Background: The chemomechanical coupling scheme of the rotary motor V1-ATPase is incompletely understood.
Results: Enterococcus hirae V1-ATPase (EhV1) showed 120° steps of rotation without substeps, as commonly seen with F1-ATPase.
Conclusion: The basic properties of rotary dynamics of EhV1 are similar to those of Thermus thermophilus V1-ATPase.
Significance: This study revealed the common properties of V1-ATPases as rotary molecular motors, distinct from those of F1-ATPases.
Keywords: Bioenergetics, Enzyme Mechanisms, Molecular Motors, Single-molecule Biophysics, Vacuolar ATPase, V1-ATPase, V0V1
Abstract
V-ATPases are rotary molecular motors that generally function as proton pumps. We recently solved the crystal structures of the V1 moiety of Enterococcus hirae V-ATPase (EhV1) and proposed a model for its rotation mechanism. Here, we characterized the rotary dynamics of EhV1 using single-molecule analysis employing a load-free probe. EhV1 rotated in a counterclockwise direction, exhibiting two distinct rotational states, namely clear and unclear, suggesting unstable interactions between the rotor and stator. The clear state was analyzed in detail to obtain kinetic parameters. The rotation rates obeyed Michaelis-Menten kinetics with a maximal rotation rate (Vmax) of 107 revolutions/s and a Michaelis constant (Km) of 154 μm at 26 °C. At all ATP concentrations tested, EhV1 showed only three pauses separated by 120°/turn, and no substeps were resolved, as was the case with Thermus thermophilus V1-ATPase (TtV1). At 10 μm ATP (⪡Km), the distribution of the durations of the ATP-waiting pause fit well with a single-exponential decay function. The second-order binding rate constant for ATP was 2.3 × 106 m−1 s−1. At 40 mm ATP (⪢Km), the distribution of the durations of the catalytic pause was reproduced by a consecutive reaction with two time constants of 2.6 and 0.5 ms. These kinetic parameters were similar to those of TtV1. Our results identify the common properties of rotary catalysis of V1-ATPases that are distinct from those of F1-ATPases and will further our understanding of the general mechanisms of rotary molecular motors.
Introduction
V-ATPase is a rotary molecular motor that couples ion transport to ATP hydrolysis and synthesis. The main function of V-ATPase in eukaryotes is to transport protons across a membrane by using the energy derived from ATP hydrolysis (1–3). V-ATPase also catalyzes ATP synthesis, harnessing the energy of proton flow in certain eubacteria such as Thermus thermophilus. V-ATPases are composed of V1-ATPase (V1),6 a water-soluble moiety that hydrolyzes and synthesizes ATP, and a membrane-embedded moiety (V0) that translocates ions. The V1 and V0 domains are connected by a rotary shaft and peripheral stalks (1–3). The V1 complex is composed of A, B, D, and F subunits, in which the three A and three B subunits are alternately arranged, forming a hexameric stator A3B3 ring (4–7). ATP hydrolysis and synthesis occur on the catalytic sites that are located at the interfaces of the A and B subunits, with the majority of the catalytic residues residing in the A subunits. The rotary shaft is composed of D and F subunits penetrating into the central cavity of the A3B3 ring (6, 7).
The rotation of V1 has been visualized using optical microscopy by attachment of a probe to the rotary shaft (8–11). V1 of T. thermophilus (TtV1), which functions as an ATP synthase, rotates stepwise in a counterclockwise direction (8). The basic step size is 120°, and similar to F1-ATPase (F1), the water-soluble moiety of F0F1-ATP synthase (12), each step is coupled to the consumption of a single ATP molecule (10). Although no substeps have yet been resolved in the rotation of TtV1 (10, 11), the 120° steps of F1 from the thermophilic Bacillus PS3 (TF1) and Escherichia coli (EF1) have been shown to be further divided into 80° and 40° substeps and into 85° and 35° substeps, respectively (13–15). The 80° and 85° substeps are triggered by ATP binding and ADP release, whereas the 40° and 35° substeps are known to occur after ATP cleavage and release of inorganic phosphate. Accordingly, the pauses before the 80° and 85° substeps are referred to as ATP-binding (ATP-waiting) pauses, and those prior to the 40° and 35° substeps are known as catalytic pauses. As described above, the chemomechanical coupling scheme of TtV1 appears to be distinct from that of F1. However, to date, the stepping rotations of V1 complexes other than TtV1 have not been described, and the chemomechanical coupling scheme of V1 remains unclear (9).
Enterococcus hirae V-ATPase functions as a primary ion pump, similar in nature to eukaryotic V-ATPases (16, 17). We recently solved the crystal structures of the V1 component of E. hirae V-ATPase (EhV1) and proposed a model of its rotation mechanism (6). In this study, to characterize the stepping rotation of EhV1, we analyzed and compared the basic properties of EhV1 rotary dynamics with those of TtV1, TF1, and EF1. As was the case with TtV1, no substeps were resolved in the rotation of EhV1, suggesting that 120° stepping rotation without substeps is a common property of V1 complexes.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant EhV1 and AviTag-tagged EhV1 Expressed in E. coli
The EhV1 holocomplex (A3B3DF) was expressed in E. coli using the expression plasmid pTR19-FABD. We synthesized a DNA fragment containing the ntpF, ntpA, ntpB, and ntpD genes (in this order) and optimized its codon usage for E. coli expression. This fragment was then cloned into plasmid pTR19, the expression vector for the F0F1-ATP synthase of thermophilic Bacillus PS3 (18), after which a His6 tag was introduced at the N terminus of the A subunit by PCR to obtain plasmid pTR19-FABD. For the rotation assay, we used the pTR19-FABD-Avi, in which the AviTag biotinylation sequence (GLNDIFEAQKIEWHE) (19) was inserted between Gly-121 and Tyr-122 of the D subunit by PCR-based mutagenesis. E. coli BL21(DE3) cells were transformed with pTR19-FABD or pTR19-FABD-Avi and cultured in Super Broth (32 g/liter Tryptone, 20 g/liter yeast extract, and 5 g/liter sodium chloride) containing 100 μg/ml ampicillin and 2 mm isopropyl β-d-thiogalactopyranoside at 37 °C for 20 h. Cells were suspended in buffer A (20 mm potassium Pi (pH 7.0), 230 mm NaCl, and 20 mm imidazole) and disrupted by sonication. After removal of the cell debris by centrifugation at 81,000 × g for 20 min at 4 °C, the solution was applied to a nickel-nitrilotriacetic acid column (Ni-NTA Superflow, Qiagen). After washing with 10 column volumes of buffer A, recombinant EhV1 or AviTag-tagged EhV1 (AviTag-EhV1) was eluted with buffer B (20 mm potassium Pi (pH 7.0), 50 mm NaCl, and 200 mm imidazole). The eluted fractions were concentrated with an Amicon Ultra-10K unit (Merck Millipore) and then passed through a Superdex 200 gel-filtration column (GE Healthcare) equilibrated with buffer C (20 mm MES-NaOH (pH 6.5), 100 mm NaCl, and 10% glycerol). The purified proteins were flash-frozen in liquid nitrogen and stored at −80 °C until used.
