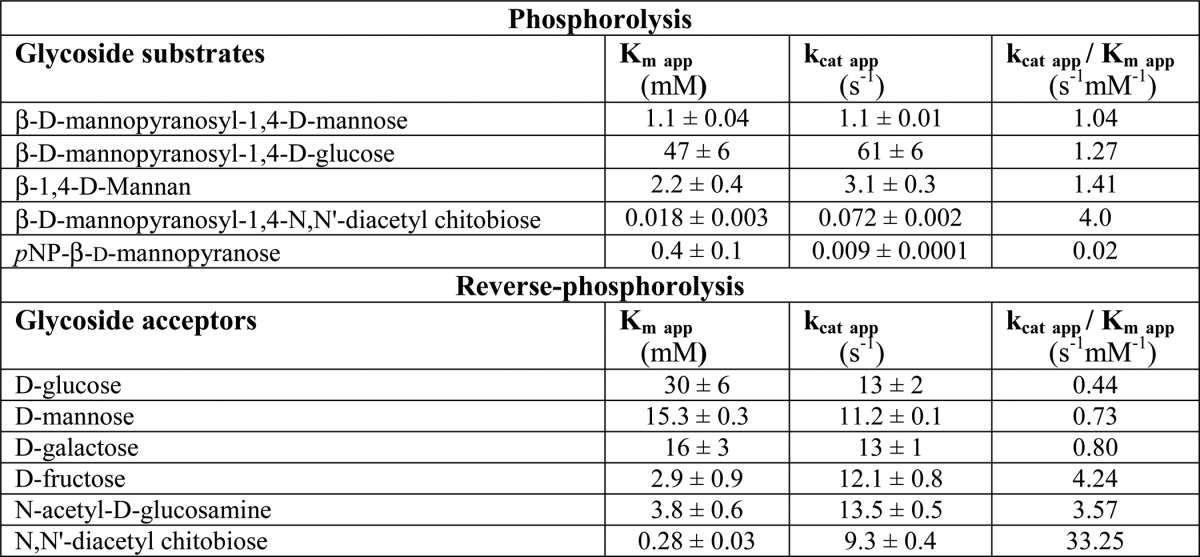
FIGURE 2.
Dependence of UhgbMP-specific activity with the DP of the phosphorolyzed β-1,4-d-manno-oligosaccharides.
Finally, no trace of oligosaccharide-phosphate was visible, at any reaction time, on HPAEC-PAD chromatograms of products obtained during phosphorolysis reactions, irrespective of the carbohydrate substrate. UhgbMP is thus an exo-acting enzyme, able to breakdown only the first β-mannosidic linkage at the nonreducing end of oligosaccharides.
α-d-Mannopyranose 1-phosphate was therefore tested as a glycosyl donor for reverse phosphorolysis. First, even without any carbohydrate acceptor, UhgbMP produces mannose and, furthermore, manno-oligosaccharides of DP ranging from 1 to 12, indicating that water itself plays the role of first acceptor at the beginning of the reaction. The β-1,4 regio-specific synthesis of manno-oligosaccharides was characterized by 1H and 13C NMR (supplemental Fig. 2). Various carbohydrates or polyols were tested as acceptors (Table 1). d-GlcpNAc and β-d-GlcpNAc-1,4-d-GlcpNAc were the best recognized acceptors, given that the UhgbMP Km values for these compounds are 4- and 55-fold lower than for d-mannose, respectively. Starting from α-d-mannopyranose 1-phosphate and d-GlcpNAc or β-d-GlcpNAc-1,4-d-GlcpNAc, UhgbMP generates series of manno-oligosaccharides containing d-GlcpNAc or β-d-GlcpNAc-1,4-d-GlcpNAc at their reducing end, with a DP of up to 4. d-Glucose, d-mannose, d-galactose, and d-fructose are also recognized as acceptors.
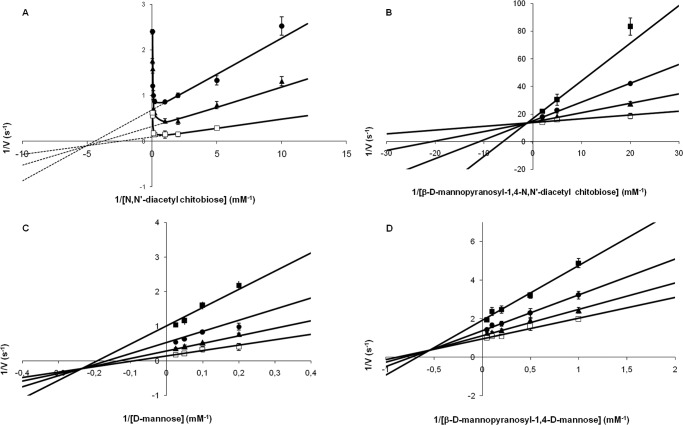
The kinetic mechanism of UhgbMP was investigated by determining the initial reaction velocities for β-d-Manp-1,4-d-Manp and β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc phosphorolysis and synthesis at various initial concentrations of substrates. First, double-reciprocal plots of 1/v versus 1/[β-d-Manp-1,4-d-Manp] at various concentrations of inorganic phosphate and of 1/v versus 1/[d-Manp] at various concentrations of α-d-mannopyranose 1-phosphate crossed at a certain point, on the left of the 1/v axis (Fig. 3). UhgbMP thus catalyzes the β-d-Manp-1,4-d-Manp phosphorolysis and synthesis through a sequential Bi Bi mechanism involving the formation of a ternary complex. The equilibrium constant was calculated from the Haldane relationship for a sequential random Bi Bi mechanism (39) (the most probable mechanism fitting with this kinetic dataset) as shown in Equation 2,
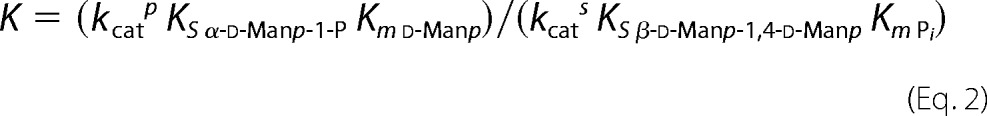 |
(kcats KS β-D-Manp1,4-D-Manp Km Pi), kcatp and kcats being kcat for phosphorolytic and synthetic reactions, respectively (Table 2). The value of 2.1 indicates that reaction equilibrium favors phosphorolysis. The kinetic parameters calculated for β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc phosphorolysis and synthesis are clearly different. β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc synthesis is inhibited by β-d-GlcpNAc-1,4-d-GlcpNAc concentrations higher than 1 mm (Fig. 3). For β-d-GlcpNAc-1,4-d-GlcpNAc and β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc concentrations lower than 1 and 0.5 mm, respectively, the most probable mechanism is also a mixed-type sequential random Bi Bi mechanism. The K value calculated in these conditions is 0.32, a value similar in magnitude to the value 0.658 obtained for d-Manp-1,4-β-d-Glcp phosphorolysis by RaMP2 (34). Further investigations of UhgbMP mechanism will be necessary to determine the order of substrate binding and product release and to better understand how this enzyme works on its natural substrates, including β-d-Manp-1,4-d-GlcpNAc.
