Background: The GEFs Rabin8 and GRAB are activators of the vesicular trafficking regulator Rab8.
Results: The catalytic mechanism of Rabin8/GRAB in Rab8 has been elucidated in biophysical and structural detail.
Conclusion: Rabin8 and GRAB are catalytically moderately efficient enzymes and act by disturbing Mg2+ binding and Rab8-guanine base interactions.
Significance: Obtaining snapshots of the nucleotide exchange reaction is crucial to understanding the mechanism of Rab GEFs.
Keywords: Enzyme Mechanisms, Guanine Nucleotide Exchange Factor (GEF), Intracellular Trafficking, Rab Proteins, Small GTPases, Intracellular Trafficking
Abstract
Small G-proteins of the Ras superfamily control the temporal and spatial coordination of intracellular signaling networks by acting as molecular on/off switches. Guanine nucleotide exchange factors (GEFs) regulate the activation of these G-proteins through catalytic replacement of GDP by GTP. During nucleotide exchange, three distinct substrate·enzyme complexes occur: a ternary complex with GDP at the start of the reaction (G-protein·GEF·GDP), an intermediary nucleotide-free binary complex (G-protein·GEF), and a ternary GTP complex after productive G-protein activation (G-protein·GEF·GTP). Here, we show structural snapshots of the full nucleotide exchange reaction sequence together with the G-protein substrates and products using Rabin8/GRAB (GEF) and Rab8 (G-protein) as a model system. Together with a thorough enzymatic characterization, our data provide a detailed view into the mechanism of Rabin8/GRAB-mediated nucleotide exchange.
Introduction
One of the hallmarks of eukaryotic cells is the intracellular movement of vesicles that transport material and allow communication between cellular compartments. The spatial and temporal regulation of vesicular trafficking is achieved by proteins of the Rab subfamily of small GTPases (1, 2). Rab proteins are molecular switches and cycle between inactive GDP-bound and active GTP-bound states. When inactive, Rab proteins exist in the cytosol in complex with the GDP dissociation inhibitor but are localized to a distinct membrane when in the active state. To exert their function, Rab proteins need to be activated by a process requiring guanine nucleotide exchange factors (GEFs).6 These enzymes accelerate GDP release from and allow the binding of GTP to a Rab protein. Rab proteins can interact with effector proteins that preferentially bind the active GTP but not the GDP state. GTPase-activating proteins stimulate the very low intrinsic GTPase activity of Rab proteins and thus convert them back into the inactive form.
The Rab subfamily consists of ∼60 members in humans, and each family member has a specific intracellular localization (2). The correct activation of a certain Rab requires the action of a cognate Rab GEF at the proper location and at the appropriate time (3). Consequently, GEFs have evolved to have mechanisms that guarantee their correct membrane targeting as well as the specific recognition of one distinct Rab protein over structurally and sequentially similar family members.
The Rab protein Rab8 is involved in events such as the delivery of secretory vesicles to the plasma membrane and polarized membrane transport in epithelial cells (4, 5). Rab8 also regulates cell shape, and interaction with its GEF Rabin8 appears to be crucial to this function (6). Rabin8 and Rab8 appear to be important in cilium formation by acting in concert with the Bardet-Biedl syndrome complex (7–9). Rabin8 is a 460-amino acid protein that consists of several domains, only two of which are functionally characterized (5, 10, 11). Rabin8 contains a central Sec2 domain with GEF activity toward Rab8 (5). Amino acid sequence and structure comparison with the yeast homolog Sec2 predicts that the central domain of Rabin8 consists of a homodimeric parallel coiled coil. Rabin8 is thought to be recruited to its target location by active Rab11, which interacts with a C-terminal Rab11 effector domain (10). Another factor possessing a Sec2-like GEF domain is the 382-amino acid protein GRAB. Despite a high sequence homology of the Sec2 domain of GRAB to Rabin8, it has previously been reported that GRAB is a GEF for Rab3A rather than Rab8 (12). However, a recent study that analyzed the activity profiles of various Rab GEFs (with a focus on the DENN domain family, which is structurally unrelated to GRAB) indicated that GRAB has GEF activity toward Rab8 but not Rab3 (13).
The mode of action of GTPases involved in signal transduction or regulation includes activation of GDP release catalyzed by a specific GEF in almost all cases known. The basic mechanistic feature is the weakening of the otherwise very tight binding of GDP (Kd values in the nanomolar to picomolar range) by interaction with GEFs, which also bind with similarly high affinity to their cognate GTPases (14). The effect is both thermodynamic and kinetic, implying the formation of a ternary complex between GEF, GTPase, and GDP, with a dramatic reduction in the affinities of both GEF and GDP in the ternary complex in comparison with the respective binary complexes. Several GTPase-GEF interactions have been examined thoroughly at the kinetic level, and a large number of GTPase·GEF complexes have been characterized structurally.