Preparation of the A3B3 Subcomplex
The A3B3 subcomplex was expressed in E. coli BL21(DE3) cells harboring the expression plasmid pTR19-AB. Plasmid pTR19-AB was constructed by removing the ntpF and ntpD genes from pTR19-FABD by PCR. Expression and purification of the A3B3 subcomplex were performed using the same procedure as described for the EhV1 holocomplex, and purified proteins were stored at −80 °C until used.
Preparation of the DF Subcomplex
An E. coli cell-free protein expression system was employed to synthesize the DF subcomplex using a mixture of plasmids containing the corresponding genes. The expressed DF subcomplex was purified as described previously (6). The homogeneity of each purified subcomplex was judged by SDS-PAGE analysis. After purification, cysteine residues in the DF subcomplex were biotinylated with a 3 m excess of biotinylation reagent (biotin-PEAC5-maleimide, Dojindo) in 20 mm MOPS-KOH (pH 7.0) and 150 mm NaCl at room temperature for 20 min. The reaction was quenched using 10 mm DTT. We used a mutant DF subcomplex containing two engineered cysteine residues in its D subunit (T60C/R131C), substituted using a QuikChange site-directed mutagenesis kit (Agilent Technologies), and a single endogenous cysteine residue in both D (Cys-153) and F (Cys-54) subunits. A maximum of three of these residues can be expected to react with the biotinylation reagent. Specific biotinylation of the D subunit was confirmed by Western blotting with streptavidin-alkaline phosphatase conjugate (see Fig. 1).
FIGURE 1.
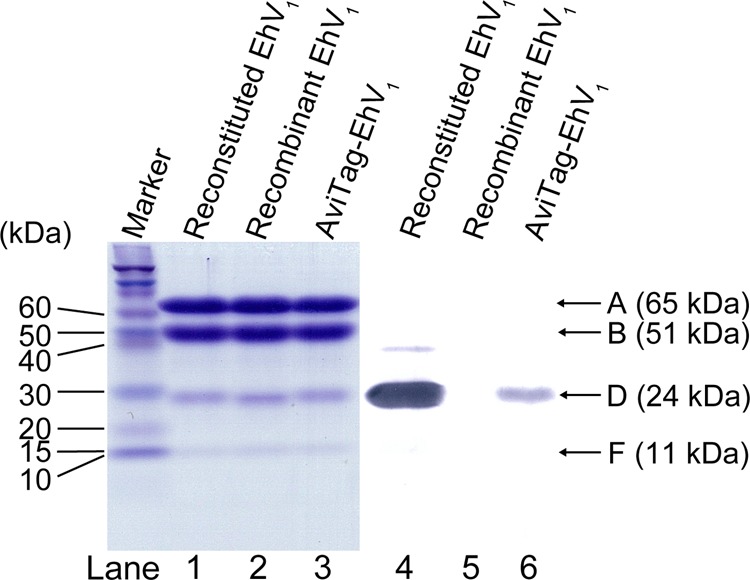
Gel electrophoresis. Lanes 1–3, SDS-PAGE of reconstituted EhV1, recombinant EhV1, and AviTag-EhV1. A 16% gel was used; 12 pmol of protein was loaded in each lane. The molecular masses of the A, B, D, and F subunits are 65, 51, 24, and 11 kDa, respectively. Proteins were stained with Coomassie Brilliant Blue. Lanes 4–6, immunoblots stained by streptavidin-alkaline phosphatase conjugates, showing biotin labeling of the D subunit. Lane 4, reconstituted EhV1 containing the biotinylated D subunit. Lane 5, non-biotinylated recombinant EhV1. Lane 6, AviTag-EhV1 containing the biotinylated D subunit.
Preparation of Reconstituted EhV1
Reconstituted EhV1 was prepared as follows. Purified A3B3 and biotinylated DF were mixed at a 1:2 molar ratio and incubated at room temperature for 2 h. Reconstituted EhV1 was purified using a Superdex 200 gel-filtration column equilibrated with buffer C, flash-frozen in liquid nitrogen, and stored at −80 °C until used.
Biochemical Assay
The protein concentration of EhV1 was determined based on UV absorbance using a molar extinction coefficient of 310,910 m−1 cm−1 calculated from its amino acid sequence (ProtParam tool, ExPASy). The ATP hydrolysis rate of EhV1 was measured using an ATP-regenerating system. The reaction mixture contained 50 mm MES-KOH (pH 6.5), 50 mm KCl, 5 mm MgCl2, 2.5 mm phosphoenolpyruvate, 0.2 mg/ml NADH, 0.1 mg/ml pyruvate kinase, and 0.1 mg/ml lactate dehydrogenase in addition to various concentrations of ATP. The rate of ATP hydrolysis was monitored as the rate of NADH oxidation at times ranging from 0 to 10 s after the addition of V1, which was measured as a decrease in the absorbance at 340 nm. All measurements were carried out at 25 ± 1 °C.
Rotation Assay
To observe ATP hydrolysis-driven rotation of EhV1, the A3B3 stator ring was immobilized on a Ni-NTA-coated glass surface via a His6 tag introduced at the N terminus of the A subunit. Streptavidin-coated 40-nm gold colloid was then attached to the biotinylated cysteine or AviTag in the rotor DF subunit as a probe. The gold colloids were prepared as described previously (20). The rotations of the gold colloid were observed at 26 ± 1 °C by an objective-type total internal reflection dark-field microscope constructed on an Olympus IX-71 inverted microscope (20). The images were recorded with a high-speed CMOS camera (FASTCAM 1024PCI, Photron) at 1000–10,000 frames/s (fps). The flow cell was assembled from a Ni-NTA-coated glass (24 × 36 mm2) and an uncoated cover glass (18 × 18 mm2) separated by two spacers of ∼50 μm thickness. First, buffer D (5 mg/ml BSA, 20 mm potassium Pi (pH 7.0), 230 mm NaCl, and 20 mm imidazole) was infused into the flow cell to prevent nonspecific binding of EhV1 and gold colloid. After incubation for 10 min, EhV1 (0.5–2 nm in buffer D) was infused into the flow cell. After incubation for 5 min, unbound EhV1 was washed out with buffer D, after which gold colloid suspended in buffer D was infused. After 10 min, unbound gold colloid was washed out. Observation of rotation was initiated after infusion of buffer E (50 mm MES-KOH (pH 6.5), 50 mm KCl, and 5 mm MgCl2) containing ATP (10 μm to 3 mm) or MgATP (4 and 40 mm) and an ATP-regenerating system.