FIGURE 3.
Double-reciprocal plots for β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose synthesis (A) and phosphorolysis (B) and for β-d-mannopyranosyl-1,4-d-mannose synthesis (C) and phosphorolysis (D). The initial velocities of α-d-mannose 1-phosphate consumption and release were measured at various concentrations of substrates. A and C, concentrations of α-d-mannose 1-phosphate were 0.5 mm (filled square), 1 mm (filled circle), 2 mm (filled triangle), and 5 mm (open square). B and D, concentrations of inorganic phosphate were 0.5 mm (filled square), 1 mm (filled circle), 2 mm (filled triangle) and 5 mm (open square). Data are the mean values ± S.D. for three to five independent experiments.
TABLE 2.
Kinetic parameters for the phosphorolysis and synthesis of β-d-mannopyranosyl-1,4-d-mannose and β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose
Substrate concentrations were as follows: β-d-mannopyranosyl-1,4-d-mannose phosphorolysis, 1–20 mm β-d-mannopyranosyl-1,4-d-mannose, and 0.5–5 mm inorganic phosphate; β-d-mannopyranosyl-1, 4-d-mannose synthesis, 5–40 mm d-mannose, and 0.5–5 mm α-d-mannopyranose 1-phosphate; β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose phosphorolysis, 0.05–0.5 mm β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose, and 0.5–5 mm inorganic phosphate; β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose synthesis, 0.1–1 mm N,N′-diacetylchitobiose, and 1–5 mm α-d-mannopyranose 1-phosphate.
Finally, with all other compounds tested as acceptors, UhgbMP synthesizes the same (Man)n series as when α-d-mannopyranose 1-phosphate was the sole substrate. This shows that none of the other tested molecules were good acceptors, because water and additional mannose and manno-oligosaccharide units were used preferentially by the enzyme. However, the presence of cellobiose, d-fucose, l-rhamnose, l- and d-xylose, N-acetyl-d-galactosamine (d-GalpNAc), d-altrose, d-allose, xylitol, d-lyxose, or d-mannitol decreased enzyme-specific activity, in some cases quite markedly (Table 3). But concentrations of these compounds did not decrease during reaction, and no significant additional product was produced compared with reaction in the presence of α-d-mannopyranose 1-phosphate as sole substrate. These carbohydrates thus act as UhgbMP inhibitors. Various sugar-phosphates were also tested as glycosyl donors for reverse phosphorolysis as follows: α-d-fructose-1- and -6-phosphate, d-ribose 1-phosphate, α-d-galactosamine 1-phosphate, α-d-glucosamine 1-phosphate, d-mannose 6-phosphate, and α-d-glucopyranosyl 1- and -6-phosphates. None of them was consumed by UhgbMP, and no reaction product appeared on HPAEC-PAD chromatograms after 24 h at 37 °C.
TABLE 3.
Percentage of UhgbMP inhibition by carbohydrates and polyols, quantified by measuring α-d-mannopyranose 1-phosphate consumption rate from 10 mm α-d-mannopyranose 1-phosphate as glycosyl donor, with and without 10 mm of carbohydrates or polyols
| Inhibitor | % of inhibition |
|---|---|
| 0 | |
| l-Arabinose | 2 ± 0.4 |
| d-Cellobiose | 37 ± 1 |
| d-Fucose | 12 ± 2 |
| l-Fucose | 11 ± 4 |
| l-Rhamnose | 31 ± 2 |
| Xylitol | 29.4 ± 0.4 |
| d-Lyxose | 19 ± 2 |
| l-Xylose | 58 ± 1 |
| d-Mannitol | 37 ± 4 |
| d-Altrose | 84 ± 3 |
| d-Xylose | 86.2 ± 0.2 |
| d-Allose | 94 ± 3 |
Key Residues Involved in Mannoside Phosphorolysis Mechanism
To investigate the UhgbMP catalytic mechanism, we first built a three-dimensional model of the enzyme using the atomic coordinates of the TM1225 protein from T. maritima MSB8 (PDB accession code 1VKD) as a structural template. The enzyme, which adopts a five-bladed β-propeller fold (Fig. 4A), was identified from structural genomics initiatives. Of the four proteins of known structure that share the same fold, TM1225 aligns with the highest sequence identity to UhgbMP (59%), but it has yet to be functionally characterized.
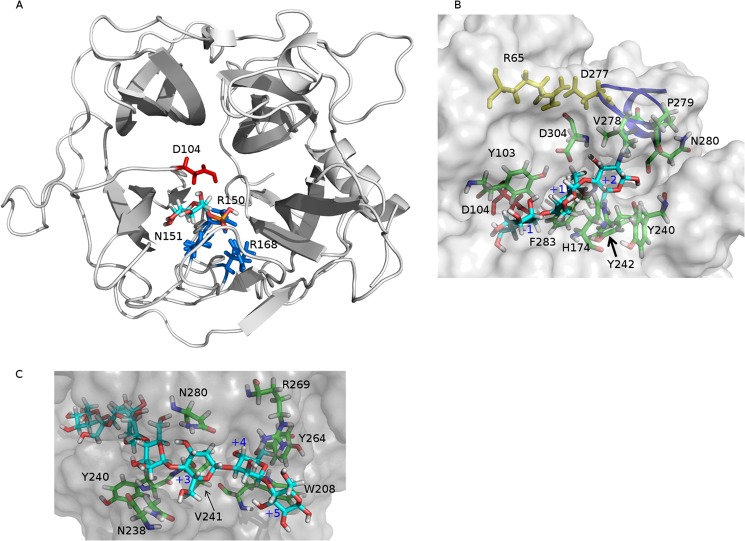
FIGURE 4.
A, α-d-mannopyranose 1-phosphate docked into the three-dimensional model of UhgbMP. The most probable proton donor, Asp-104, is colored in red. Also shown in blue are the three phosphate-stabilizing residues, namely Arg-150, Asn-151, and Arg-168. B, β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose (β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc) docked into the three-dimensional model of UhgbMP. Tyr-103, Asp-304, His-174, Tyr-240, and Phe-283 are residues involved in recognition of the glycosyl residue in the +1 subsite. Asp-304, Tyr-242, Pro-279, Asn-280, and Val-278 are residues involved in binding of the glycosyl residue in the +2 subsite. Yellow shows Arg-65 and Asp-277 residues that form a salt bridge locking the Cys-274–Val-281 loop, represented as a blue line. C, β-1,4-d-mannoheptaose docked into the three-dimensional model of UhgbMP. Tyr-240, Asn-238, and Val-241 are residues involved in the substrate accomodation at the +3 subsite. Asn-280 and Asn-238 are residues involved in oligosaccharide binding at the +4 subsite. Tyr-264, Trp-208, and Arg-269 are residues shaping the +5 subsite.