In this work, we have examined the interaction of the Ras superfamily protein Rab8 with two structurally highly similar GEF molecules (Rabin8 and GRAB) by kinetic and structural methods. GRAB is a GEF for Rab8 with almost identical structural properties to Rabin8. The work presented led to the identification of several intermediates in the overall GDP/GTP exchange of Rab8 in the presence of Rabin8.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human Rab8a(1–184) and Rab8a(6–176) were expressed in Escherichia coli and purified as described previously (15). Expression and purification of Rab3 were performed as described (16). The protein-encoding sequences of full-length GRAB and the coiled-coil domains of Rabin8/GRAB were cloned into a modified pET19 vector containing an N-terminal His6 tag followed by a tobacco etch virus protease cleavage sequence (17). In the case of GRAB, a synthetic codon-optimized gene was used. GRAB and Rabin8 variants were expressed in E. coli BL21(DE3)RIL by induction with 0.5 mm isopropyl β-d-thiogalactopyranoside at 20 °C for 18 h and purified by nickel-nitrilotriacetic acid affinity chromatography. After removal of the His6 tag with tobacco etch virus protease, the GRAB and Rabin8 variants were further purified by nickel-nitrilotriacetic acid affinity chromatography followed by size exclusion chromatography. Preparative loading of Rab8a(6–176) with GppNHp was performed as described previously (18). To obtain a nucleotide-free Rab8a(1–184)·Rabin8(157–232) complex, Rab8·GDP was mixed with Rabin8 at 1:4 molar ratio in buffer containing 20 mm HEPES (pH 7.5), 50 mm NaCl, 150 mm (NH4)2SO4, 50 μm ZnCl2, and 3 mm DTT. Alkaline phosphatase was added to hydrolyze GDP, and the mixture was incubated for 10 h at 4 °C. GDP hydrolysis was monitored by reversed phase HPLC (19), and Rab8·Rabin8 was purified by size exclusion chromatography with buffer containing 25 mm HEPES (pH 7.5), 40 mm NaCl, and 5 mm DTT after GDP was completely hydrolyzed.
Crystallization and Structure Determination
All crystals in these studies were obtained by mixing 1 μl of protein and 1 μl of reservoir solution at 20 °C using the hanging drop approach. Crystals of Rab8a(6–176)·GDP were obtained by mixing the protein (20 mg/ml; buffer containing 25 mm HEPES (pH 7.5), 40 mm NaCl, 1 mm MgCl2, 10 μm GDP, and 5 mm β-mercaptoethanol) with a reservoir solution consisting of 16% (w/v) PEG 4000, 0.1 m CaAc2, and 0.1 m HEPES (pH 7.0). The crystal was protected with cryo solution containing 30% (w/v) PEG 4000, 0.1 m CaAc2, and 0.1 m HEPES (pH 7.0) before data collection. Rab8a(6–176)·GppNHp crystals (15 mg/ml; buffer containing 25 mm HEPES (pH 7.5), 40 mm NaCl, 1 mm MgCl2, 10 μm GppNHp, and 5 mm β-mercaptoethanol) were produced in 15% (w/v) PEG 8000, 7.5% (v/v) 2-methyl-2,4-pentanediol, and 0.1 m HEPES (pH 6.8). Nucleotide-free Rab8a(1–184)·Rabin8(157–232) (10 mg/ml)) was crystallized in 18% (w/v) PEG 3350, 0.1 m Li2SO4, and 0.1 m MES (pH 6.6). Before data collection, the complex crystal was protected with cryo solution containing 30% (w/v) PEG 3350, 0.1 m Li2SO4, and 0.1 m MES (pH 6.6). To produce nucleotide-bound forms of Rab8·Rabin8 complexes, the nucleotide-free Rab8·Rabin8 crystals were soaked with cryo solution containing 30% (w/v) PEG 3350, 0.1 m Li2SO4, and 0.1 m MES (pH 6.6) and the respective nucleotide GDP/GTP (1 mm) for 1 h at 4 °C. Rab8·GRAB (10 mg/ml) was crystallized in solution containing 1.6 m ammonium sulfate and 0.1 m sodium acetate (pH 5.3). The crystals were protected with cryo solution containing 20% glycerol in the reservoir solution before data collection. All diffraction data were collected at 100 K at beamline X10SA of the Swiss Light Source (Villigen, Switzerland). Data were processed with XDS (20). The structure was determined by molecular replacement with PHASER (21) of the CCP4 suite using Sec4 in the case of Rab8·GDP/GppNHp and Sec2·Sec4 in the case of Rab8·Rabin8/GRAB complexes as a search model. The model was then corrected by alternating rounds of refinement in REFMAC5 (22) and manual adjustment in Coot (23). The nucleotide was added in the final rounds of refinement. Full data collection and refinement statistics are summarized in supplemental Table S1.
GEF Activity Measurements
Fluorescence measurements were carried out at 25 °C in buffer containing 50 mm HEPES (pH 7.5), 50 mm NaCl, 5 mm MgCl2, and 5 mm dithioerythritol. Rab8a was loaded with the fluorescent GDP/GTP derivatives (N-methylanthraniloyl (mant)-GDP/mant-GppNHp) as described for Sec4 (24). The fluorescence of mant was excited at 365 nm and detected using a 420-nm cutoff filter in an Applied Photophysics stopped-flow apparatus.