RESULTS
Recombinant EhV1
We first tried to carry out a rotation assay using recombinant EhV1 (A3B3DF complex) expressed in E. coli. To observe the rotation, the rotation probe must be attached to the rotor DF subunits through biotin-streptavidin linkage. Because three endogenous cysteine residues in the stator A subunit (Cys-28, Cys-174, and Cys-259) may react with the biotinylation reagent, we substituted these residues with serine or alanine (C28A/C174S/C259S) and measured the activity of this mutant. The ATP hydrolysis rate of the EhV1 mutant, measured by a biochemical assay, was <10% of that of wild-type EhV1 (data not shown), indicating that the effect of substitution was significant.
Reconstituted EhV1
Because the rotor DF subunit can be biotinylated separately from the stator A3B3 ring prior to reconstitution, we next used reconstituted EhV1 in the rotation assay. Reconstituted EhV1 has also been used recently for crystal structural analysis requiring a pure homogeneous sample (6). No differences were observed in the subunit composition for reconstituted EhV1 and recombinant EhV1 (Fig. 1), indicating a high reconstitution efficiency. Furthermore, reconstituted EhV1 had an ATP hydrolysis rate comparable to that of recombinant EhV1 at all ATP concentrations used, ranging from 10 μm to 4 mm (Fig. 2). The expected rotation rates of reconstituted EhV1 and recombinant EhV1, calculated as one-third of the ATP hydrolysis rates, followed Michaelis-Menten kinetics. The maximal rotation rate (Vmax) and Michaelis constant (Km) were 73 ± 2 revolutions/s (rps) and 221 ± 17 μm, respectively, for reconstituted EhV1 and 72 ± 1 rps and 246 ± 16 μm (mean ± S.E. of fitting), respectively, for recombinant EhV1 (Fig. 2B, upper panel, red and blue dashed lines). These results indicate that the kinetic parameters of reconstituted EhV1 are almost identical to those of recombinant EhV1, suggesting that the catalytic properties of EhV1 are not affected by the reconstitution process. Therefore, we decided to use reconstituted EhV1 for the rotation assay.
FIGURE 2.
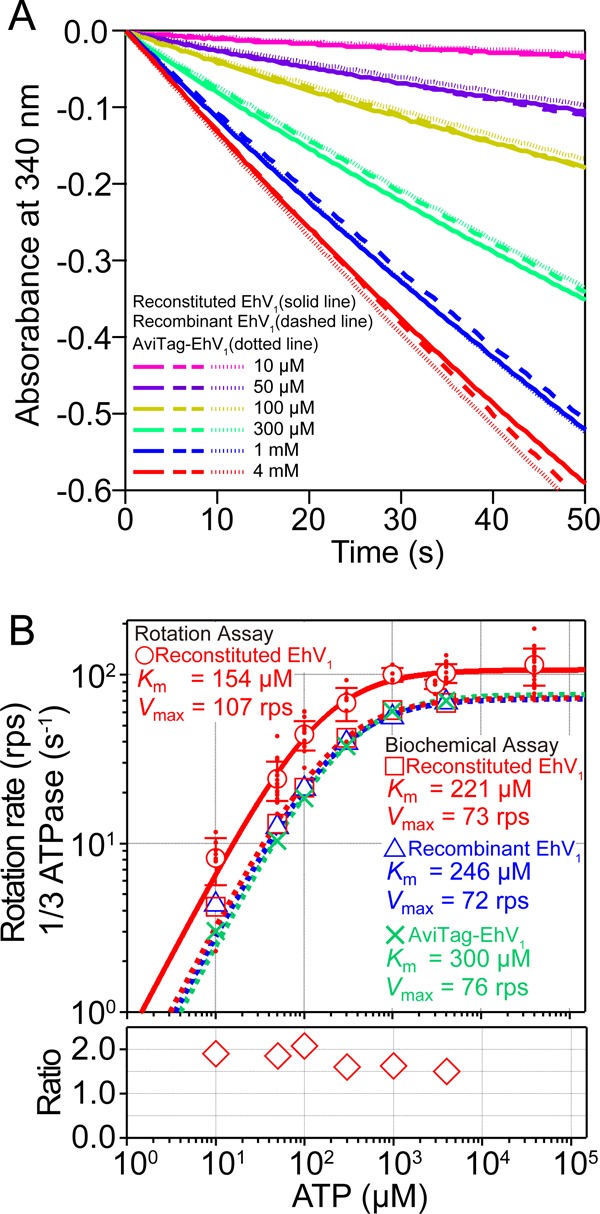
ATP dependence of ATP hydrolysis rate and rotation rate. A, time course of ATP hydrolysis by reconstituted EhV1 (solid lines), recombinant EhV1 (dashed lines), and AviTag-EhV1 (dotted lines) at 25 ± 1 °C with 10 μm, 50 μm, 100 μm, 300 μm, 1 mm, and 4 mm ATP. ATP hydrolysis was monitored as NADH oxidation in the ATP-regenerating system. The reaction was initiated by the addition of EhV1 (final concentration of 10 nm) at 0 s. The ATP hydrolysis rate of EhV1 was estimated from the slope of 0–10 s. B, upper panel, red dots indicate rotation rates determined by single-molecule rotation assay of reconstituted EhV1. Open red circles indicate average rotation rates (n ≥ 3). Error bars represent S.D. Open red squares, open blue triangles, and green crosses indicate the average of one-third of the ATP hydrolysis rates determined by the biochemical assay of reconstituted EhV1, recombinant EhV1, and AviTag-EhV1, respectively (n ≥ 3). In biochemical assay, S.D. is smaller than the size of the symbols. The solid and dashed lines indicate fits with the Michaelis-Menten equation: V = Vmax × [ATP]/(Km + [ATP]). Vmax = 107 ± 5 rps and Km = 154 ± 33 μm (mean ± S.E. of fitting) were obtained by the single-molecule rotation assay for reconstituted EhV1. Vmax = 73 ± 2 rps and Km = 221 ± 17 μm were obtained by the biochemical assay for reconstituted EhV1, Vmax = 72 ± 1 rps and Km = 246 ± 16 μm for recombinant EhV1, and Vmax = 76 ± 1 rps and Km = 300 ± 19 μm (mean ± S.E.) for AviTag-EhV1. The apparent binding constant for ATP (3 × Vmax/Km) was estimated as (2.2 ± 0.4) × 106 m−1 s−1 from the rotation assay. Lower panel, ratio of the rotation rate determined by the rotation assay to one-third of the ATP hydrolysis rate determined by the biochemical assay of reconstituted EhV1.