To identify putative catalytic amino acid residues, we analyzed the multiple alignment of the 369 protein sequences referenced in CAZy family GH130 (January 2013) (supplemental Fig. 3). The most conserved amino acid residue positions were then mapped onto the three-dimensional model of UhgbMP. Of these, Asp-104 and Asp-304 were found to be part of a narrow groove putatively considered to be the active site. Another conserved amino acid residue, Glu-273, present in a short α-helix turn section, was also identified in the groove.
These three amino acid residues were individually mutated to investigate their potential role in the UhgbMP catalytic mechanism. Results showed that mutation of Asp-104 into an asparagine completely abolished UhgbMP activity, although D304N and E273Q mutations retained only 3.9 ± 0.5 and 0.2 ± 0.1% of the native catalytic activity, respectively. These three amino acids were thus considered as candidate catalytic residues. To further elucidate the functional roles of these residues, molecular modeling techniques were used to dock α-d-mannopyranose 1-phosphate into the putative active site groove.
Of the identified docking modes, only one appeared compatible with spatial constraints provided by the UhgbMP reaction mechanism (Fig. 4A) as follows: (i) provision for the specific recognition of mannose at the −1 subsite; (ii) substrate interaction with at least one of the (Asp-104, Asp-304, or Glu-273) putative catalytic acidic residue side chains; (iii) presence of a favorable phosphate-binding site allowing reverse phosphorolysis to take place; (iv) presence of binding subsites able to accommodate β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc and β-1,4-d-mannan chains in catalytically productive binding modes with respect to active site residue(s) implicated in the chemical reaction mechanism (Fig. 4, B and C).
In this binding mode, where the α-d-mannopyranosyl 1-phosphate ring structure is stabilized in the −1 subsite through stacking interactions with Tyr-103, the Asp-104 residue located on the β-face of the catalytic chiral center can act as the unique proton donor during catalysis, whereas Arg-150, Arg-168, and Asn-151 amino acid residues can favorably assist phosphate group positioning consistent with inversion of configuration at C-1 of mannose (Fig. 4A). Given the high conservation of these amino acid residues within the GH130 family alignment, we suggest that enzymes contained in this family should share the same single displacement mechanism described for GH-like inverting phosphorylases. In such a mechanism, the aspartic acid corresponding to Asp-104 in UhgbMP would be the sole catalytic residue, having the role of proton donor (Fig. 5). The phosphate group, stabilized through ionic interactions with highly conserved Arg-150, Arg-168, and Asn-151 residues in UhgbMP, would then act as the nucleophile (Fig. 4A).
FIGURE 5.
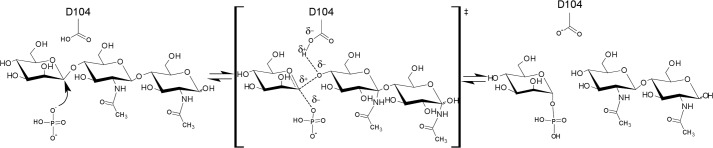
Proposed catalytic mechanism of β-d-mannopyranosyl-1,4-N,N′-diacetylchitobiose phosphorolysis by UhgbMP, with Asp-104 acting as the proton donor.
Molecular Basis of Substrate Specificity of GH130 Enzymes
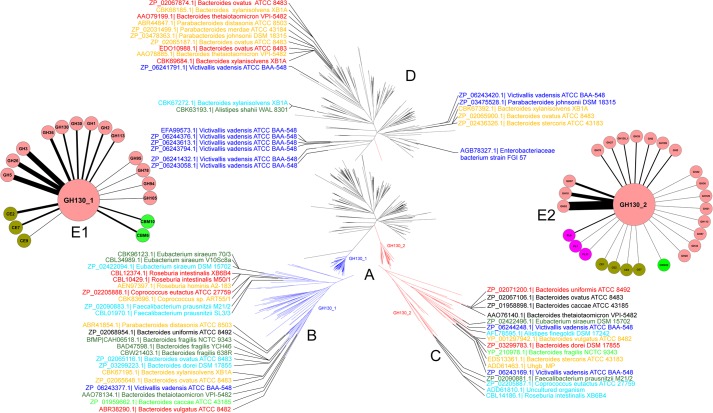
The biochemical data from the kinetic characterization of BfMP, RaMP1, RaMP2, BT1033, and UhgbMP revealed marked specificity differences among the five enzymes. The BfMP and RaMP1 + 1 subsite is highly specific for glucose, whereas RaMP2, BT1033, and UhgbMP display looser specificity both toward carbohydrate substrates for phosphorolysis and acceptors for synthetic reactions. We therefore investigated whether the five characterized proteins could represent different GH130 subfamilies. Indeed, in most of the cases, enzymes classified in a CAZy subfamily share the same substrate and/or product specificity, reflecting a high degree of conservation in their active site (46, 47). A phylogenetic tree was constructed, based on the multiple alignments of the GH130 enzyme sequences. Three clusters of sequences clearly appeared. BfMP and RaMP1 are contained in subfamily GH130_1 (79 sequences, Fig. 6 and supplemental Table 1). UhgbMP, RaMP2, and BT1033 are contained in subfamily GH130_2 (42 sequences), together with TM1225 (PDB accession code 1VKD). The GH130_NC cluster contains 248 sequences of as yet uncharacterized proteins that are too heterogeneous to permit the creation of univocal subfamilies. This cluster contains the other three proteins of known structure, namely the BDI_3141 protein from P. distasonis ATCC 8503, the BT_4094 protein from B. thetaiotamicron VPI-5482, and the BACOVA_03624 protein from B. ovatus ATCC 8483. However, a key residue position occupied by Tyr-103 in UhgbMP allows the discrimination of the GH130_NC sequences from those of the two other subfamilies. The tyrosine residue at position 103 in UhgbMP, which lies close to the phosphate group binding site in the three-dimensional model of the docked complex of the enzyme with α-d-mannopyranose 1-phosphate, is strictly conserved in subfamilies GH130_1 and _2 but is replaced by a glutamic acid in 234 of the 248 sequences of the GH130_NC group (supplemental Fig. 3). Moreover, residues Arg-150, Arg-168, and Asn-151 that could favorably assist phosphate group positioning in UhgbMP active site are not conserved in the GH_130 NC cluster, although they are perfectly conserved in the GH130_1 and GH130_2 sequences (supplemental Fig. 3). We therefore suspect that the majority of the enzymes classified as GH130_NC are hydrolases and not phosphorylases. The Glu residue corresponding to Tyr-103 in UhgbMP is expected to act as the second catalytic residue, taking on the role of base. To validate this hypothesis, we attempted to transform UhgbMP, which possesses low intrinsic hydrolase activity when assayed in the absence of inorganic phosphate, into a hydrolase, through the replacement of Tyr-Y103 by a glutamic acid. In the presence of 10 mm Pi, the pNP-β-d-mannopyranoside breakdown activity of the wild-type enzyme was 6.3 × 10−3 ± 3 × 10−4 μmol·min−1·mg−1, a value similar to that obtained for mutant Y103E (5.7 × 10−3 ± 1 × 10−4 μmol·min−1·mg−1). In contrast, without any phosphate, the hydrolytic activity of the wild-type was 1.4 × 10−3 ± 1 × 10−4 μmol·min−1·mg−1, although it was 3.8 × 10−3 ± 3 × 10−4 μmol·min−1·mg−1 for Y103E. Nevertheless, the Y103E mutant pNP release curve versus time reached a plateau after only 10 min reaction at 37 °C with and without phosphate, although less than 5% of substrate was consumed (supplemental Fig. 4). This phenomenon was also observed for the wild-type enzyme without phosphate. This indicates that inorganic phosphate is probably involved in maintaining the UhgbMP active site conformation and that the Y103E mutation may alter this conformation. Because the product yields are so weak without phosphate, it remains difficult to conclude that the Y103E mutant is really a hydrolase. Functional and structural investigations will thus be needed to confirm that the GH130_NC cluster contains mannoside hydrolases.