RESULTS
Exchange Activities of the GEF Domains of Rabin8 and GRAB
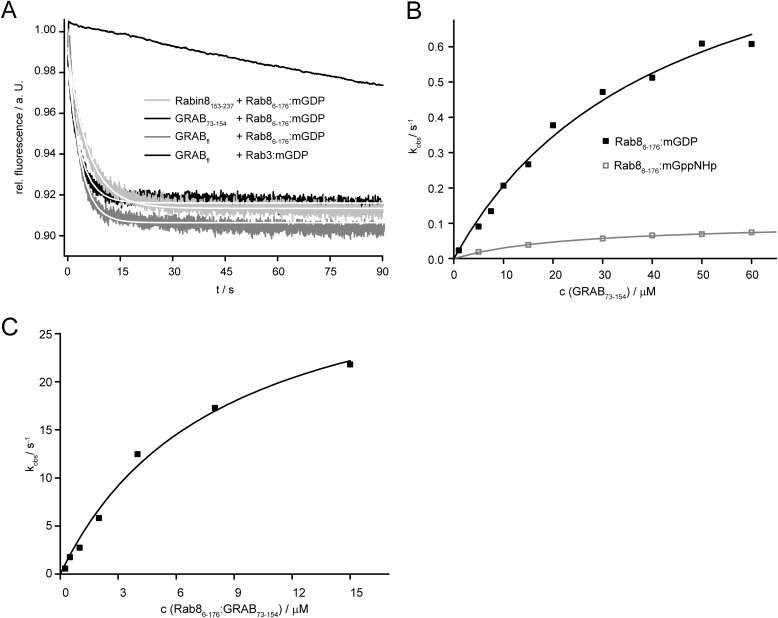
Using human Rab8(6–176), we investigated the GEF properties of Rabin8 and GRAB in detail. The displacement of a fluorescent GDP analog (mant-GDP) from its complex with Rab8 catalyzed by Rabin8(153–237) in the presence of excess GDP or GTP (see discussion below concerning the choice of this fragment) showed a marked acceleration with respect to the intrinsic dissociation rate (Fig. 1A). A similar effect was seen when Rabin8 was replaced by GRAB(73–154) or full-length GRAB. The difference between the observed rate constants of Rabin8(153–237) (kobs = 0.171 s−1) and GRAB(73–154) (kobs = 0.207 s−1) is ∼20% at 10 μm GEF. The small differences in the kobs values are not enough to exclude the possibility that they arise from errors in the determination of concentrations the GEF domains of GRAB and Rabin8. Thus, we concluded that the catalytic activities of the two GEFs are similar, and more detailed kinetic investigations were carried out with the GEF domain of GRAB. Interestingly, no acceleration of mant-GDP release from Rab3a could be observed even at an elevated GRAB concentration (10 μm) (Fig. 1A). We concluded from this that GRAB is actually a GEF for Rab8 rather than Rab3a, at least under in vitro conditions. A similar observation has been made recently by Yoshimura et al. (13).
FIGURE 1.
Kinetic analysis of the Rab8-GRAB interaction. A, comparison of full-length GRAB (GRABfl) GEF activity with the GRAB and Rabin8 GEF domains (10 μm) interacting with Rab8·mant-GDP (mGDP) in the presence of 100 μm GDP. Rab3·mant-GDP did not interact with full-length GRAB (10 μm). rel., relative; a. U., arbitrary units. B, dependence of the observed rate constant of mant-GDP/mant-GppNHp release from Rab8 on the concentration of GRAB. Plots of the observed rate constants for mant-GDP (black squares)/mant-GppNHp (gray squares) were hyperbolic. Fitting the hyperbolic curve leads to the values for KD3 and k″−4. C, dependence of the observed rate constants for the association reaction on the concentration of nucleotide-free Rab8·GRAB complex.
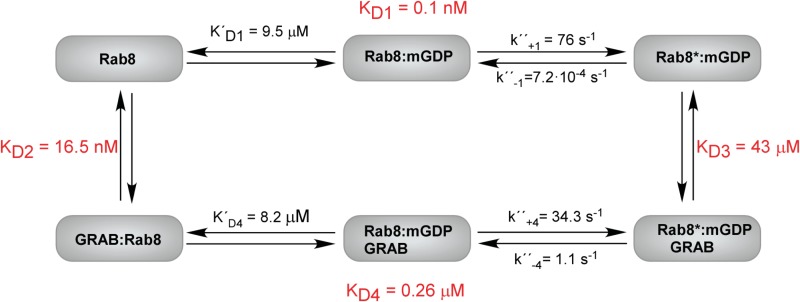
The observed rate constant of mant-GDP dissociation from Rab8·mant-GDP showed a hyperbolic dependence on the GRAB concentration (Fig. 1B), allowing the determination of KD3 and k″−4 (Scheme 1). KD3 represents the dissociation constant defining the interaction of GRAB with Rab8·mant-GDP (KD3 = 43 μm), whereas k″−4 is the maximal rate constant for mant-GDP dissociation (k″−4 = 1.1 s−1). Thus, the overall catalytic efficiency of GRAB toward Rab8 can be determined as kcat/Km = k″−4/KD3 = 2.6 104 m−1 s−1.
SCHEME 1.
Scheme of GRAB-Rab8-nucleotide interactions. Kinetic and equilibrium constants corresponding to Rab8-nucleotide interactions (KD1, K′D1, k″+1, and k″−1) have been reported previously (25). The value of KD2 has been calculated according to the relationship KD2 = (KD1 × KD3)/KD4 (14).
In addition, we analyzed the reverse reaction, i.e. the association of the nucleotide-free Rab8·GRAB complex with mant-nucleotides. The observed pseudo-first-order rate constants of the association of mantdeoxy-GDP with the Rab8·GRAB complex were fitted to a hyperbolic function, yielding K′D4 = 8.2 μm and k″+4 = 34.3 s−1 (Fig. 1C). Together with the constants already derived for the interaction of mant-GDP with Rab8 (25), these data led to a complete description of the interactions in the Rab8, GRAB, and mant-GDP system. The general trend of GEFs is also seen here: a marked reduction in GDP affinity in the ternary complex is caused predominantly by an increased rate constant for GDP release (k″−4 ≫ k−1). The kcat/Km value (k″−4/K′D3 in Scheme 1), giving a measure of the catalytic efficiency of the GEF for a specific substrate, is ∼2.6 × 104 m−1 s−1. This is similar to the value of 4.7 × 104 m−1 s−1 for the interaction of Ras with Cdc25 (26) but lower than the highly efficient Rab GEFs Sec2 (2 × 105 m−1 s−1 (24)) and DrrA from Legionella pneumophila (2.2 × 105 m−1 s−1 (27)). One interesting aspect of the constants derived is the relatively high affinity of GDP in the ternary complex (KD4 = 0.26 μm), suggesting the possibility of generating the ternary complex for structural studies (see below).