EhV1 Has Two Distinct Rotational States
The rotary motion of reconstituted EhV1 was observed in a single-molecule assay using streptavidin-coated 40-nm gold colloid as a load-free probe at a rate of 1000–10,000 fps (Fig. 3). We found that the reconstituted EhV1 complexes exhibited two distinct reversible states of rotation, namely clear and unclear (Fig. 4, A–C). In the clear rotational state, the majority of the centroids of gold colloid in each frame were distributed in three positions separated by 120° and were remote from the rotation center. Moreover, the time course showed clear unidirectional rotation in a counterclockwise direction (Fig. 4A). In contrast, in the unclear state, the centroids showed wide fluctuations toward the rotation center (Fig. 4B). Because the rotation rate in the unclear state seems to be similar to that in the clear state, the complex appears to rotate unidirectionally, although the measured rotation rate in the unclear state may not be accurate.
FIGURE 3.
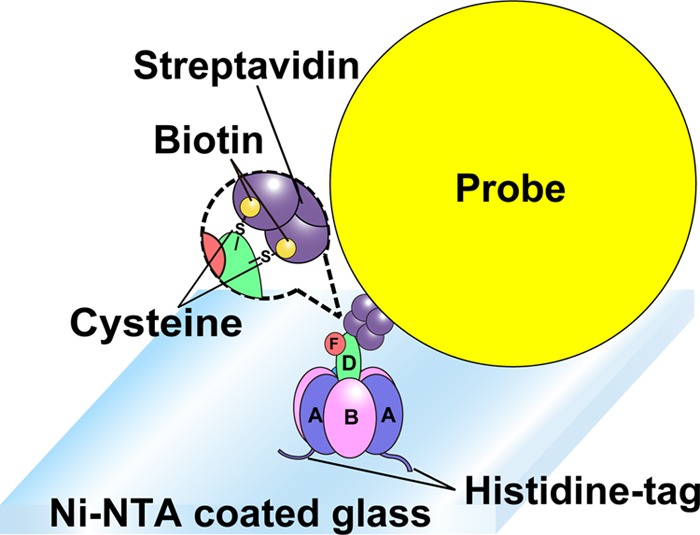
Schematic image of the rotation assay. Shown is the experimental setup for single-molecule rotation assay of reconstituted EhV1. The stator A3B3 ring of EhV1 was fixed on the glass surface with His6 tag at the N termini of the A subunits. Streptavidin-coated 40-nm gold colloid was attached to biotinylated cysteine residues in the rotor DF subunit.
FIGURE 4.
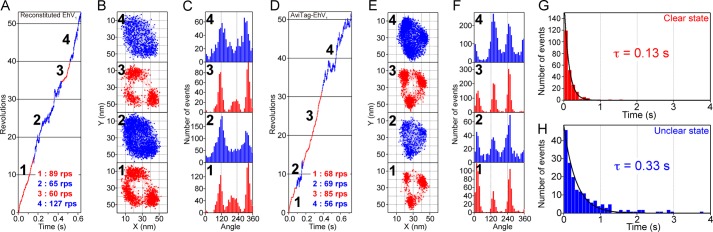
Two distinct states in the rotation of EhV1. A and D, time courses of rotation, including two reversible states of single molecule-reconstituted EhV1 (3 mm ATP) and AviTag-EhV1 (2 mm ATP). The rotations in the clear and unclear states are highlighted in red and blue, respectively. B and E, The x-y trajectories of the centroid of a rotating gold colloid shown in A and D. C and F, distributions of rotary angles shown in A and D. The numbers in A–C and D–F indicate the corresponding parts. G and H, distributions of duration times of clear and unclear states of reconstituted EhV1. The data at various ATP concentrations (from 10 μm to 40 mm) were analyzed collectively. The bin width was 0.1 s. The solid curves show the fit with single-exponential decay: constant × (exp(−t/τ)), where τ = 0.13 ± 0.003 s (mean ± S.E. of fitting, 238 events from 58 molecules) and 0.33 ± 0.009 s (mean ± S.E., 199 events from 58 molecules) for the clear (G) and unclear (H) states, respectively.
To assess whether the two rotational states are observed only in reconstituted EhV1, the D subunit of recombinant EhV1 was fused to AviTag, a 15-amino acid sequence that is subject to biotinylation by biotin ligase in E. coli (19). No differences were observed in the subunit compositions of AviTag-EhV1 and reconstituted or recombinant EhV1 (Fig. 1). Moreover, the ATP hydrolysis rate of AviTag-EhV1 was comparable to that of reconstituted and recombinant EhV1 in the biochemical assay (Fig. 2). Importantly, as was the case in the rotation assay with reconstituted EhV1, AviTag-EhV1 also exhibited two reversible rotational states (Fig. 4, D–F). This result clearly shows that the two rotational states are not an artifact of damage or inactivity caused by the reconstitution procedure but instead represent an intrinsic property of EhV1. Given that the efficiency of biotinylation (Fig. 1) and the frequency of rotating probes for AviTag-EhV1 were significantly lower than those for reconstituted EhV1, we used reconstituted EhV1 in the subsequent experiments.
Next, we analyzed the duration of the clear and unclear states for 10 μm to 40 mm ATP. Because we did not find a clear dependence of the duration on ATP concentration, we analyzed the data at various ATP concentrations collectively. The distributions of the duration times fit well to a single-exponential decay function, suggesting a single rate-limiting step in the transition between the clear and unclear states. The time constants were 0.13 ± 0.003 s (mean ± S.E. of fitting, 238 events from 58 molecules) for the clear state and 0.33 ± 0.009 s (mean ± S.E. of fitting, 199 events from 58 molecules) for the unclear state (Fig. 4, G and H). The ratio of the clear state to the total observation time was ∼0.3 (=0.13/(0.13 + 0.33)).
To date, it has been unclear why transitions occur between the clear and unclear states. A study reported that although unusual fluctuations have been reported in the rotation of TtV1 (11), the behaviors are not entirely identical; in that study (11), the authors attributed the fluctuating state to the probe adopting two orientations relative to the D subunit and excluded these data from the analysis. Alternatively, the unclear state may be due to less stable interactions between the rotor and stator in V1 compared with those in F1. Because V0 and V1 are connected not only by the rotor but also by the two peripheral stalks composed of the E and G subunits, this unstable interaction would be anticipated to occur only in the isolated V1 complex and not in the physiological V0V1 complex. We nevertheless concluded that the tight chemomechanical coupling of EhV1 is achieved at least in the clear state and, accordingly, restricted our analysis in the remainder of the study to that state.