FIGURE 6.
A, phylogenetic tree of the GH130 family. In this radial phylogram, branches corresponding to subfamilies GH130_1 and GH130_2 are colored in blue and red, respectively. Branches in black correspond to proteins that are not classified into any subfamily (GH130_NC). B–D, detailed views of the phylogentic tree of the GH130_1 and GH130_2 subfamilies and of not assigned sequences (GH130_NC). Only sequences corresponding to proteins from human gut bacteria are shown. Label color is relative to their prevalence in the human gut metagenome of 301 different individuals as follows. Blue, sequences found in the metagenome of 0–5 individuals; light blue, sequences found in the metagenome of 24–40 individuals; green, sequences found in the metagenome of 44–59 individuals; light green, sequences found in the metagenome of 62–76 individuals; orange, sequences found in the metagenome of 83–118 individuals; red, sequences found in the metagenome of 121–153 individuals; and black, sequences found in the metagenome of 173–197 individuals. E, cytoscape representations of CAZy encoding genes surrounding GH130_1 (E1) and GH130_2 (E2) encoding ones. This representation summarizes the number of co-presence of 65 known GH130 encoding genes and others CAZy ones in operon-like multigenic clusters of human gut bacteria genomes. Each gene encoding for a CAZy family found in a cluster containing a GH130 encoding gene is represented by a node. The width of the edges connecting the nodes is proportional to the number of times the co-presence of the two CAZymes encoding genes is observed in all the genomes analyzed.
Our attempts to check correct folding of the Y103E variant, as well as that of the wild-type enzyme and of variants D104N, D304N, and E273Q by circular dichroïsm using a J-815 UV Spectrum spectropolarimeter (Jasco) failed, because of the high absorbance of Tween 80 at wavelengths between 200 and 290 nm (48).
To study the active site conservation in family GH130, we projected the results of the multiple alignments of the GH130 protein sequences onto the UhgbMP three-dimensional model, in which β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc or β-1,4-linked d-mannoheptaose (Fig. 7) was docked to enable the mapping of putative binding carbohydrate subsites. Clearly, residues lining the UhgbMP catalytic furrow are highly conserved only among the GH130_2 sequences. The +1 putative subsite appears delimited by Tyr-103, Asp-304, His-174, Tyr-240, and Phe-283 residues that are specifically conserved in the GH130_2 family (Fig. 4B and supplemental Fig. 5). In the presence of β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc, Tyr-103 and Asp-304 residues can form H-bond interactions with the N-acetyl-d-glucosamine positioned in the +1 subsite. In the presence of β-1,4-linked d-mannoheptaose, the Tyr-103 can interact with the mannosyl residue located in the +1 subsite, whereas Asp-304 establishes a hydrogen bond with the mannosyl moiety at +2 subsite. Asp-304 thus probably plays a key role in substrate binding at +1 and +2 subsites, explaining why its mutation dramatically alters UhgbMP activity.
FIGURE 7.
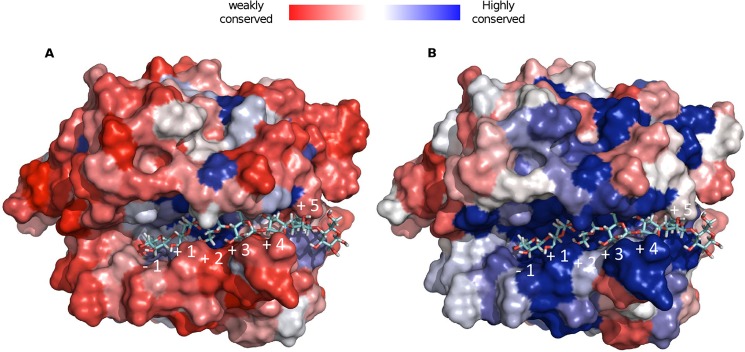
Structural conservation of GH130 (A) and GH130_2 (B) enzymes, mapped onto the UhgbMP three-dimensional model, in which β-1,4-d-mannoheptaose has been docked.
The Tyr-242 and Phe-283 residues provide additional staking platforms at +1 subsite to stabilize the bound glycosyl unit. At the putative +2 subsite, the N-acetyl-d-glucosamine residue at the reducing end of the β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc is found stabilized through hydrogen bonding interactions with Tyr-242, Pro-279, Asn-280, and Asp-304, while establishing van der Waals interactions with Val-278. Interestingly, in our three-dimensional model of the active site, the N-acetyl groups of the d-GlcpNAc moieties docked at +1 and +2 subsites are found to fit nicely into a pocket formed by the loop Pro-271–Pro-284, which is locked in a “closed conformation” via a salt bridge ionic interaction between Asp-277 and Arg-65 residues (Fig. 4B and supplemental Fig. 5). The conformation of this loop may be modified by the E273Q mutation, thus altering oligosaccharide accommodation in the active site. Sequence analysis indicates that such a loop is very well conserved within the subfamily GH130_2 but less within GH130_1 (supplemental Fig. 3). This could thus suggest an involvement of this loop in the specificity determination of GH130_2 enzymes toward manno-oligosaccharides longer than DP2.
The +3 to +6 putative subsites have been mapped using the docked mannoheptosaccharide (Figs. 4C and 7 and supplemental Fig. 5). Surprisingly, very few aromatic residues able to provide stacking interactions with the manno-oligosaccharide chain have been found. However, a dense network of likely hydrogen bonding interactions and van der Waals contacts could explain the increase of UhgbMP catalytic efficiency with the polymerization degree of manno-oligosaccharides. In the +3 putative subsite, the mannosyl residue can be stabilized by interactions with Tyr-240, Asn-238, and Val-241 residues, although at the putative +4 subsite, the sugar moiety is rather engaged in interactions with Asn-280 and Asn-238 residues. Residues Tyr-264, Trp-208, and Arg-269 are seen to establish interactions with the mannosyl moiety located at subsite +5. The length of the funnel binding site is found to accommodate up to +5 subsites. Outside this funnel, the +6 subsite has been putatively defined as sitting on the aromatic Trp-208 residue, which is almost perfectly conserved among the GH130_2 subfamily and not in GH130_1.
