Background: The V-ATPase has been suggested to function in tumor cell invasion.
Results: Invasion by MCF10CA1a cells is inhibited by knockdown of the a3 isoform, whereas a3 overexpression increases invasion and plasma membrane localization of V-ATPases in MCF10a cells.
Conclusion: Invasion and plasma membrane V-ATPase localization can be controlled by expression of a3.
Significance: a3 is a possible therapeutic target to limit metastasis.
Keywords: Bioenergetics, Breast Cancer, Invasion, Proton Pumps, Proton Transport, Vacuolar ATPase, a Subunit Isoforms
Abstract
The vacuolar H+ ATPases (V-ATPases) are ATP-driven proton pumps that transport protons across both intracellular and plasma membranes. Previous studies have implicated V-ATPases in the invasiveness of various cancer cell lines. In this study, we evaluated the role of V-ATPases in the invasiveness of two closely matched human breast cancer lines. MCF10a cells are a non-invasive, immortalized breast epithelial cell line, and MCF10CA1a cells are a highly invasive, H-Ras-transformed derivative of MCF10a cells selected for their metastatic potential. Using an in vitro Matrigel assay, MCF10CA1a cells showed a much higher invasion than the parental MCF10a cells. Moreover, this increased invasion was completely sensitive to the specific V-ATPase inhibitor concanamycin. MCF10CA1a cells expressed much higher levels of both a1 and a3 subunit isoforms relative to the parental line. Isoforms of subunit a are responsible for subcellular localization of V-ATPases, with a3 and a4 targeting V-ATPases to the plasma membrane of specialized cells. Knockdown of either a3 alone or a3 and a4 together using isoform-specific siRNAs inhibited invasion by MCF10CA1a cells. Importantly, overexpression of a3 but not the other a subunit isoforms greatly increased the invasiveness of the parental MCF10a cells. Similarly, overexpression of a3 significantly increased expression of V-ATPases at the plasma membrane. These studies suggest that breast tumor cells employ particular a subunit isoforms to target V-ATPases to the plasma membrane, where they function in tumor cell invasion.
Introduction
The metastatic dissemination of tumor cells is responsible for the majority of cancer mortalities (1, 2). Metastasis is a complex, multistep process that includes intravasation of tumor cells out of a primary tumor site and extravasation of cells into a secondary site within the body (1). Both intravasation and extravasation require that cells develop an invasive phenotype, which includes the ability to degrade basement membrane and extracellular matrices (3). Typical cellular pH gradients are disrupted in metastatic cancer cells so that the intracellular pH is more alkaline and the extracellular pH is more acidic in cancer cells when compared with normal cells. Multiple reports have demonstrated that an acidic extracellular pH enhances tumor cell invasiveness (4, 5). Determining the mechanisms by which pH is regulated in metastatic cancer cells may result in the identification of novel therapeutic targets to inhibit metastasis.
The V-ATPase2 is a multisubunit proton pump expressed in lysosomes, endosomes, secretory vesicles, clathrin-coated vesicles, and on the plasma membranes of some specialized cells (6–8). The V-ATPase is composed of two domains. In mammals, the peripheral V1 domain is composed of eight subunits (A-H) and is responsible for ATP hydrolysis, whereas the membrane-embedded V0 domain is composed of five subunits (a, c, c′′, d, and e) and is responsible for proton translocation (6). The 100-kDa a subunit is composed of a 50-kDa membrane-embedded C-terminal domain involved in proton transport and a 50-kDa hydrophilic N-terminal domain located on the cytoplasmic side of the membrane (9). Targeting information is located in the N-terminal domain of subunit a (10).
In mammalian cells, subunit a is expressed as four isoforms (a1-a4) (11). V-ATPases containing the a1 and a2 isoforms appear to localize primarily to intracellular compartments. a1 localizes to synaptic vesicles (12), whereas a2 is present in endosomal compartments and the Golgi (13, 14). a3 localizes to lysosomes in pre-osteoclasts (13) and insulin-containing secretory vesicles in pancreatic β cells (15). Both a3 and a4 target V-ATPases to the plasma membranes of specialized cell types. a3 targets to the plasma membrane of osteoclasts, where V-ATPase activity plays a critical role in bone resorption (13, 16–18). a4 targets to the apical membrane of renal intercalated cells, where V-ATPases are responsible for acid secretion into the urine (8, 19–21). a4 also localizes to the apical membrane of epididymal clear cells, where acid secretion facilitates sperm maturation (22).
A number of studies have suggested that V-ATPase activity is important for cancer cell invasion. Highly invasive MDA-MB231 human breast cancer cells express much higher levels of V-ATPase at the plasma membrane than poorly invasive MCF7 cells (23). Moreover, in vitro invasion by MB231 but not MCF7 cells is inhibited by the specific V-ATPase inhibitors bafilomycin and concanamycin (23). More recent studies from our laboratory have shown that MB231 cells express much higher levels of the a3 and a4 isoforms relative to MCF7 cells and that siRNA knockdown of both a3 and a4 inhibits invasion by MB231 cells (24). Knockdown of a4 in these cells appears to significantly reduce plasma membrane localization of V-ATPases. However, a limitation of these studies is that MB231 and MCF7 cells are independently derived cell lines that differ in many phenotypic and genetic properties (25, 26).