ATP Dependence of Rotation
The rotation rates of reconstituted EhV1 were measured at various concentrations of ATP, ranging from 10 μm to 40 mm (Fig. 2B, upper panel, red dots and open circles). Below 100 μm ATP, the rotation rates were almost proportional to the ATP concentration, indicating that ATP binding is rate-limiting in this range. Above 1 mm ATP, the rotation rate was essentially constant. The rotation rates followed Michaelis-Menten kinetics with a Vmax of 107 ± 5 rps and a Km of 154 ± 33 μm (mean ± S.E. of fitting) (Fig. 2B, upper panel, solid red line). The second-order binding rate constant for ATP (kon(ATP)) determined from 3 × Vmax/Km was (2.2 ± 0.4) × 106 m−1 s−1 under the assumption that three ATP molecules were hydrolyzed per turn.
The value of Km determined by the single-molecule rotation assay was slightly lower than that determined by the biochemical assay. Furthermore, the value of Vmax estimated by the single-molecule rotation assay was ∼50% greater than that deduced from the biochemical assay. This ratio was essentially constant at each ATP concentration (Fig. 2B, lower panel). This result may arise from inaccuracy in protein concentration determination based on the molar absorbance coefficient calculated from the amino acid sequence and/or imply that the ATP hydrolysis rate during the unclear state is slightly lower than that during the clear state. Furthermore, in the biochemical assay, the ATP hydrolysis rate gradually decreased during measurement (Fig. 2A). It is well known that the ATP hydrolysis of TtV1, which functions as ATP synthase, is strongly regulated by MgADP inhibition to prevent wasteful ATP consumption (21). The ATP hydrolysis activity of TtV1 is inhibited rapidly and irreversibly in the presence of ATP. Because the decrease in the ATP hydrolysis rate of EhV1 was much slower than that of TtV1 (Fig. 2A) and some EhV1 molecules showed reversible and irreversible long pauses in the rotation assay, unknown inhibited states of EhV1 other than MgADP inhibition may exist.
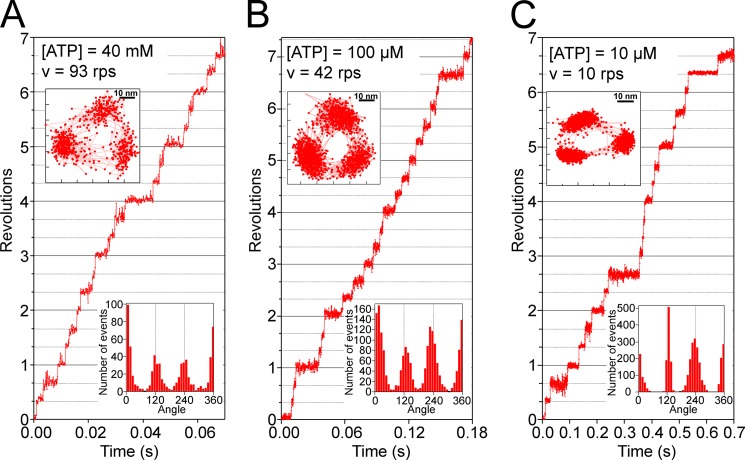
Typical examples of rotation at 40 mm, 100 μm, and 10 μm ATP are shown in Fig. 5. At all ATP concentrations, EhV1 exhibited stepwise rotation with three intervening pauses separated by 120°, as shown in the x-y trajectories and the distribution of the rotary angle (Fig. 5, insets). Because the 120° steps were completed within 0.2 ms (one to two frames captured at 10,000 fps), the rotation rate was determined primarily by the duration of the intervening pause. At 40 mm ATP, a concentration considerably higher than the Km, the intervening pauses would represent the catalytic pauses (Fig. 5A) because the expected binding time constant for ATP (0.011 ms, estimated from 1/(40 × 10−3 m × 2.2 × 106 m−1 s−1)) was 100-fold smaller than the duration times (3.1 ms) of pauses expected from the Vmax (1/(107 × 3)). At 10 μm ATP, a concentration substantially lower than the Km, the intervening pauses would correspond to the ATP-waiting pauses because ATP binding is rate-limiting under these conditions (Fig. 5C).
FIGURE 5.
Steps and pauses in the rotation of reconstituted EhV1. Shown are typical time courses of rotation of reconstituted EhV1 at various ATP concentrations. A, rotation at 40 mm ATP, captured at 10,000 fps. B, rotation at 100 μm ATP, captured at 10,000 fps. C, rotation at 10 μm ATP, captured at 5000 fps. The upper left insets show the x-y trajectories of the centroid of a rotating gold colloid. The lower right insets show the distributions of the rotary angle.
Even at 100 μm ATP, a concentration near the Km and at which the duration of the ATP-waiting pause approaches that of the catalytic pause, EhV1 exhibited only three pauses separated by 120° (Fig. 5B). This finding suggests that there are no substeps in the rotation of EhV1, as is the case for TtV1 (10, 11).
Buffer-exchange Experiment
To further confirm that the angles of the ATP-waiting pauses observed at low ATP concentrations correspond to those of the catalytic pauses observed at saturating ATP concentrations, we next conducted a buffer-exchange experiment. After recording the stepwise rotation of EhV1 pausing every 120° at 10 μm ATP, we increased the ATP concentration to 3 mm by infusing buffer containing 3 mm ATP into the flow cell. After buffer exchange, although the rotation rate increased significantly, EhV1 continued to rotate with discrete 120° steps, pausing at almost the same angles (Fig. 6, A and B). The distributions of the rotary angles indicated three peaks corresponding to three pauses in the rotation (Fig. 6B). To assess the difference in the angular position (Δθ) between 10 μm and 3 mm ATP, the positions of the peaks were determined by fitting the histogram with Gaussian functions and then comparing them with the nearest ones (Fig. 6B). The distribution of Δθ showed a single peak around 0°, with a mean value of 3.2 ± 12° (mean ± S.D., 21 events from seven molecules) (Fig. 6C). On the basis of these data, we confirmed again that the rotation of EhV1 contains no substeps.
FIGURE 6.