Finally, two important differences between GH130_1 and GH130_2 subfamilies, identified by sequence alignment analysis, could explain why GH130_1 enzymes seem to exhibit a very narrow specificity toward β-d-Manp-1,4-d-Glcp, as observed for RaMP1 and BfMP, although GH130_2 enzymes would be able to act on longer manno-oligosaccharides. First, the UhgbMP loop Gly-121–Gly-125, which defines the extremity of the −1 subsite, is very well conserved within the GH130_2 subfamily, whereas an insertion of 12 residues is observed in all GH130_1 sequences (supplemental Fig. 3). This longer loop could thus prevent the accommodation of long oligosaccharides in the negative subsite of GH130_1 enzymes. Second, we observed that the His-174 residue, which is not conserved in the GH130_1 subfamily and is contained in the UhgbMP Pro-169–Asp-179 loop, interacts with the sugar moiety bound at the +1 subsite. Interestingly, a motif containing five glycine residues was rather found within the GH130_1 subfamily in place of His-174 observed in GH130_2 enzymes that could play a major role in substrate accommodation within the active site (supplemental Fig. 3).
Genomic Context of GH130 Encoding Genes, Focus on Human Gut Bacteria
Differences in substrate specificity of enzymes belonging to the GH130_1 and GH130_2 subfamilies may illustrate different roles in the microbial ecosystem in which they are produced. 23 of the 79 GH130_1, 17 of the 42 GH130_2, and 25 of the 248 GH130_NC enzymes belong to human gut bacteria. To assess how globally prevalent these GH130 encoding genes are in gut microbiomes, we compared the 369 GH130 sequences with the human fecal metagenome sequences currently available, sampled from 162 individuals of the MetaHit cohort (3, 49) and 139 individuals of the NIH Human Microbiome Project (50). No less than 15 GH130_1, 10 GH130_2, and 14 GH130_NC sequences were detected in the fecal metagenome of at least 50 of the 301 individuals (Fig. 6). With the exception of four (one each from pig and chicken gut bacteria and two from cow rumen bacteria), these sequences all belong to human gut bacteria. The UhgbMP sequence was detected in the metagenomes of 93 of the 301 considered individuals. This indicates that it is not a rare gene and that it probably plays a critical role in mannose foraging in the gut. The highest occurrence values (sequences found in 173 to 197 individuals) were found for the GH130_1 sequence of Bacteroides uniformis ATCC 8492 and the GH130_2 sequences of B. ovatus ATCC 8483, Bacteroides caccae ATCC 43185, and B. thetaiotamicron VPI-5482 (BT1033 sequence) (GenBankTM accession numbers ZP_02068954.1, ZP_02067106.1, ZP_01958898.1, and AAO76140.1, respectively), which can be considered as common genes according to the Qin et al. definition (3), as they were found in more than 50% of the individuals. Interestingly, the mean number of GH130 BLAST hits per Mbp of sequence, obtained against the metagenomes of the 27 IBD patients (suffering either with Crohn disease or ulcerative colitis) of the MetaHit cohort, was 12, 28, and 58% higher than that obtained from the data sampled from the 135 other individuals (healthy or obese individuals), for GH130_1, GH130_2, and GH130_NC respectively (supplemental Table 2). This indicates a higher prevalence of GH130 encoding genes, and particularly of GH130_2 and GH130_NC, in the gut microbiome of IBD patients. This prevalence difference is thin, but it could be significant, considering that the gut microbiome of IBD patients harbors, on average, 25% fewer genes than that of healthy individuals (3).
To explore this question in more depth, we extracted from the GOLD database the 28 public genomes of human gut bacteria presenting sequences contained in the GH130_1, GH130_2, and GH130_NC subfamilies. We analyzed the genomic context of these 65 GH130 encoding genes, with a particular focus on the CAZy-encoding genes surrounding them. For each of these genomes, we highlighted operon-like multigenic clusters encoding CAZymes that act synergistically to break down complex carbohydrate structures, such as those contained in hemicellulose and human N-glycans. Interestingly, the 23 GH130_1 sequences are mainly surrounded by sequences of GH3, GH5, and GH26 (Fig. 6). These families contain enzymes that target hemicellulose, in particular β-mannanases and β-mannosidases. In contrast, the 17 GH130_2 sequences are mainly flanked by GH18 and GH92 and, to a lesser extent, GH97 sequences (Fig. 6 and supplemental Fig. 6). GH18 and GH92 contain the endo-β-N-acetylglucosaminidases and α1,2-, α1,3-, or α1,6-mannosidases that are implicated in the hydrolysis of human N-glycans (16, 17, 19, 20), although the only three characterized GH97 enzymes present α-glucosidase (EC 3.2.1.20) or α-galactosidase (EC 3.2.1.22) activity. Finally, only two multigenic clusters were found to encode both a GH130_2 enzyme and a GH2 enzyme, the GH2 family containing β-mannosidases capable of hydrolyzing the disaccharide β-d-Manp-1,4-d-GlcNAc (17).
The multigenic cluster containing UhgbMP encoding gene and that from B. stercoris ATCC 43183 containing its homolog BACSTE_03540 (accession number EDS13361.1) are clear examples of this gene organization with several GH92, GH18, and GH97 sequences surrounding a GH130_2 encoding gene (Fig. 1 and supplemental Fig. 6). In these cases, the results of in silico detection of signal peptide or transmembrane topology indicate that UhgbMP and BACSTE_03540 are probably intracellular, although the glycoside-hydrolases encoded by the same multigenic clusters would rather be secreted.
DISCUSSION
The biochemical characterization of UhgbMP, a representative of the GH130 enzyme family, revealed its flexibility toward carbohydrate substrates. UhgbMP is capable of catalyzing the phosphorolysis of β-d-mannopyranosyl-1,4-d-glucopyranose, β-1,4-d-manno-oligosaccharides of DP >5 and mannan, as well as the N-glycan core oligosaccharide β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc. Based on a sequence analysis of the GH130 family and on a structural model of UhgbMP, supported by the experimental findings presented here and elsewhere by Senoura et al. (32), Nihira et al. (33), and Kawahara et al. (34), we propose the creation of two GH130 subfamilies, of which the members probably share the same single displacement mechanism involving a single catalytic acidic residue (corresponding to UhgbMP D104) acting as the proton donor. Subfamily GH130_1 gathers together enzymes (including BfMP and RaMP1) exhibiting narrow specificity toward β-d-Manp-1,4-d-Glc. Conversely, enzymes of the GH130_2 subfamily show high promiscuity toward their substrates and products. Their active sites would be sufficiently extensible to accommodate complex carbohydrate structures such as β-d-mannan and β-d-manno-oligosaccharides, with or without β-d-GlcpNAc-1,4-d-GlcNAc, d-GlcpNAc, or d-glucose at their reducing end. The resolution of crystallographic structures of representatives of each of GH130 clusters in complex with substrates and products will, however, be necessary to confirm the role of the key residues identified here and to deepen our understanding of the reaction mechanisms operating in these enzymes.