The involvement of V-ATPases in invasiveness is not limited to breast cancer cells. In human pancreatic ductal adenocarcinoma, high V-ATPase expression correlates with increasing cancer grade, and V-ATPases localize to the plasma membrane of the invasive Panc-1 pancreatic cancer cell line (27). Furthermore, blocking V-ATPase activity inhibits pancreatic cancer cell invasion and reduces matrix metalloproteinase 9 activity (27). The highly metastatic mouse melanoma cell line B16-F10 expresses more a3 than the less metastatic B16 cell line (28). B16-F10 cells also localize V-ATPases to the plasma membrane, and knockdown of a3 suppresses invasion. Importantly, administration of a V-ATPase inhibitor blocks bone metastasis of B16-F10 (28). A recent report has demonstrated that inhibition of V-ATPases blocks invasion of prostate cancer cell lines as well (29).
It is currently unknown which a subunit isoforms are expressed at the plasma membrane in breast cancer cells and whether plasma membrane V-ATPase expression is directly involved in the invasive phenotype. To better examine whether expression of particular a subunit isoforms is critical to invasiveness of breast tumor cells, two closely related breast cancer cell lines have been examined. MCF10a cells are a non-invasive, immortalized human cell line, and MCF10CA1a cells are a highly invasive, H-Ras-transformed derivative of MCF10a cells selected for their ability to form metastases in mice (30, 31). We compared the invasiveness of these lines and their dependence on V-ATPases for invasion using V-ATPase inhibitors and knockdown of specific a subunit isoforms. We also investigated the effect of overexpression of particular a subunit isoforms on invasiveness and plasma membrane localization of V-ATPases. The results suggest a role for the a3 isoform in both plasma membrane localization and invasiveness of human breast cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF10a cells were purchased from the ATCC. MCF10CA1a cells were provided by Dr. Yibin Kang (Princeton University). MCF10a and MCF10CA1a cells were cultured as described previously (32) in DMEM/F12 medium (Invitrogen) containing 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (Peprotech), 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma), 60 μg/ml penicillin, and 125 μg/ml streptomycin (Invitrogen). Cells were grown in a 95% air, 5% CO2 humidified environment at 37 °C.
Real-time Reverse Transcription PCR
Quantitative real-time reverse transcription PCR was conducted as described previously (24). Cells were harvested and lysed, and RNA was isolated using an RNeasy® mini kit (Qiagen). After RNA isolation, mRNA was isolated with the MicroPoly(A) PuristTM kit from Ambion. Total RNA or mRNA concentration was quantified using Quant-iT RiboGreen RNA reagent (Molecular Probes). One-step quantitative RT-PCR was performed in a 96-well format on a Stratagene MX-3000P QPCR system using Brilliant SYBR Green QRT-PCR master mix kit (Stratagene). The PCR cycling sequence consisted of 30 min at 50 °C to allow for reverse transcription and then a heat inactivation and denaturation step for 10 min at 95 °C. This was followed by 40 cycles of 30 s at 95 °C, 1 min at 55 °C, and 30 s at 72 °C to allow for denaturation, annealing, and extension, respectively. To quantitate the results, cDNA clones were obtained from the ATCC (a1 and a2 isoforms), Open Biosystems (a3 and a4 isoforms), and Thermo Scientific (A). The cDNA sequences were verified by sequencing. Plasmid DNA for each isoform isolated from Escherichia coli was quantitated by measuring the absorbance at 260 nm. Serial dilutions were made, and these DNA standards were used to facilitate quantification of the initial mRNA levels for each experimental sample by use of a standard curve.
RNA Interference
siRNA pools specific for the a1, a2, a3, or a4 isoform were purchased from Dharmacon. Each pool contained four siRNAs specific for the appropriate a subunit isoform. MCF10CA1a cells were plated in 60-mm dishes at 4 × 105 cells/dish in the media described above, without antibiotics, and incubated overnight. Cells were transfected with 20 nm siRNA directed against a1 or a2, 100 nm siRNA directed against a3, or 10 nm siRNA directed against a4 according to the directions of the manufacturer. Briefly, the siRNA was mixed 1:1 with Opti-MEMTM (Invitrogen), allowed to incubate for 5 min, and then mixed with Dharmafect 1 transfection reagent (Dharmacon). The siRNA/transfection reagent mixture was incubated for 20 min at room temperature and then added to the appropriate volume of DMEM/F12 + serum, and 4 ml was added to each dish. After incubation of cells with siRNA for 24 h, the media were changed to siRNA-free media, and cells were incubated for an additional length of time (depending upon the assay) prior to harvesting. For all experiments, data were collected 96 h post-transfection. To quantitate reduction in the a subunit isoform mRNA levels, quantitative RT-PCR was performed as described above using RNA isolated from cells after siRNA treatment. To confirm the specificity of knockdown, we used primers specific for each isoform tested. The effect of siRNA treatment on cell viability was assessed by trypan blue exclusion. Insignificant staining with trypan blue was observed with all siRNA treatments, indicating that siRNA knockdown of a isoforms did not reduce cell viability.
Plasmid Transfection
cDNAs for each human a subunit isoform were cloned separately into the pTracerTM-CMV/Bsd plasmid (Invitrogen). In all cases, insertion was verified by sequencing. 15 μg of the resultant plasmids, as well as the empty vector, was transfected separately into MCF10a cells using Lipofectamine 2000 (Invitrogen) in accordance with the recommendations of the manufacturer. Stable transfection was achieved by treatment of cells with 7.5 μg/ml blasticidin S (Invitrogen) for 21 days, beginning 3 days post-transfection. After selection for stable transfection, overexpression of the appropriate a subunit isoform was verified using quantitative RT-PCR as described above.