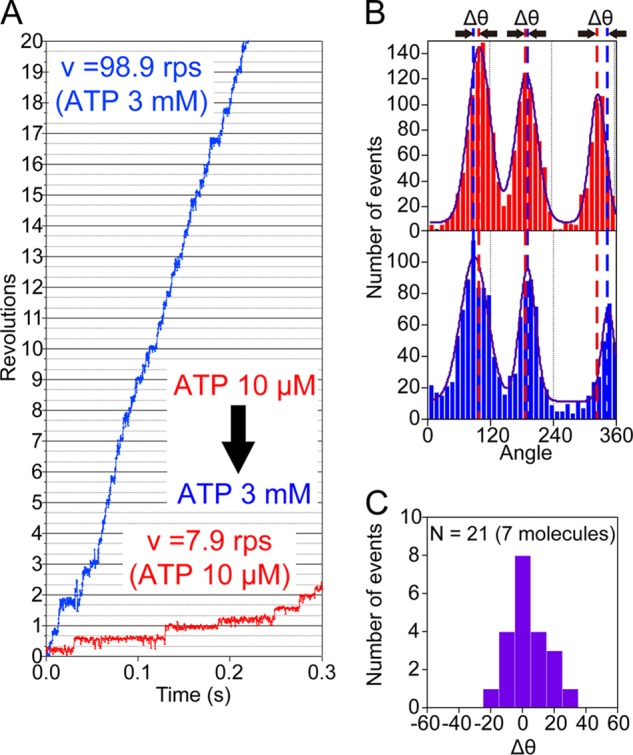
Buffer-exchange experiments. A, time courses of rotation of the same molecule at 10 μm ATP (red) and 3 mm ATP (blue). The ATP concentration was increased from 10 μm to 3 mm. B, distributions of the rotary angle at 10 μm ATP (upper) and 3 mm ATP (lower) shown in A. Δθ represents the angular differences between the pause angles (peaks) before and after buffer exchange. C, distribution of Δθ. The mean value was 3.2 ± 12° (mean ± S.D., 21 events from seven molecules).
Dwell Time Analysis
To obtain the time constants and kinetic parameters for elementary reaction steps such as ATP binding, ATP cleavage, and product release, we analyzed the duration of the pauses. On analyzing four molecules at saturating ATP concentrations (40 mm), all distributions of the durations of the catalytic pauses showed a convex shape (Fig. 7, A–D). At 40 mm ATP, the expected time constant for ATP binding (0.011 ms) was too short to be resolved. Therefore, three elementary reaction steps could occur during the catalytic pauses, namely ATP cleavage, ADP release, and phosphate release. We first attempted to fit the distributions with a model of three consecutive reactions. This failed to improve the fits compared with a model of two consecutive reactions with two time constants. The average time constants from four molecules were 2.4 ± 1.1 and 0.6 ± 0.2 ms (mean ± S.D.) (Fig. 7F, open red circle and square). These values are consistent with the time constants of 2.6 ± 0.1 and 0.5 ± 0.02 ms (mean ± S.E. of fitting, 2097 events) determined by reproducing the accumulated distribution of the pause duration time from all four molecules (Fig. 7E). In addition, at 4 mm ATP (the expected time constant for ATP binding is 0.11 ms), similar accumulated distribution and time constants of 2.4 ± 0.05 and 0.5 ± 0.02 ms (mean ± S.E. of fitting, 4303 events from nine molecules) were obtained (Fig. 7, G and H), consistent with the saturation of the rotation rate at this concentration (Fig. 2B).
FIGURE 7.
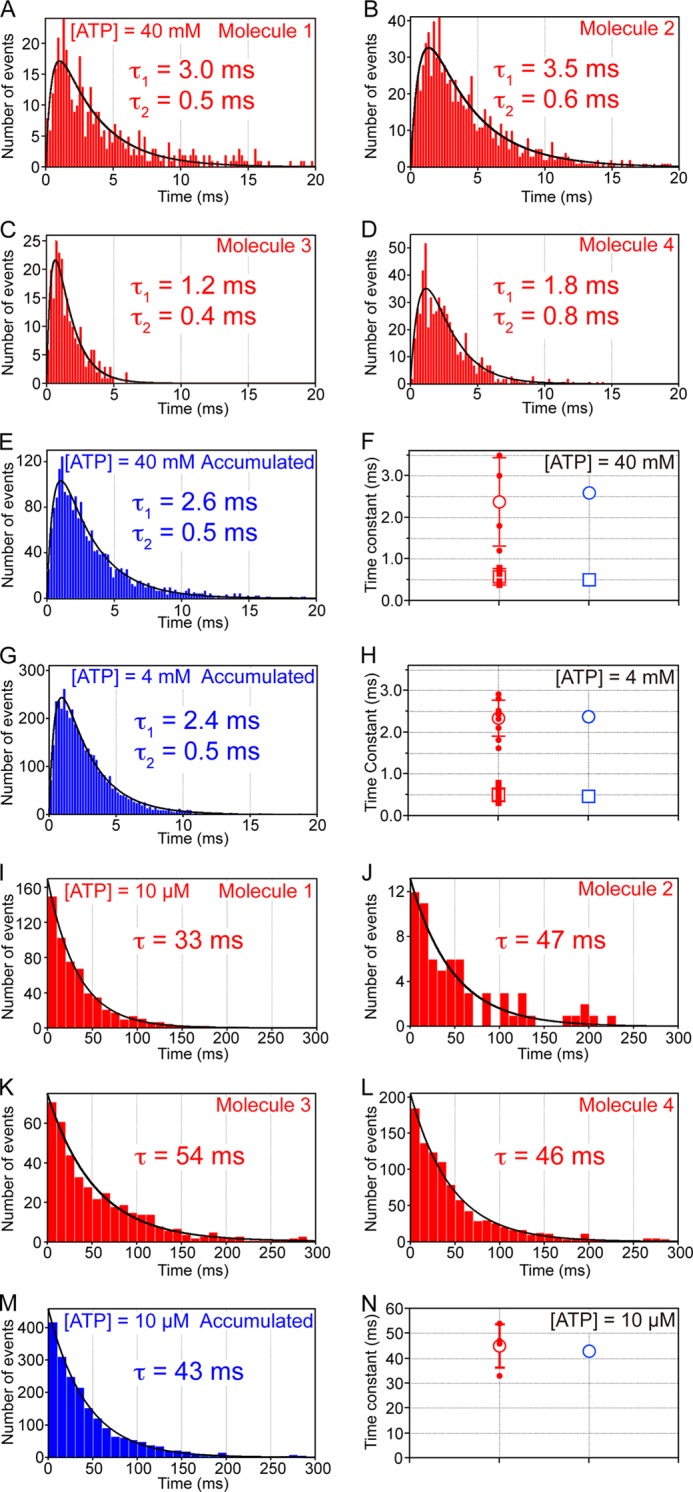
Distributions of duration times of the pauses. A–D, distributions of duration times of four single molecules at 40 mm ATP with a 0.2-ms bin width captured at 10,000 fps. The solid curves show fits with a model of two consecutive reactions: constant × (exp(t/τ1) − exp(t/τ2)), where τ1 = 3.0 ± 0.2 ms and τ2 = 0.5 ± 0.1 ms (mean ± S.E. of fitting, 398 events) (A), 3.5 ± 0.1 ms and 0.6 ± 0.04 ms (863 events) (B), 1.2 ± 0.2 ms and 0.4 ± 0.1 ms (228 events) (C), and 1.8 ± 0.3 ms and 0.8 ± 0.2 ms (608 events) (D). E, accumulated distribution of duration times of all four molecules at 40 mm. The solid curve shows a fit with a model of two consecutive reactions with time constants of 2.6 ± 0.1 and 0.5 ± 0.02 ms (2097 events). F, red dots and square dots indicate τ1 and τ2, respectively, shown in A–D. The open red circle and square indicate average τ1 (2.4 ± 1.1 ms) and τ2 (0.6 ± 0.2 ms) (mean ± S.D.) from four molecules. Error bars represent S.D. The open blue circle and square indicate τ1 and τ2, respectively, shown in E. G, accumulated distribution of duration times (nine molecules) at 4 mm ATP with a 0.2-ms bin width captured at 10,000 fps. The solid curve shows a fit with a model of two consecutive reactions: constant × (exp(t/τ1) − exp(t/τ2)), where τ1 = 2.4 ± 0.05 ms and τ2 = 0.5 ± 0.02 ms (mean ± S.E. of fitting, 4303 events). H, red circles and square dots indicate τ1 and τ2 determined by fitting the individual distributions of duration times of nine single molecules with a model of two consecutive reactions, respectively (each distribution is not shown). The open red circle and square indicate average τ1 (2.3 ± 0.5 ms) and τ2 (0.4 ± 0.2 ms) (mean ± S.D.) from nine molecules. Error bars represent S.D. The open blue circle and square indicate τ1 and τ2, respectively, shown in G. I–L, distributions of duration times of four single molecules at 10 μm ATP with a 10-ms bin width captured at 5000 or 2000 fps. The solid curves show fits with single-exponential decay: constant × (exp(−t/τ)), where τ = 33 ± 1 ms (mean ± S.E. of fitting, 581 events) (I), 47 ± 6 ms (67 events) (J), 54 ± 1 ms (432 events) (K), and 46 ± 1 ms (978 events) (L). M, accumulated distribution of duration times of all four molecules at 10 μm. The solid curve shows a fit with single-exponential decay with a time constant of 43 ± 1 ms (2058 events). N, the red dots indicate τ shown in I–L. The open red circle indicates average τ (45 ± 9 ms) (mean ± S.D.). Error bars represent S.D. The open blue circle indicates τ shown in M.