Structure of Rab8·Nucleotide Complexes
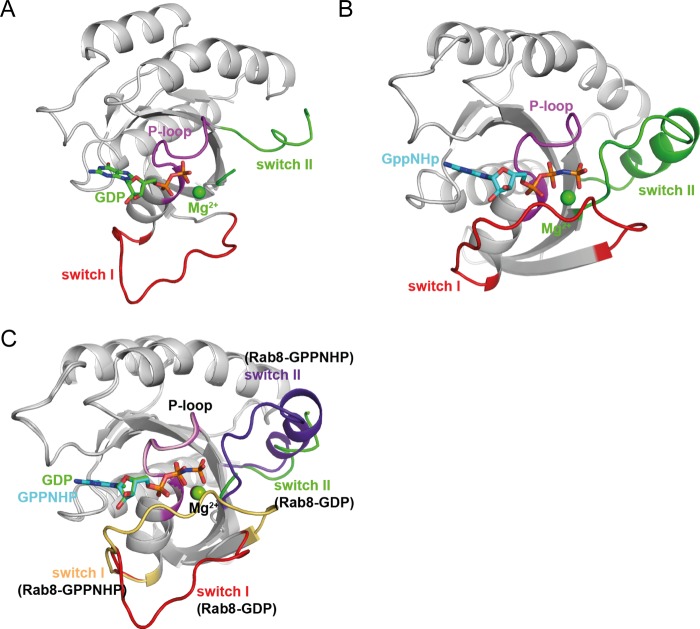
To obtain insight into the exchange mechanism of GRAB and Rabin8, we first determined the crystal structures of active and inactive Rab8 truncated at the N and C termini (amino acids 6–176, Rab8(6–176)) (Fig. 2, A and B). In both cases, there were five Rab8 molecules in the asymmetric unit (supplemental Fig. S1, A and B). The structures of the flexible regions, including switch I and particularly switch II in the GDP state, varied significantly between the five molecules (supplemental Fig. S1, C and D). As seen in many other instances of GTPases from the Ras superfamily, only switch I and switch II showed significant structural differences between active and inactive Rab8 (Fig. 2C). The switch I and switch II regions were largely disordered in the GDP state but ordered in the active GppNHp state. Details of the protein-nucleotide interactions are shown in (supplemental Fig. S1, E and F).
FIGURE 2.
Structures of Rab8 in its inactive GDP-bound and active GppNHp-bound forms. A, structure of Rab8·GDP. B, structure of Rab8·GppNHp. C, structural comparison of Rab8 in its inactive and active states. Superposition indicates that in its inactive GDP-bound form, the switch domains adopt unfolded conformations or ordered conformations that depend on crystal packing. Conversely, in its active GppNHp-bound form, the switch domains adopt well defined conformations.
Structure of the Rab8·Rabin8 Complex
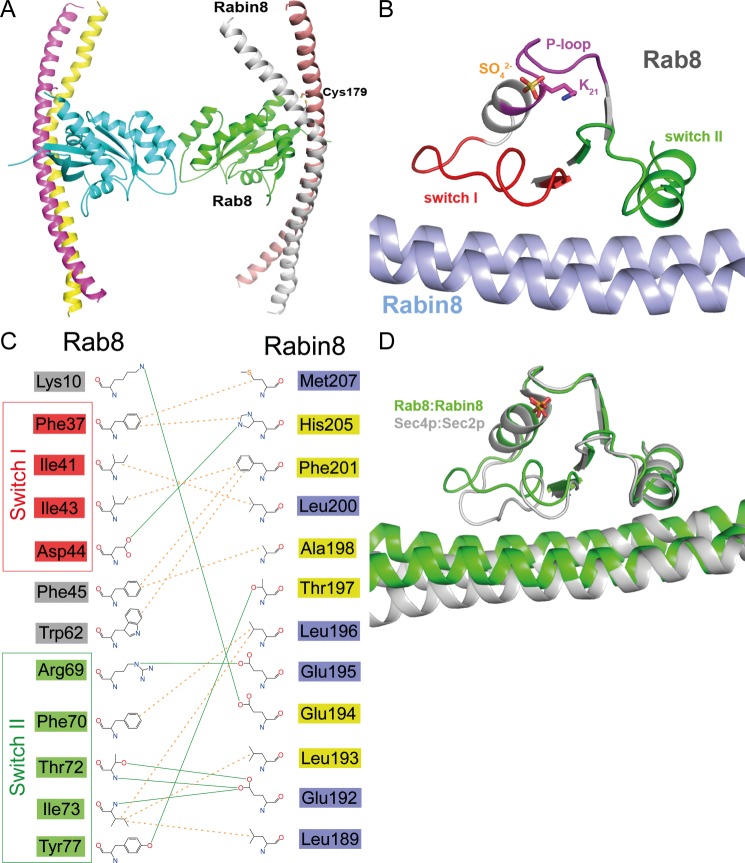
Next, we aimed to determine the structures of the Rab8·GRAB complex and its homolog Rab8·Rabin8. Both Rabin8 and GRAB contain predicted coiled-coil domains (residues 149–247 in Rabin8 and residues 70–160 in GRAB). These align well with highly similar regions in Sec2, which is a GEF for the yeast Rab8 homolog Sec4 (Fig. 3). The recently determined structure of the Sec2·Sec4 complex reveals that the interaction occurs in a central region of the Sec2 coiled coil (28). Several constructs of Rabin8 and GRAB were tested, and this led to the identification of Rabin8(157–232) and GRAB(79–149) as the minimal constructs with full GEF activity. Complexes with Rab8(6–176) were produced, and their crystal structures were determined. There were two complexes (corresponding to four molecules of Rabin8(157–232)/GRAB(79–149) and two molecules of Rab8) in the asymmetric unit (Fig. 4A), resulting in two identical complexes. As in the Sec2·Sec4 structure, the GEF region of Rabin8 adopts a parallel homodimeric structure with a length of ∼180 Å, and the two helices can be structurally distinguished from each other as sharply bent and moderately bent (28). There is an intermolecular disulfide bond between the Cys-179 residues in the individual Rabin8 chains. There is a presumed sulfate ion in Rab8 at the position normally occupied by the β-phosphate of GDP or GTP (Fig. 4B), as seen in many other structures of GTPases and ATPases in the absence of nucleotides (29, 30). Also, Rabin8 interacts mainly with the switch regions of Rab8 (Fig. 4B). An overview of the interactions between individual amino acids demonstrates that half of the Rab8-Rabin8 interactions are with one chain of the Rabin8 coiled coil, whereas the other half are with the other chain in the homodimeric structure (Fig. 4C). Rab8·Rabin8 shows a mode of binding reminiscent of the Sec2·Sec4 structure (Fig. 4D) (28). In line with this observation, Sec2 can also function as a GEF for Rab8 in vitro (data not shown).