Among the 369 enzymes archived in the GH130 family (January, 2013), 65 belong to human gut bacteria. The strong prevalence of the corresponding genes, in particular that coding for UhgbMP, in the human gut metagenome suggests that these enzymes have a major role for foraging mannose in this ecosystem. Here, we have demonstrated that UhgbMP, and probably also the 16 other enzymes in the proposed GH130_2 subfamily produced by gut bacteria (supplemental Table 1), including BT1033, are able to participate in breaking down the host mannosylated glycoproteins lining the intestinal epithelium.
In vivo, incorporation of GH reaction products into metabolic pathways is energy-consuming, although direct metabolism of the glycoside phosphates synthesized by GPs does not require ATP. Phosphorolysis reactions mediated by GPs would thus be more advantageous than hydrolysis under certain physiological conditions, for example during intensive use of carbohydrate resources or in anoxic environments such as the gastrointestinal tract, where ATP cannot be efficiently produced by the respiratory chain-linked phosphorylation process (51).
In common with other GPs, especially those previously identified in microorganisms having a facultatively anaerobic lifestyle like Bifidobacterium sp., Lactobacillus sp., or Clostridium sp (50), enzymes in the proposed GH130_1 and GH130_2 families can therefore be considered as catabolic enzymes, working in tandem with catabolic GHs in dietary and host mannoside breakdown.
Based on the analysis of the genomic context of GH130_2 encoding genes in gut bacteria, and accordingly with Nihira et al. (33), we propose that mannoside phosphorolysis catalyzed by GH130_2 enzymes acts in concert with GH activities of enzymes in the GH18 and GH92 families, to completely break down N-glycans, as proposed in Fig. 1. The role of the GH97s whose genes belong to the same multigenic systems as those coding for the proposed GH130_2 enzymes (for example, in B. stercoris ATCC 43183) is less clear. Family GH97 contains only three characterized enzymes, of which two are α-glucosidases with α-1,4-link specificities (52, 53). However, the functional diversity of GH97 enzymes has not yet been thoroughly explored, and if these enzymes were previously thought to contribute to dietary carbohydrates in the human gut, they may well also be active in the breakdown of N-glycans, for example by liberating, like the GH99 enzymes (18), the α-1,3-linked glucosyl residue at the nonreductive end of the immature N-glycans. In the particular case of the unknown gut bacterium producing UhgbMP and of B. stercoris ATCC 43183 producing its homolog BACSTE_03540, and based on the results of in silico detection of signal peptides, we assume that the UhgbMP and BACSTE_03540 physiological role would be the intracellular phosphorolysis of short oligosaccharides that can be internalized in the cell (di- or trisaccharides (54)), like the β-d-Manp-1,4-d-GlcpNAc disaccharide or the β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc trisaccharide resulting from extracellular hydrolysis of N-glycans by glycoside hydrolases belonging to the GH18, GH92, and maybe also GH97 families. This deglycosylation arsenal thus allows one to deprotect the glycoproteins of the intestinal epithelium. Their protein part could also thus be degraded by the endopeptidases, similarly to those putatively encoded by the genes ADD61465.1 and ADD61469.1, which belong to the same metagenomic DNA fragment than the UhgbMP encoding gene, so as to perforate the intestinal epithelium (Fig. 1). However, further studies will be needed to confirm the physiological role of these enzymes, like metabolomic and transcriptomic analyses of gut bacteria producing GH130_2 enzymes (like B. stercoris ATCC 43183) in the presence of N-glycans as carbon source.
The results presented here show that in vitro, UhgbMP is also able to effectively degrade manno-oligosaccharides and mannan itself. This is also probably the case for other gut bacterial enzymes of the proposed GH130_2 subfamily, which show a similar active site topology, allowing the accommodation of long oligosaccharides. The intestinal bacteria that produce GH130_2 enzymes would thus be able to use dietary hemicelluloses and their hydrolysis products as carbon sources if needed, which would give them a competitive advantage with respect to the other bacteria, to colonize and maintain themselves in the intestinal tract. The use of mannose-rich carbohydrates (plant or yeast mannans, linear β-1,4-linked manno-oligosaccharides, or β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc) as functional foods could thus allow the metabolism of the N-glycan-degrading bacteria to be bypassed and, eventually, may reduce the degradation of the epithelial barrier for therapeutic applications, especially for patients suffering from IBD. Indeed, in their gut metagenome, the known GH130_2 and GH130_NC encoding genes seem highly prevalent, even if enzyme assays on fecal samples, as well as metatranscriptomic and metaproteomic studies, should be performed as evidence that GH130 could be biomarkers of IBDs. However, this approach presents a risk of overfeeding this specific type of bacteria and increasing their prevalence in the gut, and thus the potential for N-glycan breakdown, in the case of reduced intake of exogenous mannose-rich carbohydrates. The utilization of GH130 enzyme inhibitors, like d-altrose, d-xylose, and d-allose for UhgbMP or suicide inhibitors that could be specifically designed thanks to the available or future structural data, would thus be the preferred strategy in therapeutic contexts.
Finally, we have shown here that the relaxed specificity displayed by UhgbMP toward acceptor substrates can be used for the stereo- and region-selective synthesis of original glyco-conjugates and oligosaccharides. In particular, this enzyme is able to synthesize β-1,4-linked d-manno-oligosaccharides, which have been reported to present prebiotic properties (55), even if their use as functional food may be risky, from our point of view. UhgbMP is also highly effective for production of N-glycan core oligosaccharides, such as β-d-Manp-1,4-d-GlcNAc and β-d-Manp-1,4-β-d-GlcpNAc-1,4-d-GlcpNAc, whose commercial price today exceeds $10,000 per mg. UhgbMP-based β-mannoside synthesis processes appear as highly attractive compared with those based on mannosyltransferases, which use expensive activated sugar nucleotides as donors (56, 57), or on transmannosylation catalyzed by native or engineered mannosidases or mannanases (44, 58–62). Indeed, a two-step UhgbMP-based process would allow one to use phosphate and β-mannan as substrates to first catalyze phosphorolysis and second to reverse phosphorolysis in the presence of hydroxylated acceptors to synthesize mannosylated products. UhgbMP is thus an enzymatic tool with high potential for synthesizing molecules like N-glycan core oligosaccharides, of major importance for studying, and potentially also for controlling interactions between host and gut microbes.
Acknowledgments
We cordially thank Nelly Monties for technical assistance. MetaToul (Metabolomics & Fluxomics Facitilies, Toulouse, France) is gratefully acknowledged for access to NMR facilities. MetaToul is supported by grants from the Région Midi-Pyrénées, the European Regional Development Fund, the SICOVAL, the Infrastructures en Biologie Sante et Agronomie (IBiSa, France), the CNRS, and the Institut National de la Recherche Agronomique.
This work was supported by the French Ministry of Higher Education and Research and by the French National Institute for Agricultural Research (INRA, CEPIA division and the “Meta-omics of Microbial Ecosystems” research program).

This article contains supplemental Figs. S1–S6 and Tables 1 and 2.