These time constants would correspond to (i) ATP cleavage and (ii) ADP and/or phosphate release, although it is currently unclear which time constant corresponds to which elementary reaction step. At a low ATP concentration (10 μm), as discussed above, the duration of the pauses corresponded to that of ATP waiting. Analysis of the four molecules showed that the distributions of the ATP-waiting duration time followed single-exponential decay (Fig. 7, I–L), indicating that EhV1 consumed one ATP molecule/120° step. The average time constant from four molecules was 45 ± 9 ms (mean ± S.D.) (Fig. 7N, open red circle), which is consistent with a time constant of 43 ± 1 ms (mean ± S.E. of fitting, 2058 events) determined by reproducing the accumulated distribution of the duration time from all four molecules (Fig. 7M). This value corresponds to a kon(ATP) of (2.3 ± 0.03) × 106 m−1 s−1, which is consistent with that determined by 3 × Vmax/Km ((2.2 ± 0.4) × 106 m−1 s−1), shown in Fig. 2B.
DISCUSSION
In this study, using a single-molecule assay, we have shown that EhV1 is a rotary molecular motor. To our knowledge, this is the first report showing that a eubacterial V1 functions as an ATP-driven ion pump under physiological conditions. EhV1 exhibited two rotational states, namely clear and unclear (Fig. 4). Assuming that the clear rotational state represents the tight chemomechanical coupling of EhV1, we analyzed this state to elucidate the basic rotational properties of EhV1. Our hypothesis that the unclear state is caused by unstable interactions between the rotor and stator of EhV1 must be examined by rotation assay of the entire E. hirae V-ATPase complex, in which the interactions between the rotor and stator are stabilized by two peripheral stalks. To perform this study, we are currently designing an E. coli expression system in which an appropriately tagged recombinant V-ATPase complex can be produced for a rotation assay.
In the clear rotational state, at all ATP concentrations ranging from below to above the Km, EhV1 rotated unidirectionally in a counterclockwise direction, exhibiting three pauses separated by 120° (Fig. 5). No substeps were resolved, as has been reported for TtV1 (10, 11). In contrast, in the region of their respective Km values, TF1 and EF1 have been reported to rotate with six pauses/turn (13–15). Recently, the overall crystal structures of TtV1 and EhV1 were shown to be similar (4–7), especially with respect to the interaction sites between the rotor and stator. These structures are distinct from the structure of F1 (22), although many amino acid residues associated with catalysis in the binding pocket are conserved between V1 and F1. These results imply that the degree of similarity in the interactions between the rotor and stator determines the presence or absence of substeps in the rotation.
Table 1 contains a comparison of the kinetic parameters determined by the single-molecule assay for EhV1, showing values for TtV1, TF1, and EF1. Despite the difference in physiological function between EhV1 and TtV1 and notwithstanding the large difference (>30 °C) in the optimal growth temperatures between E. hirae and T. thermophilus, the values for EhV1 are closer to those for TtV1 than for TF1 and EF1. This result implies that the basic properties of rotary dynamics are determined by their overall structures and that the difference in the physiological function derives from regulatory mechanisms such as MgADP inhibition.
TABLE 1.
Kinetic parameters of V1-ATPases and F1-ATPases from different sources
| Protein/measurement temperature | kon(ATP)a | kon(ATP) (3 × Vmax/Km)a | τ1/τ2a | Ref. |
|---|---|---|---|---|
| m−1 s−1 | m−1 s−1 | ms | ||
| EhV1d | ||||
| 26 ± 1 °C | (2.3 ± 0.03) × 106 | (2.2 ± 0.4) × 106 | 2.6 ± 0.1/0.5 ± 0.02e | This work |
| TtV1 | ||||
| 23 °C | 1.5 × 106 | 0.84 × 106 | 2.8/2.8 | 11 |
| 24 ± 1 °C | 1.39 × 106 | 21 | ||
| TF1 | ||||
| 23 °C | 3.0 × 107 | 2.6 × 107 | 1.6/0.71 | 13 |
| 25 ± 1 °C | 1.34/0.29 | 20 | ||
| 25 ± 1 °C | 2.2 × 107 | 27 | ||
| EF1 | ||||
| 23 °C | 4.7 × 107 | 6.4 × 107 | 0.41/0.29 | 15 |
a The second-order binding rate constant for ATP (kon(ATP)) was determined from the distribution of the duration of the ATP-waiting pause.
b The second-order binding rate constant for ATP (kon(ATP)) was determined from 3 × Vmax/Km.
c Time constants were determined from the distribution of the duration of the catalytic pause, which corresponds to ATP cleavage and either ADP or phosphate release (or both).
d The values are the mean ± S.E. of fitting.
e The values were obtained at 40 mm ATP.