FIGURE 3.
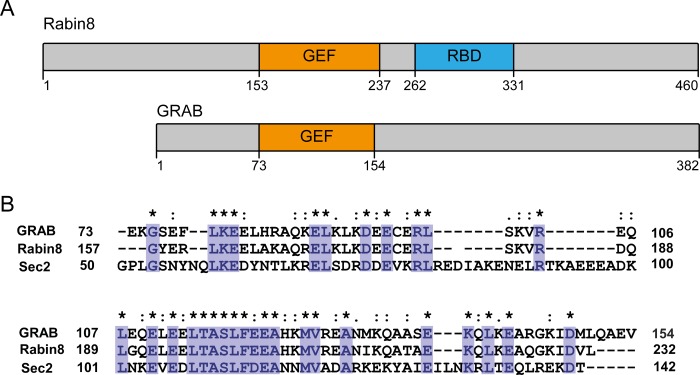
Schematic depiction of Rabin8 and GRAB. A, domain architecture of Rabin8 and GRAB. The GEF activity is confined to the Sec2 homology domain. Rabin8 contains a Rab11-binding domain (RBD) (10). B, sequence alignment of GRAB, Rabin8, and Sec2 in the GEF domain. Identical amino acids are shown in purple.
FIGURE 4.
Structure of the nucleotide-free Rab8·Rabin8 complex. A, overall structure of the nucleotide-free Rab8·Rabin8 complex. Remarkably, the Cys-179 residues of both Rabin8 chains form a disulfide bond, stabilizing the dimer. B, binding interface of the nucleotide-free Rab8·Rabin8 complex. Lys-21 interacts with a sulfate ion. The binding of Rab8 to Rabin8 is via the interaction between the switch I and switch II domains of Rab8 and residues 187–212 of Rabin8. C, schematic representation of the binding interface of Rab8·Rabin8. The residues from switch I and switch II of Rab8 are colored red and green, respectively. The residues of Rabin8 are colored yellow or blue depending on their chain ID. D, superposition of nucleotide-free Rab8·Rabin8 with the Sec2·Sec4 complex. There is no difference in switch II between these two complexes, whereas switch I binds to the GEF differently. This could be the reason for the higher GEF activity of Sec2 compared with Rabin8 and GRAB.
Structure of Rab8·Rabin8·Nucleotide Complex
Due to the relatively high affinity of the Rab8·Rabin8/GRAB complex for nucleotides (KD4 = 0.26 μm), we reasoned that a ternary GTPase·GEF·GDP/GTP complex could be amenable for protein crystallization and structure determination. We therefore soaked Rab8·Rabin8/GRAB crystals with GDP or GTP and determined their structures. The structures of Rab8(6–176)·Rabin8(157–232)·GDP and Rab8(6–176)·Rabin8(157–232)·GTP were determined at 3.1 and 3.2 Å resolution, respectively.
The overall structure of the Rab8(6–176)·Rabin8(157–232) complexes did not change significantly when GDP or GTP is bound (Fig. 5, A and B). The most important interactions with the nucleotide are between the base and Asp-124 and between the side chain of Lys-21 of the P-loop and the β-phosphate of GDP or the β- and γ-phosphates of GTP. Several backbone interactions of the P-loop with the phosphates are also observed. The dramatic loss of nucleotide affinity in the Rab8·Rabin8 complex in comparison with Rab8 results from the loss of GDP/GTP interactions with the switch regions and the interaction of the guanine base with Phe-33. The latter interaction is conserved in Ras superfamily proteins and was originally shown to contribute significantly to tight nucleotide binding in H-Ras (Phe-28 in Ras (31)). In addition, the Mg2+ ion essential for high affinity nucleotide binding is not present in either the GDP or GTP complex, contributing to the decrease in nucleotide affinity. These arguments also apply to the structure of the Rab8·GRAB complex because it is essentially identical to that of Rab8·Rabin8 (supplemental Fig. S2, A and B).
FIGURE 5.
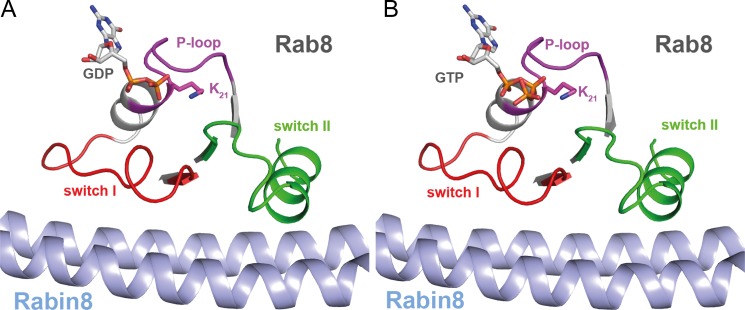
Structure of the nucleotide-bound Rab8·Rabin8 complex. A, structure of the GDP-bound Rab8·Rabin8 complex. B, structure of the GTP-bound Rab8·Rabin8 complex. Upon GDP or GTP binding, neither Rab8 nor Rabin8 changes its conformation compared with the nucleotide-free binary complex (Rab8·Rabin8).