Invasion Assay
Assays for in vitro invasion were performed using FluoroBlok inserts (BD Biosciences) with an 8-μm pore size membrane and coated with MatrigelTM (BD Biosciences) (33). A chemoattractant (FBS) was added to the trans-side of the membrane to induce invasive cells to digest the coating and invade through the pores to the trans-side. MatrigelTM was diluted in PBS to a final concentration of 0.3 μg/μl, and a total of 18 μg was coated onto the membrane in each well. The membrane was allowed to dry overnight under a vacuum at room temperature. The MatrigelTM-coated membrane was rehydrated with 60 μl of DMEM + 4.5 g/liter d-glucose without phenol red, l-glutamine, or sodium pyruvate (Invitrogen) (termed media) for at least 2 h. 500 μl of the media containing 5% FBS were added to wells in a 24-well plate to act as a chemoattractant. Cells were harvested by trypsinization and were brought to a concentration of 2 × 105 cells/ml in media. For experiments testing the effects of concanamycin on invasion, cells were resuspended in media containing either dimethyl sulfoxide or 100 nm concanamycin A in dimethyl sulfoxide and allowed to incubate for 15 min at 37 °C. 2.5 × 104 cells were seeded onto the rehydrated membrane, which was then placed onto the wells containing chemoattractant. After 15 h, membranes were placed onto wells containing 4 μg/ml calcein AM in Hanks' balanced salt solution (Invitrogen) and incubated for 30 min at 37 °C in 5% CO2. Cells that had migrated to the trans-side of the membrane were quantitated using a Zeiss Axiovert 10 fluorescence microscope. The number of invading cells was averaged over three wells (15 images/well). Invasion for treated cells was measured relative to control cells. For concanamycin- or dimethyl sulfoxide-treated samples, cell viability was assessed by trypan blue exclusion 24 h after treatment was initiated. Less than 1% of harvested cells stained with trypan blue in either treatment group, indicating minimal cell death.
Immunocytochemistry
A monoclonal antibody raised against the V-ATPase subunit A (Sigma, clone 4F5) was used to determine the in situ localization of V-ATPases. Cells were plated onto 24 × 24 mm coverslips. Approximately 24 h later, cells were washed, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Nonspecific binding was blocked by incubation with 1% bovine serum albumin in PBS for 1 h and then incubated with the anti-A subunit antibody at a 1:1000 dilution overnight. The cells were next rinsed with PBS and then incubated with Alexa Fluor® 488-conjugated goat anti-rabbit secondary antibody (1:500 dilution) and Alexa Fluor® 594 phalloidin (to stain F-actin, 1:250 dilution) (Invitrogen) in 1% bovine serum albumin/PBS. After 2 h of incubation at room temperature, the cells were rinsed with PBS. The cells were prepared for viewing using ProLong® Gold (Invitrogen) mounting medium and allowed to cure at room temperature for 24 h. The stained cells were imaged on a Leica TCS SP2 confocal microscope. To quantify plasma membrane staining, 60 cells from three separate batches of immunostained images were counted, and the percentage of cells showing plasma membrane V-ATPase localization was determined.
Cell Lysis and Western Blotting
Cells were harvested, resuspended in lysis buffer with protease inhibitors (PBS-EDTA containing 137 mm NaCl, 1.2 mm KH2PO4, 15.3 mm Na2HPO4, 2.7 mm KCl, 2 mm EDTA (pH 7.2), 2 μg/ml aprotinin, 5 μg/ml leupeptin, 0.7 μg/ml pepstatin, and 1 mm PMSF) and lysed by sonication. Cell lysates were centrifuged for 10 min at 4 °C at 15,000 × g to remove cellular debris. Protein concentrations were determined using the Lowry method. SDS sample buffer was added to the lysates and aliquots containing 20 μg of protein were separated by SDS-PAGE on 4–15% gradient acrylamide gels. The presence of subunit A or α-tubulin was detected by Western blotting using monoclonal antibodies from Sigma or GenScript, respectively, followed by a horseradish peroxidase-conjugated secondary antibody (Bio-Rad). Blots were developed using the Amersham Biosciences ECL Western blotting analysis system from GE Healthcare.
Statistical Analysis
All results are expressed as means. Error bars represent S.D. Significance was determined by analysis of variance for each experiment, followed by pairwise Student's t test to compare individual treatments to the appropriate controls.
RESULTS
The Effect of V-ATPase Inhibition on in Vitro Invasion of MCF10a and MCF10CA1a Cells
To determine the role of V-ATPases in breast tumor cell invasion, we compared V-ATPase involvement in two closely related human breast cancer cell lines. MCF10a cells are spontaneously immortalized breast epithelial cells derived from a patient with fibrocystic disease that are unable to form tumors in immunocompromised mice (28). MCF10CA1a cells are a highly invasive cell line derived from H-Ras-transformed MCF10a cells (MCF10AT) after selection for the ability to forms tumors in mice (29, 32, 34). The effect of treatment of MCF10a and MCF10CA1a cells with the specific V-ATPase inhibitor concanamycin (100 nm) on invasion was determined using an in vitro Matrigel invasion assay (33). MCF10CA1a cells are significantly more invasive than the parental MCF10a cell line (Fig. 1). Moreover, treatment with concanamycin dramatically reduced invasion by MCF10CA1a cells but did not affect invasion by MCF10a cells. Measurement of cell viability using trypan blue exclusion revealed that treatment with 100 nm concanamycin for 24 h (a period longer than the 15 h employed in the cell invasion assay) did not induce cell death in either MCF10a or MCF10CA1a cells (data not shown), indicating that inhibition of invasion by concanamycin is not a consequence of decreased cell viability. These data show that V-ATPase activity is important for in vitro invasion by MCF10CA1a cells but not MCF10a cells.