O. Rivoire, and R. Ranganathan, unpublished data.
- IBD
- inflammatory bowel disease
- GT
- glycosyltransferase
- GH
- glycoside hydrolase
- PL
- polysaccharide lyase
- GP
- glycoside phosphorylase
- DP
- degree of polymerization
- CE
- carbohydrate esterase
- BfMP
- B. fragilis NCTC 9343 mannosylglucose phosphorylase
- HPAEC-PAD
- high performance anion exchange chromatography with pulsed amperometric detection
- PDB
- Protein Data Bank
- pNP
- p-nitrophenyl.
REFERENCES
- 1. Dicksved J., Halfvarson J., Rosenquist M., Järnerot G., Tysk C., Apajalahti J., Engstrand L., Jansson J. K. (2008) Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2, 716–727 [DOI] [PubMed] [Google Scholar]
- 2. Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J. (2006) Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J. M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., MetaHIT Consortium (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erickson A. R., Cantarel B. L., Lamendella R., Darzi Y., Mongodin E. F., Pan C., Shah M., Halfvarson J., Tysk C., Henrissat B., Raes J., Verberkmoes N. C., Fraser C. M., Hettich R. L., Jansson J. K. (2012) Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS ONE 7, e49138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ottman N., Smidt H., de Vos W. M., Belzer C. (2012) The function of our microbiota: who is out there and what do they do? Front. Cell Infect. Microbiol. 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y.-Y., Keilbaugh S. A., Bewtra M., Knights D., Walters W. A., Knight R., Sinha R., Gilroy E., Gupta K., Baldassano R., Nessel L., Li H., Bushman F. D., Lewis J. D. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koropatkin N. M., Cameron E. A., Martens E. C. (2012) How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Englyst K. N., Englyst H. N. (2005) Carbohydrate bioavailability. Br. J. Nutr. 94, 1–11 [DOI] [PubMed] [Google Scholar]
- 9. Brett C. T., Waldron K. W. (1996) in Physiology and Biochemistry of Plant Cell Walls (Black M., Charlwood B. V., eds) p. 222–238, Chapman and Hall, London [Google Scholar]
- 10. Moran A. P., Gupta A., Joshi L. (2011) Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 60, 1412–1425 [DOI] [PubMed] [Google Scholar]
- 11. Molinari M. (2007) N-Glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 12. Sheng Y. H., Hasnain S. Z., Florin T. H., McGuckin M. A. (2012) Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 27, 28–38 [DOI] [PubMed] [Google Scholar]
- 13. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreira L. R., Filho E. X. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178 [DOI] [PubMed] [Google Scholar]
- 16. Zhu Y., Suits M. D., Thompson A. J., Chavan S., Dinev Z., Dumon C., Smith N., Moremen K. W., Xiang Y., Siriwardena A., Williams S. J., Gilbert H. J., Davies G. J. (2010) Mechanistic insights into a Ca2+-dependent family of α-mannosidases in a human gut symbiont. Nat. Chem. Biol. 6, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tailford L. E., Money V. A., Smith N. L., Dumon C., Davies G. J., Gilbert H. J. (2007) Mannose foraging by Bacteroides thetaiotaomicron structure and specificity of the β-mannosidase, BtMan2A. J. Biol. Chem. 282, 11291–11299 [DOI] [PubMed] [Google Scholar]
- 18. Thompson A. J., Williams R. J., Hakki Z., Alonzi D. S., Wennekes T., Gloster T. M., Songsrirote K., Thomas-Oates J. E., Wrodnigg T. M., Spreitz J., Stütz A. E., Butters T. D., Williams S. J., Davies G. J. (2012) Structural and mechanistic insight into N-glycan processing by endo-α-mannosidase. Proc. Natl. Acad. Sci. U.S.A. 109, 781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I. (2003) A genomic view of the human Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 [DOI] [PubMed] [Google Scholar]
- 20. Martens E. C., Chiang H. C., Gordon J. I. (2008) Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deleted in proof [Google Scholar]
- 22.Deleted in proof [Google Scholar]
- 23. Nakai H., Kitaoka M., Svensson B., Ohtsubo K. (2013) Recent development of phosphorylases possessing large potential for oligosaccharide synthesis. Curr. Opin. Chem. Biol. 17, 1–9 [DOI] [PubMed] [Google Scholar]
- 24. Desmet T., Soetaert W. (2012) Broadening the synthetic potential of disaccharide phosphorylases through enzyme engineering. Process Biochem. 47, 11–17 [Google Scholar]
- 25. Han S.-E., Kwon H.-B., Lee S.-B., Yi B.-Y., Murayama I., Kitamoto Y., Byun M. O. (2003) Cloning and characterization of a gene encoding trehalose phosphorylase (TP) from Pleurotus sajor-caju. Protein Expr. Purif. 30, 194–202 [DOI] [PubMed] [Google Scholar]
- 26. Van der Borght J., Chen C., Hoflack L., Van Renterghem L., Desmet T., Soetaert W. (2011) Enzymatic properties and substrate specificity of the trehalose phosphorylase from Caldanaerobacter subterraneus. Appl. Environ. Microbiol. 77, 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rathore R. S., Garg N., Garg S., Kumar A. (2009) Starch phosphorylase: role in starch metabolism and biotechnological applications. Crit. Rev. Biotechnol. 29, 214–224 [DOI] [PubMed] [Google Scholar]
- 28. Aerts D., Verhaeghe T. F., Roman B. I., Stevens C. V., Desmet T., Soetaert W. (2011) Transglucosylation potential of six sucrose phosphorylases toward different classes of acceptors. Carbohydr. Res. 346, 1860–1867 [DOI] [PubMed] [Google Scholar]
- 29. Nakai H., Hachem M. A., Petersen B. O., Westphal Y., Mannerstedt K., Baumann M. J., Dilokpimol A., Schols H. A., Duus J. Ø., Svensson B. (2010) Efficient chemoenzymatic oligosaccharide synthesis by reverse phosphorolysis using cellobiose phosphorylase and cellodextrin phosphorylase from Clostridium thermocellum. Biochimie 92, 1818–1826 [DOI] [PubMed] [Google Scholar]
- 30. Nakajima M., Kitaoka M. (2008) Identification of lacto-N-biose I phosphorylase from Vibrio vulnificus CMCP6. Appl. Environ. Microbiol. 74, 6333–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hidaka M., Honda Y., Kitaoka M., Nirasawa S., Hayashi K., Wakagi T., Shoun H., Fushinobu S. (2004) Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyltransferase family 36, has a clan GH-L-like (α/α)6 barrel fold. Structure 12, 937–947 [DOI] [PubMed] [Google Scholar]
- 32. Senoura T., Ito S., Taguchi H., Higa M., Hamada S., Matsui H., Ozawa T., Jin S., Watanabe J., Wasaki J., Ito S. (2011) New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem. Biophys. Res. Commun. 408, 701–706 [DOI] [PubMed] [Google Scholar]