The structural changes that occur upon interaction of Rab8 with Rabin8 are illustrated in Fig. 6 and (supplemental Fig. S1, E and F). Starting from a disordered state of switch II in Rab8·GDP, a new α-helix is generated by the interaction with Rabin8 in the GTPase·GEF complex. There is a large conformational change in switch I that has at least two specific effects on nucleotide binding: First, the side chain of Ile-38 of Rab8 moves into the site commonly occupied by Mg2+, thus leading to displacement of the metal ion. Second, the conserved interaction between Phe-33 of Rab8 and the guanine base is disrupted by Rabin8-mediated displacement of the amino acid side chain. The structure of the Rab8·Rabin8·GTP complex (Fig. 6B) is almost identical to that of the GDP complex, but a more extensive change in the conformation of switch I is required to generate the final conformation starting from the Rab8·GppNHp structure. In the latter, the switch I region closes over the nucleotide due to the interaction of the Thr-40 side chain hydroxyl group and the backbone NH group with the Mg2+ ion and the γ-phosphate, respectively. The change in switch II conformation is less profound than in the transition from Rab8·GDP to the Rab8·Rabin8 complex, but a movement of the N-terminal region of switch II away from the nucleotide is still apparent. No complex structure containing both nucleotide and Mg2+ could be obtained because the addition of nucleotides in the presence of Mg2+ ions led to rapid loss of diffraction power and destruction of the crystals.
FIGURE 6.
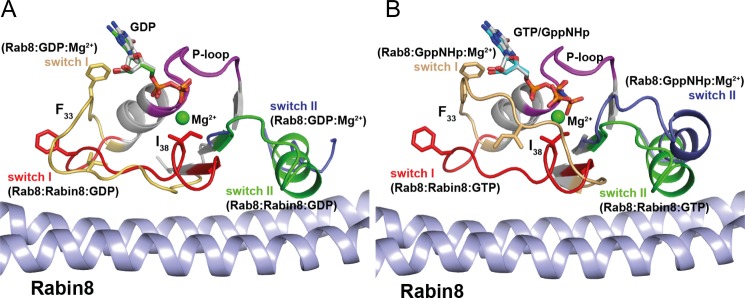
Conformational changes in Rab8·GDP (A) or Rab·GTP (B) upon binding of Rabin8. Superposition of Rab8·GDP/GppNHp and Rab8·Rabin8·GDP/GTP revealed that upon Rabin8 binding, the switch domains adopt highly ordered structures. The driving forces for nucleotide release are as follows. 1) The interaction of Rabin8 sterically occludes the magnesium-binding site by Ile-38 of switch I. The repositioned Ile-38 residue sterically interferes with Mg2+ binding. The release of the Mg2+ ion leads to a reduction in the affinity of the nucleotide for Rab8. 2) Upon Rabin8 binding, Phe-33 undergoes a conformational change. The repositioning of this conserved residue in switch I, which interacts with the guanine base, is part of the driving force in reducing the affinity for the nucleotide.
DISCUSSION
In this work, we have investigated the GEF reactions of Rabin8 and GRAB with their substrate Rab8 in biochemical and structural detail. Kinetic analysis of the GEF Sec2 domains of GRAB and Rabin8 revealed that they possess a moderately efficient catalytic activity. Unexpectedly, the affinity of the Rab·GEF complex for nucleotides is relatively high (KD4 = 0.26 μm for GDP), allowing the generation of ternary Rab8·Rabin8/GRAB·nucleotide complexes with GDP and GTP. The structures of these complexes allow us to describe the nucleotide exchange in detail at the molecular level.
The mechanism of GDP displacement in the Rab8/Rabin8 system can be described as a disturbance of the structure of the nucleotide-binding regions of Rab8, in particular switch I and switch II. In previously determined structures of GTPase·GEF complexes, the direct interaction of the GEF with switch II appears to be a universal feature. Contacts with switch I occur in many (but not all) complexes, whereas contacts with the P-loop are less common but are seen in some instances. In Rab8·Rabin8, interaction of the C-terminal part of switch II leads to the formation of an α-helix and displacement of Asp-63 from its position near to the nucleotide-binding site, where it is involved in an indirect (via a water molecule) interaction with Mg2+ in the Rab8·GDP and Rab8·GppNHp structures. This interaction is highly conserved in the Ras superfamily. In the absence of GEFs, the highly conserved Gly-66 residue interacts via its backbone NH with the γ-phosphate of GTP but not GDP. In the GDP displacement mechanism, the disturbance of the Mg2+ coordination presumably contributes to destabilization of the metal ion at this site. Further destabilization arises from structural changes induced in switch I, leading to displacement of Mg2+ by Ile-38. Similar effects are seen in other GTPase·GEF complexes, with the residue displacing Mg2+ coming either from the GTPase itself (32, 33) or from the GEF (34, 35). As determined in earlier work on Rab7, removal of Mg2+ leads to an acceleration of a factor of ∼250 for GDP dissociation (36).