FIGURE 1.
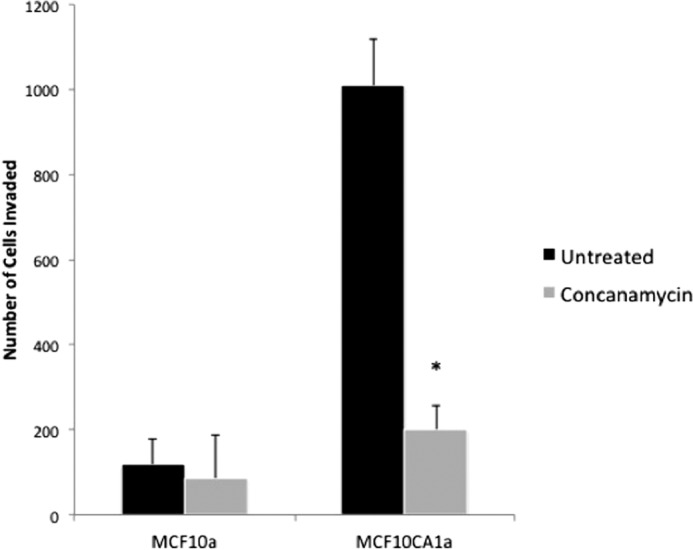
In vitro invasion of MCF10a and MCF10CA1a cells after concanamycin treatment. In vitro invasion was assayed using MatrigelTM-coated FluoroBlokTM inserts as described under “Experimental Procedures.” Cells were treated with or without 100 nm concanamycin, allowed to invade toward a chemoattractant, and stained with calcein-AM. An equal amount of solvent (dimethyl sulfoxide) was added to untreated cells. Cells that had migrated to the trans-side were counted, with three wells analyzed per sample and 15 images analyzed per well. Values are the mean ± S.D. of three independent experiments. *, p < 0.01 compared with the untreated control.
mRNA Levels of a Subunit Isoforms in MCF10a and MCF10CA1a cells
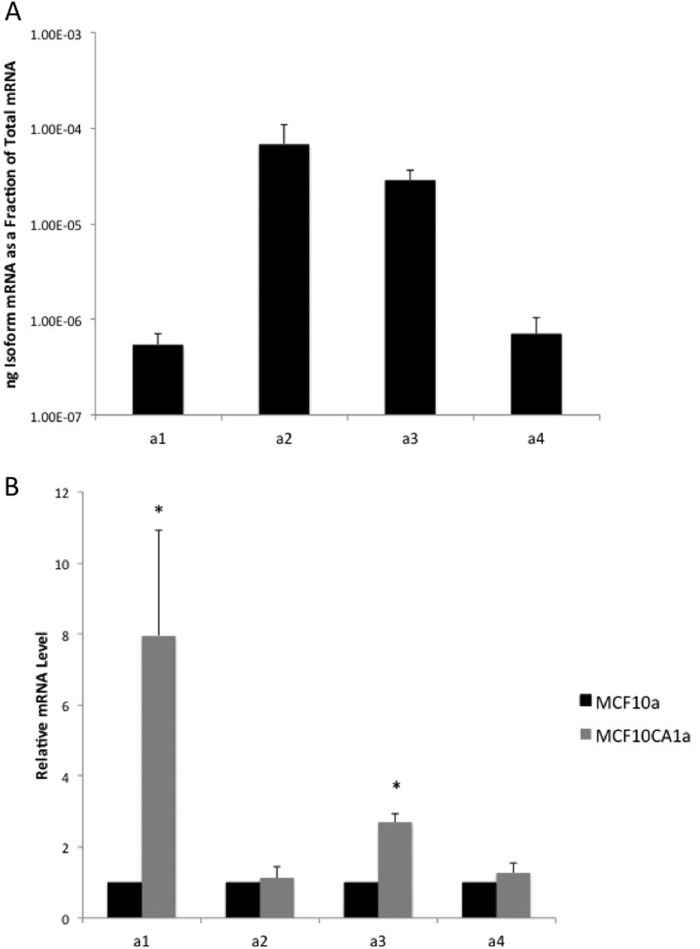
We next compared the expression levels of isoforms of subunit a in MCF10a and MCF10CA1a cells using quantitative RT-PCR. We demonstrated previously that the highly invasive MDA-MB231 cells expressed higher levels of both the a3 and a4 isoforms of subunit a as compared with the poorly invasive MCF7 cells (24). The significance of these results was limited, however, by the fact that MB231 and MCF7 cells are independently derived lines and, thus, likely differ in many functional and genetic properties. The parental MCF10a cells expressed the highest levels of mRNA encoding the a2 and a3 isoforms (Fig. 2A). Relative to MCF10a cells, MCF10CA1a cells expressed 8- and 3-fold higher levels of the a1 and a3 mRNA, respectively (Fig. 2B). We are not able, at present, to confirm the relative protein expression of a subunit isoforms because of the lack of antibodies that are specific for particular a subunit isoforms.