- 33. Nihira T., Suzuki E., Kitaoka M., Nishimoto M., Ohtsubo K., Nakai H. (2013) Discovery of β-1,4-d-mannosyl-N-acetyl-d-glucosamine phosphorylase involving in the metabolism of N-glycans. J. Biol. Chem. 288, 27366–27374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawahara R., Saburi W., Odaka R., Taguchi H., Ito S., Mori H., Matsui H. (2012) Metabolic mechanism of mannan in a ruminal bacterium, Ruminococcus albus, involving two mannoside phosphorylases and cellobiose 2-epimerase discovery of a new carbohydrate phosphorylase, β-1,4-manno-oligosaccharide phosphorylase. J. Biol. Chem. 287, 42389–42399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berman H., Henrick K., Nakamura H., Markley J. L. (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35, D301–D303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tasse L., Bercovici J., Pizzut-Serin S., Robe P., Tap J., Klopp C., Cantarel B. L., Coutinho P. M., Henrissat B., Leclerc M., Doré J., Monsan P., Remaud-Simeon M., Potocki-Veronese G. (2010) Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 20, 1605–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 38. Motomura K., Hirota R., Ohnaka N., Okada M., Ikeda T., Morohoshi T., Ohtake H., Kuroda A. (2011) Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol. Lett. 320, 25–32 [DOI] [PubMed] [Google Scholar]
- 39. Segel, Irwin H. (1975) Enzyme Kinetics–Behaviour and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 274–320, John Wiley & Sons, Inc., New York [Google Scholar]
- 40. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huson D. H., Scornavacca C. (2012) Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 43. Süel G. M., Lockless S. W., Wall M. A., Ranganathan R. (2002) Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Mol. Biol. 10, 59–69 [DOI] [PubMed] [Google Scholar]
- 44. Dilokpimol A., Nakai H., Gotfredsen C. H., Baumann M. J., Nakai N., Abou Hachem M., Svensson B. (2011) Recombinant production and characterisation of two related GH5 endo-β-1,4-mannanases from Aspergillus nidulans FGSC A4 showing distinctly different transglycosylation capacity. Biochim. Biophys. Acta 1814, 1720–1729 [DOI] [PubMed] [Google Scholar]
- 45. Dey P. M. (1978) in Advances in Carbohydrate Chemistry and Biochemistry (Tipson R. S., Horton D., eds) pp. 341–376, Elsevier, New York [Google Scholar]
- 46. Stam M. R., Danchin E. G., Rancurel C., Coutinho P. M., Henrissat B. (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: toward improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 19, 555–562 [DOI] [PubMed] [Google Scholar]
- 47. Aspeborg H., Coutinho P. M., Wang Y., Brumer H., 3rd, Henrissat B. (2012) Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu L., Zhang N., Yang G., Zhang J. (2011) Effects of Tween-80 on the dissolution properties of daidzein solid dispersion in vitro. Int. J. Chem. 3, 68–73 [Google Scholar]
- 49. Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., Fernandes G. R., Tap J., Bruls T., Batto J. M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H. B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E. G., Wang J., Guarner F., Pedersen O., de Vos W. M., Brunak S., Doré J., MetaHIT Consortium, Antolín M., Artiguenave F., Blottiere H. M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., (2011) Enterotypes of the human gut microbiome. Nature 473, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. NIH HMP Working Group, Peterson J., Garges S., Giovanni M., McInnes P., Wang L., Schloss J. A., Bonazzi V., McEwen J. E., Wetterstrand K. A., Deal C., Baker C. C., Di Francesco V., Howcroft T. K., Karp R. W., Lunsford R. D., Wellington C. R., Belachew T., Wright M., Giblin C., David H., Mills M., Salomon R., Mullins C., Akolkar B., Begg L., Davis C., Grandison L., Humble M., Khalsa J., Little A. R., Peavy H., Pontzer C., Portnoy M., Sayre M. H., Starke-Reed P., Zakhari S., Read J., Watson B., Guyer M. (2009) The NIH Human Microbiome Project. Genome Res. 19, 2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luley-Goedl C., Nidetzky B. (2010) Carbohydrate synthesis by disaccharide phosphorylases: reactions, catalytic mechanisms and application in the glycosciences. Biotechnol. J. 5, 1324–1338 [DOI] [PubMed] [Google Scholar]
- 52. Hughes C. V., Malki G., Loo C. Y., Tanner A. C., Ganeshkumar N. (2003) Cloning and expression of α-d-glucosidase and N-acetyl-β-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus). Oral Microbiol. Immunol. 18, 309–312 [DOI] [PubMed] [Google Scholar]
- 53. Gloster T. M., Turkenburg J. P., Potts J. R., Henrissat B., Davies G. J. (2008) Divergence of catalytic mechanism within a glycosidase family provides insight into evolution of carbohydrate metabolism by human gut flora. Chem. Biol. 15, 1058–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heinken A., Sahoo S., Fleming R. M., Thiele I. (2013) Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 4, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Salinardi T. C., Rubin K. H., Black R. M., St-Onge M.-P. (2010) Coffee manno-oligosaccharides, consumed as part of a free-living, weight-maintaining diet, increase the proportional reduction in body volume in overweight men. J. Nutr. 140, 1943–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Revers L., Bill R. M., Wilson I. B., Watt G. M., Flitsch S. L. (1999) Development of recombinant, immobilised β-1,4-mannosyltransferase for use as an efficient tool in the chemoenzymatic synthesis of N-linked oligosaccharides. Biochim. Biophys. Acta 1428, 88–98 [DOI] [PubMed] [Google Scholar]
- 57. Zhao Y., Thorson J. S. (1999) Chemoenzymatic synthesis of the Salmonella group E1 core trisaccharide using a recombinant β-(1→4)-mannosyltransferase. Carbohydr. Res. 319, 184–191 [DOI] [PubMed] [Google Scholar]
- 58. Ishimizu T., Sasaki A., Okutani S., Maeda M., Yamagishi M., Hase S. (2004) Endo-β-mannosidase, a plant enzyme acting on N-glycan: purification, molecular cloning, and characterization. J. Biol. Chem. 279, 38555–38562 [DOI] [PubMed] [Google Scholar]
- 59. Usui T., Suzuki M., Sato T., Kawagishi H., Adachi K., Sano H. (1994) Enzymic synthesis of the trisaccharide core region of the carbohydrate chain of N-glycoprotein. Glycoconj. J. 11, 105–110 [DOI] [PubMed] [Google Scholar]
- 60. Murata T., Usui T. (1997) Preparation of oligosaccharide units library and its utilization. Biosci. Biotechnol. Biochem. 61, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 61. Sasaki A., Ishimizu T., Hase S. (2005) Substrate specificity and molecular cloning of the lily endo-β-mannosidase acting on N-glycan. J. Biochem. 137, 87–93 [DOI] [PubMed] [Google Scholar]
- 62. Eneyskaya E. V., Sundqvist G., Golubev A. M., Ibatullin F. M., Ivanen D. R., Shabalin K. A., Brumer H., Kulminskaya A. A. (2009) Transglycosylating and hydrolytic activities of the β-mannosidase from Trichoderma reesei. Biochimie 91, 632–638 [DOI] [PubMed] [Google Scholar]