Another universally conserved effect seen upon interaction of GEFs with members of the Ras superfamily is the disturbance of the interaction of a highly conserved phenylalanine in switch I (Phe-33 in Rab8) with the guanine base. In the case of Ras, it was shown that mutation of this residue to isoleucine results in an ∼100-fold increase in the rate constant for GDP dissociation (31). Assuming an additive contribution of both of these mechanisms (i.e. removal of Mg2+ and disruption of the Phe-33 interaction) to the dissociation rate of GDP, this would imply an acceleration of ∼2.5 × 104 for Rab8·Rabin8·GDP relative to Rab8·GDP. The factor determined (k″−1/k″−4 in Scheme 1) is actually smaller (∼1.5 × 103), which could indicate that there is rate limitation for Mg2+ dissociation or Phe-33 movement. Rabin8 has a similar catalytic efficiency as other GEFs, with GDP/GTP exchange accelerations in the range of ∼102–104-fold, but some GEFs are considerably faster. Among these are Cdc25 (for Ras; acceleration factor of ∼2 × 105), DrrA from L. pneumophila (for Rab1b; ∼8 × 105-fold), and RCC1 (for Ran; ∼1.3 × 106-fold) (26, 27, 37).
It has recently been pointed out that in many GTPase·GEF complexes of the Ras superfamily, the conserved lysine of the P-loop (Lys-21 in Rab8 and Lys-16 in H-Ras), which interacts with the β-phosphate of GDP or GTP, is intermediately stabilized by a conserved glutamate in the G3 motif at the beginning of switch II (DxxGQE motif; Glu-62 in Ras) (38). In a number of cases, substitution of this glutamate by alanine results in significant impairment of GEF activity. The corresponding residue is conserved in Rab molecules but does not appear to be involved in such an interaction and is not essential for the exchange reaction (38). In this work, the corresponding Rab8 P-loop lysine (Lys-21) interacts with GTP, GDP, or sulfate in the crystal structures of Rab8·Rabin8. Because sulfate is bound in the nucleotide-free state, we do not in fact have a structure corresponding to the genuine nucleotide-free situation, so we cannot make a definitive statement concerning the possible formation of an interaction of Lys-21 and Glu-68 in the nucleotide-free state. Although we cannot present a bona fide nucleotide-free Rab8·Rabin8 structure (the sulfate presumably mimics the β-phosphate position), the ϵ-amino group of Lys-21 is close to the carboxylate of Asp-63 of the DxxGQE sequence and could thus interact with Lys-21 in a hypothetical Rab8·Rabin8 complex devoid of nucleotides. This interaction (i.e. lysine of the P-loop with aspartate of the DxxGQE motif) is seen in at least two other Rab·GEF complexes (Rabex5·Rab21 and DrrA·Rab1b), as well as in the Ran·RCC1 complex. It therefore appears that nucleotide-free states of GTPase·GEF complexes are stabilized in part by interactions of the P-loop lysine with the glutamate of the DxxGQE motif, with the aspartate, or, in some cases, with both acidic residues.
Rabin8 and GRAB are specific GEFs for Rab8, in contrast to the rather promiscuous protein MSS4, which shows GEF activity toward Rab1, Rab3, Rab10, and Rab13 in addition to Rab8 (25, 39, 40). MSS4 is a catalytically inefficient GEF with a kcat/Km value about four times lower than that of Rabin8/GRAB. More importantly, the Rab8·MSS4 complex displays a very small second-order rate constant of GTP reassociation (25); therefore, the Rab8·MSS4 complex remains in the nucleotide-free form for a considerably longer time than other GEFs, such as Rabin8/GRAB. It is believed that the weak GEF properties of MSS4 are a result of its enzyme mechanism, in which nucleotide release is induced by a local protein-unfolding reaction of Rab8 (25): in contrast to Rabin8/GRAB, MSS4 unfolds the entire switch I region together with α-helix α1 of Rab8. This mechanism requires refolding of the nucleotide-binding pocket prior to GTP rebinding, and thus, nucleotide association is impaired. However, Sec2 domain GEFs (such as Rabin8 and GRAB) act differently from MSS4 because they stabilize the nucleotide-binding pocket by keeping switch I, switch II, and the P-loop in structurally defined confirmations that allow unimpaired rebinding of nucleotides (28, 41).
The work presented describes the structures of several intermediates in the exchange mechanism of Rabin8/GRAB with respect to Rab8. What is still missing is the structure of an intermediate in which Mg2+ is bound. Such a structure has been described recently for the GEF·GTPase pair DOCK9·Cdc42 in the presence of GTP (42). Here, the amino acid homologous to Ile-38 in Rab8 (a valine from the DOCK9 molecule) is displaced from its Mg2+-occluding position in the presence of Mg2+ and GTP, but not in the presence of Mg2+ and GDP. The authors interpreted this as a nucleotide sensor, leading to an alleged preference of the GEF for the GDP state of the GTPase over the GTP form and therefore allowing an unidirectional nucleotide exchange reaction. However, a similar effect is not observed in our structure, and the mode of nucleotide binding to Rab8·Rabin8/GRAB is identical for GDP and GTP. This is in keeping with the observation that there is no general preference for GEF interaction with the GDP or GTP form of GTPases, as required on theoretical grounds (14).
Acknowledgments
We thank Nathalie Bleimling for expert support in protein production. We thank the x-ray community of Max Planck Institute of Molecular Physiology Dortmund for help with data collection and the staff of beamline X10SA at the Swiss Light Source (Paul Scherrer Institute) for generous access to the facilities. We thank Prof. Wulf Blankenfeldt for invaluable help in structure determination.
This work was supported in part by German Research Foundation (DFG) Sonderforschungsbereich SFB1035 (Projekt B05) and SFB642 (Projekt A4).

This article contains supplemental Figs. S1 and S2 and Table S1.
- GEF
- guanine nucleotide exchange factor
- GppNHp
- 5′-guanylyl imidodiphosphate
- mant
- N-methylanthraniloyl.