FIGURE 2.
mRNA levels of a subunit isoforms in MCF10a and MCF10CA1a cells. mRNA levels were determined using quantitative RT-PCR for each a subunit isoform on mRNA isolated from each cell line as described under “Experimental Procedures.” Plasmids expressing the cDNA of each a subunit isoform were used to establish a standard curve. Values were normalized to the total mRNA loaded and are the mean of four separate experiments. Error bars show standard deviation. A, a subunit isoform levels in MCF10a cells reported as the ratio of a subunit isoform mRNA to the total mRNA. B, ratio of a subunit isoform mRNA levels in MCF10CA1a cells versus MCF10a cells. *, p < 0.01 compared with the MCF10a mRNA level.
The Effect of Knockdown of a Subunit Isoforms Using Isoform-specific siRNAs on Invasion of MCF10CA1a Cells
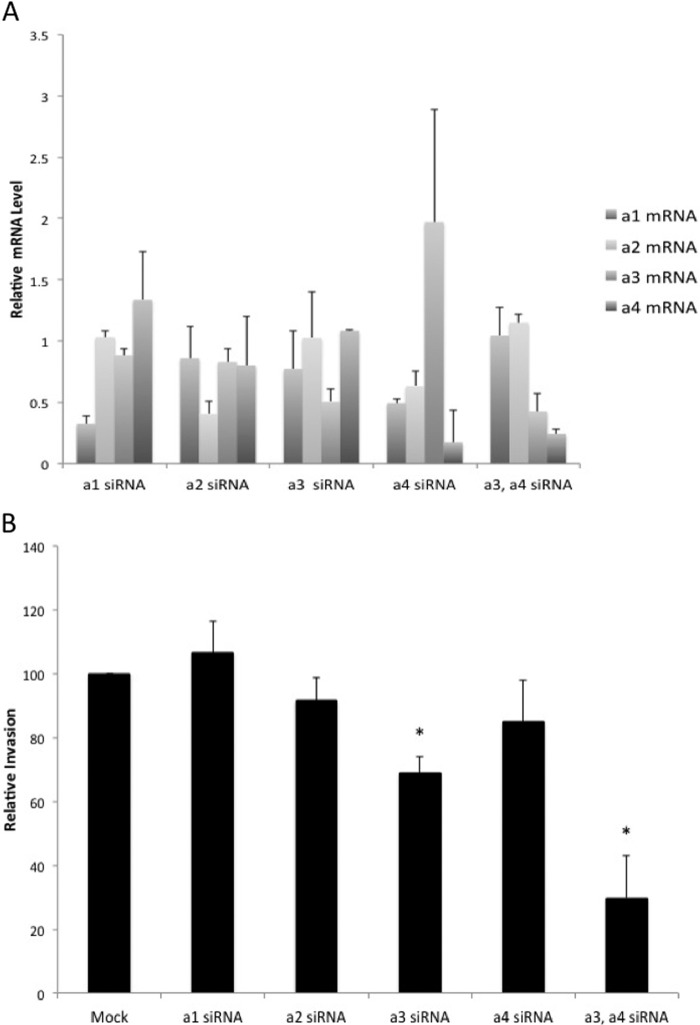
To determine the role of various a subunit isoforms in invasion by MCF10CA1a cells, cells were transfected with pools of siRNAs specific for each a subunit isoform for 81 h prior to measuring in vitro invasion as described above. The effect of siRNA treatment on mRNA levels for each isoform was assessed by quantitative RT-PCR. siRNAs targeting a1, a2, or a3 resulted in a 50% or greater reduction in mRNA levels for the corresponding isoforms without significantly affecting expression levels of the other a subunit isoforms (Fig. 3A). Treatment of cells with siRNAs targeting a4 decreased a4 subunit mRNA levels to 20% of untreated cells. However, this treatment also resulted in measurable decreases in a1 and a2 mRNA levels and a compensatory increase in a3 mRNA levels. With respect to invasion, knockdown of a3, but not a1, a2, or a4, significantly inhibited invasion by ∼30% (Fig. 3B). To determine whether the increase in a3 levels observed upon treatment with a4 siRNAs might be relevant for invasion, MCF10CA1a cells were simultaneously transfected with siRNAs targeting a3 and a4. Cotransfection of a3 and a4 siRNAs led to a decrease in expression of both a3 and a4 without affecting a1 and a2 levels (Fig. 3A). Concurrent knockdown of a3 and a4 inhibited invasion to a much greater extent than knockdown of a3 alone (Fig. 3B). These results support an important role for a3 expression in invasion by MCF10CA1a cells.
FIGURE 3.
In vitro invasion of MCF10CA1a cells after siRNA treatment. A, mRNA levels of a subunit isoforms in MCF10CA1a cells after siRNA treatment. Cells were exposed to a subunit isoform-specific siRNA pools for 96 h prior to measuring mRNA levels, as described under “Experimental Procedures.” Quantitative RT-PCR was conducted as described in the legend for Fig. 2. Knockdown is reported as the ratio of a subunit isoform mRNA in cells treated with siRNA versus a subunit isoform mRNA in untreated cells. Values are the mean of three separate experiments. Error bars indicate S.D. B, Cells were exposed to a subunit isoform-specific siRNAs for 81 h prior to measuring invasion through MatrigelTM-coated FluoroBlokTM wells. Invasion is reported as the percentage of invasion observed for siRNA-treated cells relative to untreated cells (Mock). Three wells were counted per sample, with 15 images analyzed per well. Values are the mean ± S.D. of two or three independent experiments. *, p < 0.005.