During the unclear rotational state, the centroids of the gold colloid showed wide fluctuations toward the rotation center. It should be noted that EhV1 nevertheless rotated unidirectionally, implying that even if the interactions between the rotor and stator are not perfect, EhV1 maintains unidirectional and cooperative rotary catalysis. Recently, rotary catalysis of the rotor-less stator α3β3 ring of TF1 was demonstrated by high-speed atomic force microscopy (23), and we speculate that the stator A3B3 ring also likely exhibits rotary catalysis in the absence of the rotor DF subunits.
The chemomechanical coupling scheme of TF1 has been extensively studied by advanced single-molecule techniques such as a rotation assay of hybrid molecules and single-molecule manipulation with magnetic tweezers (24–26). For a single catalytic site of TF1, after ATP binding at 0°, ATP cleavage, ADP release, and phosphate release occur at 200°, 240°, and 320°, respectively (26). Further studies on EhV1 using advanced single-molecule techniques and high-resolution structural analysis will provide details on its chemomechanical coupling scheme. Moreover, comparison of the schemes of V1 and F1 from various species will shed light on the general mechanism of rotary molecular motors.
Acknowledgments
We thank Mio Inoue and Ken Ishii for preparation of the plasmids and Shoichi Toyabe for providing the data analysis software. We also thank all members of the laboratory for valuable discussions and comments.
This work was supported in part by Grants-in-aid for Scientific Research 24651167 (to R. I.) and 23370047 (to T. M.) and the Target Proteins Research Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
- V1
- V1-ATPase
- V0
- V0-ATPase
- TtV1
- T. thermophilus V1-ATPase
- F1
- F1-ATPase
- TF1
- thermophilic Bacillus PS3 F1-ATPase
- EF1
- E. coli F1-ATPase
- EhV1
- E. hirae V1-ATPase
- Ni-NTA
- nickel-nitrilotriacetic acid
- fps
- frames/s
- rps
- revolutions/s.
REFERENCES
- 1. Nishi T., Forgac M. (2002) The vacuolar (H+)-ATPases–nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 2. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 3. Marshansky V., Futai M. (2008) The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr. Opin. Cell Biol. 20, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Numoto N., Hasegawa Y., Takeda K., Miki K. (2009) Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase. EMBO Rep. 10, 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maher M. J., Akimoto S., Iwata M., Nagata K., Hori Y., Yoshida M., Yokoyama S., Iwata S., Yokoyama K. (2009) Crystal structure of A3B3 complex of V-ATPase from Thermus thermophilus. EMBO J. 28, 3771–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arai S., Saijo S., Suzuki K., Mizutani K., Kakinuma Y., Ishizuka-Katsura Y., Ohsawa N., Terada T., Shirouzu M., Yokoyama S., Iwata S., Yamato I., Murata T. (2013) Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature 493, 703–707 [DOI] [PubMed] [Google Scholar]
- 7. Nagamatsu Y., Takeda K., Kuranaga T., Numoto N., Miki K. (2013) Origin of asymmetry at the intersubunit interfaces of V-ATPase from Thermus thermophilus. J. Mol. Biol. 425, 2699–2708 [DOI] [PubMed] [Google Scholar]
- 8. Imamura H., Nakano M., Noji H., Muneyuki E., Ohkuma S., Yoshida M., Yokoyama K. (2003) Evidence for rotation of V1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirata T., Iwamoto-Kihara A., Sun-Wada G. H., Okajima T., Wada Y., Futai M. (2003) Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J. Biol. Chem. 278, 23714–23719 [DOI] [PubMed] [Google Scholar]
- 10. Imamura H., Takeda M., Funamoto S., Shimabukuro K., Yoshida M., Yokoyama K. (2005) Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. U.S.A. 102, 17929–17933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Furuike S., Nakano M., Adachi K., Noji H., Kinosita K., Jr., Yokoyama K. (2011) Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iino R., Noji H. (2013) Operation mechanism of FoF1-adenosine triphosphate synthase revealed by its structure and dynamics. IUBMB Life 65, 238–246 [DOI] [PubMed] [Google Scholar]
- 13. Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 14. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bilyard T., Nakanishi-Matsui M., Steel B. C., Pilizota T., Nord A. L., Hosokawa H., Futai M., Berry R. M. (2013) High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kakinuma Y., Igarashi K. (1994) Purification and characterization of the catalytic moiety of vacuolar-type Na+-ATPase from Enterococcus hirae. J. Biochem. 116, 1302–1308 [DOI] [PubMed] [Google Scholar]
- 17. Murata T., Takase K., Yamato I., Igarashi K., Kakinuma Y. (1997) Purification and reconstitution of Na+-translocating vacuolar ATPase from Enterococcus hirae. J. Biol. Chem. 272, 24885–24890 [DOI] [PubMed] [Google Scholar]
- 18. Suzuki T., Ueno H., Mitome N., Suzuki J., Yoshida M. (2002) F0 of ATP synthase is a rotary proton channel. Obligatory coupling of proton translocation with rotation of c-subunit ring. J. Biol. Chem. 277, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 19. Beckett D., Kovaleva E., Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ueno H., Nishikawa S., Iino R., Tabata K. V., Sakakihara S., Yanagida T., Noji H. (2010) Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 98, 2014–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uner N. E., Nishikawa Y., Okuno D., Nakano M., Yokoyama K., Noji H. (2012) Single-molecule analysis of inhibitory pausing states of V1-ATPase. J. Biol. Chem. 287, 28327–28335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 23. Uchihashi T., Iino R., Ando T., Noji H. (2011) High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 [DOI] [PubMed] [Google Scholar]
- 24. Ariga T., Muneyuki E., Yoshida M. (2007) F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat. Struct. Mol. Biol. 14, 841–846 [DOI] [PubMed] [Google Scholar]
- 25. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 26. Watanabe R., Iino R., Noji H. (2010) Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 [DOI] [PubMed] [Google Scholar]
- 27. Tanigawara M., Tabata K. V., Ito Y., Ito J., Watanabe R., Ueno H., Ikeguchi M., Noji H. (2012) Role of the DELSEED loop in torque transmission of F1-ATPase. Biophys. J. 103, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]