REFERENCES
- 1. Cherfils J., Zeghouf M. (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 [DOI] [PubMed] [Google Scholar]
- 2. Hutagalung A. H., Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y. W., Bastiaens P., Goody R. S., Itzen A. (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200, 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peränen J., Auvinen P., Virta H., Wepf R., Simons K. (1996) Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol. 135, 153–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hattula K., Furuhjelm J., Arffman A., Peränen J. (2002) A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol. Biol. Cell 13, 3268–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hattula K., Furuhjelm J., Tikkanen J., Tanhuanpää K., Laakkonen P., Peränen J. (2006) Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 119, 4866–4877 [DOI] [PubMed] [Google Scholar]
- 7. Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., Jackson P. K. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 8. Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. (2007) Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peränen J. (2011) Rab8 GTPase as a regulator of cell shape. Cytoskeleton 68, 527–539 [DOI] [PubMed] [Google Scholar]
- 10. Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. (2010) Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westlake C. J., Baye L. M., Nachury M. V., Wright K. J., Ervin K. E., Phu L., Chalouni C., Beck J. S., Kirkpatrick D. S., Slusarski D. C., Sheffield V. C., Scheller R. H., Jackson P. K. (2011) Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. U.S.A. 108, 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo H. R., Saiardi A., Nagata E., Ye K., Yu H., Jung T. S., Luo X., Jain S., Sawa A., Snyder S. H. (2001) GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron 31, 439–451 [DOI] [PubMed] [Google Scholar]
- 13. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goody R. S., Hofmann-Goody W. (2002) Exchange factors, effectors, GAPs and motor proteins: common thermodynamic and kinetic principles for different functions. Eur. Biophys. J. 31, 268–274 [DOI] [PubMed] [Google Scholar]
- 15. Bleimling N., Alexandrov K., Goody R., Itzen A. (2009) Chaperone-assisted production of active human Rab8A GTPase in Escherichia coli. Protein Expr. Purif. 65, 190–195 [DOI] [PubMed] [Google Scholar]
- 16. Mihai Gazdag E., Streller A., Haneburger I., Hilbi H., Vetter I. R., Goody R. S., Itzen A. (2013) Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep. 14, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., Owens R. J. (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou X., Hagemann N., Schoebel S., Blankenfeldt W., Goody R. S., Erdmann K. S., Itzen A. (2011) A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 30, 1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Müller M. P., Peters H., Blümer J., Blankenfeldt W., Goody R. S., Itzen A. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itzen A., Rak A., Goody R. S. (2007) Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J. Mol. Biol. 365, 1359–1367 [DOI] [PubMed] [Google Scholar]
- 25. Itzen A., Pylypenko O., Goody R. S., Alexandrov K., Rak A. (2006) Nucleotide exchange via local protein unfolding–structure of Rab8 in complex with MSS4. EMBO J. 25, 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lenzen C., Cool R. H., Prinz H., Kuhlmann J., Wittinghofer A. (1998) Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37, 7420–7430 [DOI] [PubMed] [Google Scholar]
- 27. Schoebel S., Oesterlin L. K., Blankenfeldt W., Goody R. S., Itzen A. (2009) RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol. Cell 36, 1060–1072 [DOI] [PubMed] [Google Scholar]
- 28. Sato Y., Fukai S., Ishitani R., Nureki O. (2007) Crystal structure of the Sec4p·Sec2p complex in the nucleotide exchanging intermediate state. Proc. Natl. Acad. Sci. U.S.A. 104, 8305–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buchwald G., Friebel A., Galán J. E., Hardt W. D., Wittinghofer A., Scheffzek K. (2002) Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coureux P. D., Wells A. L., Ménétrey J., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. (2003) A structural state of the myosin V motor without bound nucleotide. Nature 425, 419–423 [DOI] [PubMed] [Google Scholar]
- 31. Reinstein J., Schlichting I., Frech M., Goody R. S., Wittinghofer A. (1991) p21 with a phenylalanine 28 → leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J. Biol. Chem. 266, 17700–17706 [PubMed] [Google Scholar]
- 32. Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J. (1998) The structural basis of the activation of Ras by Sos. Nature 394, 337–343 [DOI] [PubMed] [Google Scholar]
- 33. Worthylake D. K., Rossman K. L., Sondek J. (2000) Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408, 682–688 [DOI] [PubMed] [Google Scholar]
- 34. Béraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. (1998) A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the β-phosphate to destabilize GDP on ARF1. EMBO J. 17, 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uejima T., Ihara K., Goh T., Ito E., Sunada M., Ueda T., Nakano A., Wakatsuki S. (2010) GDP-bound and nucleotide-free intermediates of the guanine nucleotide exchange in the Rab5·Vps9 system. J. Biol. Chem. 285, 36689–36697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simon I., Zerial M., Goody R. S. (1996) Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J. Biol. Chem. 271, 20470–20478 [DOI] [PubMed] [Google Scholar]
- 37. Klebe C., Prinz H., Wittinghofer A., Goody R. S. (1995) The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry 34, 12543–12552 [DOI] [PubMed] [Google Scholar]
- 38. Gasper R., Thomas C., Ahmadian M. R., Wittinghofer A. (2008) The role of the conserved switch II glutamate in guanine nucleotide exchange factor-mediated nucleotide exchange of GTP-binding proteins. J. Mol. Biol. 379, 51–63 [DOI] [PubMed] [Google Scholar]
- 39. Wixler V., Wixler L., Altenfeld A., Ludwig S., Goody R. S., Itzen A. (2011) Identification and characterisation of novel Mss4-binding Rab GTPases. Biol. Chem. 392, 239–248 [DOI] [PubMed] [Google Scholar]
- 40. Burton J. L., Burns M. E., Gatti E., Augustine G. J., De Camilli P. (1994) Specific interactions of Mss4 with members of the Rab GTPase subfamily. EMBO J. 13, 5547–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong G., Medkova M., Novick P., Reinisch K. M. (2007) A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol. Cell 25, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D. (2009) Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science 325, 1398–1402 [DOI] [PubMed] [Google Scholar]