The Effect of a Subunit Isoform Overexpression on the Invasiveness of MCF10a Cells
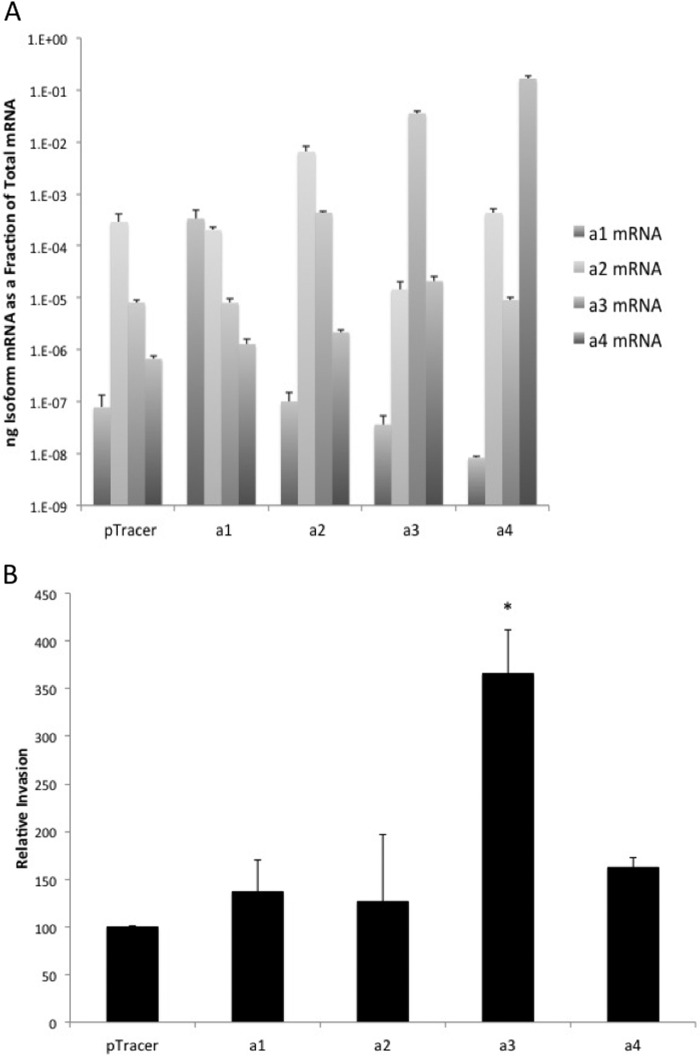
To assay the effect of overexpression of each a subunit isoform on invasion by the parental MCF10a cells, MCF10a cells were stably transfected with plasmids encoding each a subunit isoform. As a control, cells were stably transfected with the empty vector. Stable transfection of cells resulted in significant increases of the corresponding a subunit mRNA levels relative to vector-transfected cells (Fig. 4A). In the case of a4-transfected cells, there was also a measurable decrease in the levels of a1 mRNA. Overexpression of the a3 isoform, but not a1, a2, or a4, significantly increased invasion of MCF10a cells (Fig. 4B). To determine whether high expression of a3 led to increased overall V-ATPase expression, A subunit expression levels were determined using RT-PCR. No significant difference in subunit A expression was observed in a subunit isoform overexpressing MCF10a cells when compared with vector-transfected controls (data not shown). To confirm this result, Western blotting was performed on whole cell lysates of cells expressing the vector alone or the various a subunit isoforms. As seen in Fig. 5, no significant difference in subunit A protein levels was observed between these cell lines. These data suggest that up-regulation of a3 is sufficient to significantly increase the invasiveness of the parental MCF10a cell line and highlight the importance of a subunit isoform expression in breast cancer cell invasion.
FIGURE 4.
In vitro invasion assay of MCF10a cells selectively overexpressing each a subunit isoform. Each a subunit isoform was separately cloned into an overexpression vector, and the vectors were individually stably transfected into MCF10a cells. A, mRNA levels of a subunit isoforms in MCF10a cells expressing a subunit isoform overexpression vectors. mRNA levels were determined using quantitative RT-PCR for each a subunit isoform with mRNA isolated from each stable cell line. Plasmids expressing the cDNA of each a subunit isoform were used to establish a standard curve. The values reported are the ratio of isoform-specific mRNA to total mRNA. Values represent the mean ± S.D. of two or three experiments. B, in vitro invasion of MCF10a cells selectively overexpressing each a subunit isoform through MatrigelTM-coated FluoroBlokTM wells. Invasion is reported as the percentage of invasion observed for cells overexpressing particular a subunit isoforms relative to cells expressing empty vector. Three wells were counted per sample, with 15 images analyzed per well. Values are the mean ± S.D. of three independent experiments. pTracer, MCF10a cells transfected with an empty pTracer vector; a1, MCF10a cells overexpressing a1; a2, MCF10a cells overexpressing a2; a3, MCF10a cells overexpressing a3; a4, MCF10a cells overexpressing a4. *, p < 0.01 compared with pTracer.
FIGURE 5.
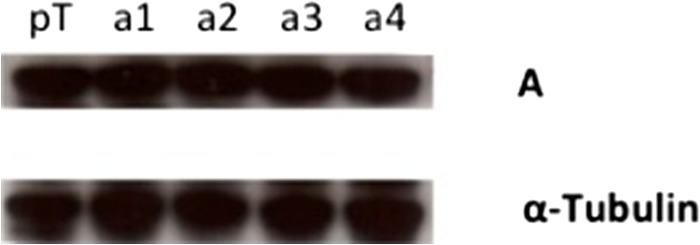
Protein levels of subunit A in MCF10a cells overexpressing various a subunit isoforms. The Western blot analysis shows protein levels of subunit A in MCF10a cells overexpressing various subunit a isoforms. pT, MCF10a cells transfected with the empty pTracer vector; a1, MCF10a cells overexpressing a1; a2, MCF10a cells overexpressing a2; a3, MCF10a cells overexpressing a3; a4, MCF10a cells overexpressing a4. The Western blot displayed is representative of data obtained from two separate experiments.
The Effect of a Subunit Expression on Cellular Distribution of V-ATPases Using Immunocytochemistry
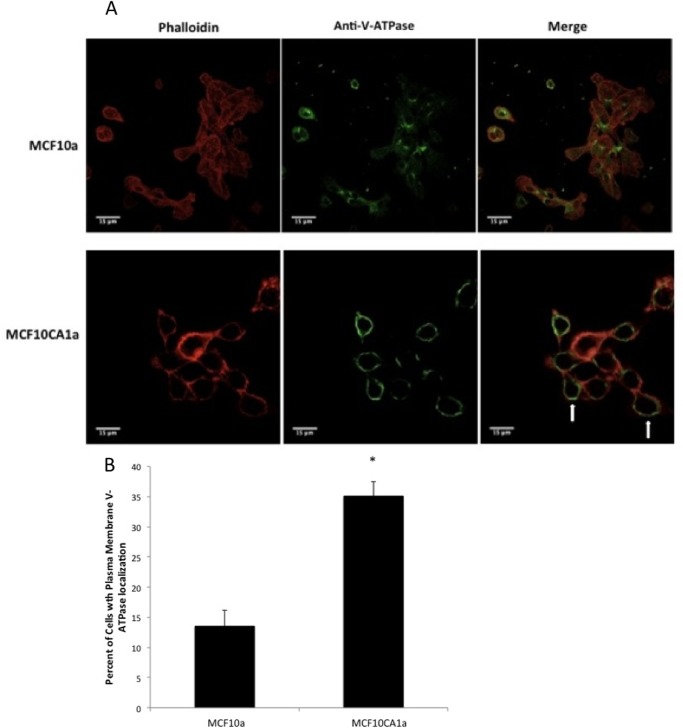
Previous studies have correlated plasma membrane V-ATPase distribution with invasive potential in breast cancer cell lines (23, 24) and migration of microvascular endothelial cells (35). To determine whether this correlation applies to the cell lines employed in this study, immunocytochemistry was performed using antibodies directed against the A subunit of the V-ATPase. This subunit is part of the V1 domain and is common to all V-ATPases in the cell (6). V-ATPases were observed at the cell periphery in MCF10CA1a cells but only rarely in the parental MCF10a cells (Fig. 6A). Phalloidin staining of F-actin was used to outline cells. V-ATPase staining observed at the periphery of the cell was interpreted as plasma membrane localization. Quantitation revealed an approximately 3-fold increase in the abundance of plasma membrane V-ATPases in MCF10CA1a cells relative to the parental line (Fig. 6B).
FIGURE 6.
Immunostaining of MCF10a and MCF10CA1a cells using an antibody against V-ATPase. MCF10a and MCF10CA1a cells were grown as a monolayer on coverslips in 6-well plates. Cells were immunostained using an antibody against subunit A of V-ATPase (part of the V1 domain, which stains all V-ATPases in the cell) as well as phalloidin to stain actin. Images were taken with identical exposure times and antibody concentrations. A, MCF10a (top row) and MCF10CA1a (bottom row) cells showing fluorescence staining of actin (left column), V-ATPase (center column), and the merge (left column). B, quantification of plasma membrane staining in cells from immunostained images. 60 cells from each of three separate batches of immunostained images were counted, and the number of cells showing plasma membrane V-ATPase localization (arrows in A) was determined. The values represent the mean percentage ± S.D. of cells displaying plasma membrane staining. *, p < 0.01.
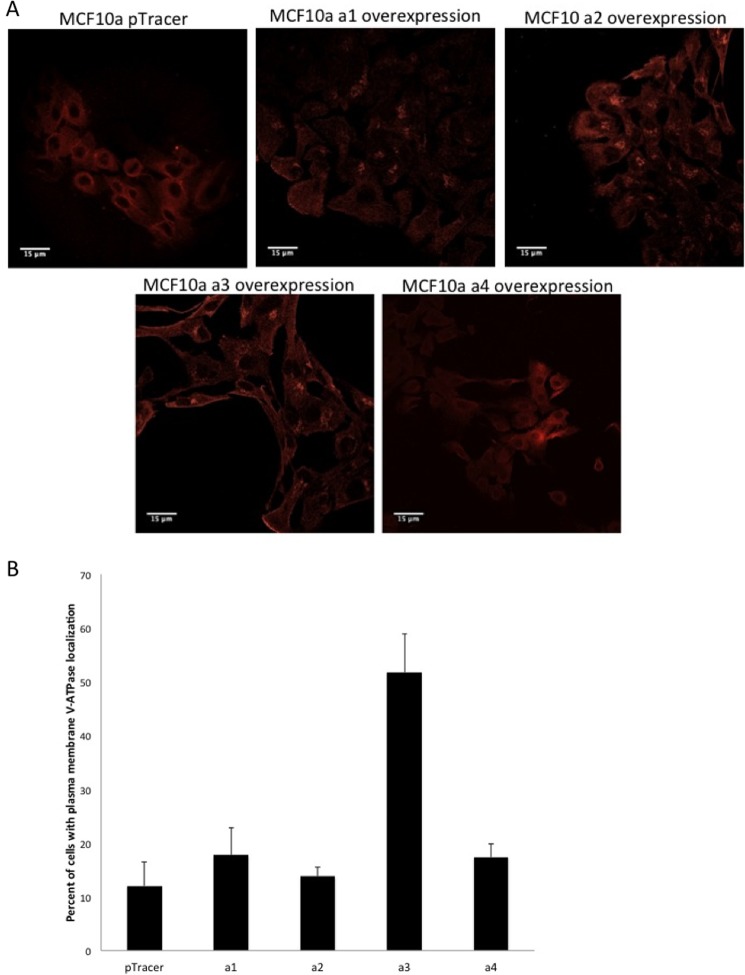
We next compared V-ATPase distribution in the MCF10a cells stably transfected with each of the a subunit isoforms (Fig. 7A). MCF10a cells transfected with the empty vector or overexpressing a4 showed a primarily diffuse pattern of staining, whereas cells overexpressing a1 or a2 showed both punctate and diffuse patterns of staining. By contrast, MCF10a cells overexpressing a3 showed not only diffuse and punctate intracellular staining but also staining at the cell periphery that we interpret as plasma membrane localization. Quantitation revealed a 2.5- to 4-fold increase in the fraction of cells displaying plasma membrane V-ATPase localization relative to cells transfected with the empty vector or overexpressing a1, a2, or a4 (Fig. 7B). These results suggest that a3 likely targets V-ATPase to the plasma membrane in a3-transfected MCF10a cells and that it is plasma membrane V-ATPases that likely contribute to the increased invasiveness of these cells.
FIGURE 7.
Immunostaining of MCF10a cells overexpressing subunit a isoforms using an antibody against V-ATPase. MCF10a cells were grown as a monolayer on coverslips in 6-well plates. Cells were immunostained using an antibody against subunit A of V-ATPase. Images were taken with identical exposure times and antibody concentrations. A, MCF10a cells expressing the empty pTracer vector or overexpressing the subunit a isoform indicated. B, quantification of plasma membrane staining in cells from immunostained images. 60 cells from each of two or three separate batches of immunostained images were counted, and the number of cells showing plasma membrane V-ATPase localization was determined. The values represent the mean percentage ± S.D. of cells displaying plasma membrane staining. a1, MCF10a cells overexpressing a1; a2, MCF10a cells overexpressing a2; a3, MCF10a cells overexpressing a3; a4, MCF10a cells overexpressing a4.
DISCUSSION
Targeting of V-ATPases to distinct cellular destinations is regulated by isoforms of subunit a (11). The role of different populations of V-ATPases containing particular isoforms of subunit a has begun to be probed in a number of cancers types, including breast cancer, melanoma, and pancreatic cancer. In the cases of melanoma and pancreatic cancer cells, high expression of the a3 isoform was found to be critical for invasion of highly metastatic cancer cells (27, 28). In the case of breast cancer cells, comparison of two unrelated cell lines (MCF-7 and MDA-MB231) showed that the more invasive MDA-MB231 cells express much higher levels of both a3 and a4 compared with the less invasive MCF-7 cell line (24). Because MDA-MB231 and MCF-7 cells were derived independently and differ in numerous characteristics (25, 26), a comparison of the expression of a subunit isoforms in more closely related cell lines was necessary. Furthermore, the effect of increasing expression of particular a subunit isoforms on tumor cell invasion has not been investigated previously. The goal of this study was to further probe the role of a subunit isoforms in the invasive potential of more closely related breast cancer cell lines.
This study focused on the MCF10a and the related MCF10CA1a cell lines (28, 29). MCF10a cells are a non-invasive, immortalized human cell line, and MCF10CA1a cells are a highly invasive, H-Ras-transformed derivative of MCF10a cells selected for their ability to form metastases in mice (29). These cell lines have been used to study genetic alterations linked to tumorigenesis. Here we demonstrated that V-ATPase activity is critical for the in vitro invasion of highly metastatic MCF10CA1a cells but not the parental MCF10a cells (Fig. 1). In addition, MCF10CA1a cells have increased expression of both the a1 and a3 isoforms relative to MCF10a cells (Fig. 2). Interestingly, siRNA knockdown of a3 (but not a1, a2, or a4) led to a significant decrease in invasion by MCF10CA1a cells (Fig. 3). These results suggest that a3 participates in invasion by MCF10CA1a cells. Because knockdown of other isoforms did not affect invasion, higher a1 expression in MCF10CA1a cells does not appear to be required for the invasive phenotype. It is possible that elevated expression of a1 in MCF10CA1a cells may confer some other advantage not tested in our invasion experiments. For example, elevated expression of a1 may be required for enhanced Golgi function associated with elevated expression of cell surface glycoproteins. Further work will be required to elucidate the function of a1 in tumor cells.
It is interesting to note that although knockdown of the a4 isoform alone did not reduce invasion, it did result in an increase in the levels of a3 mRNA (Fig. 3). The a4 isoform is expressed at low levels in MCF10a and MCF10CA1a cells compared with a3 (Fig. 3). Thus, it is surprising that knockdown of a4 in MCF10CA1a cells results in a 2-fold increase in a3 expression. It is possible that a reduction in invasion caused by the loss of a4 may have been masked by this up-regulation in a3 levels. To address this possibility, knockdown of a3 and a4 was performed concurrently. Under these conditions, although the level of a3 mRNA observed was comparable with that obtained with knockdown of a3 alone, the inhibition of invasion observed was much larger. However, it should be noted that overexpression of a4 did not significantly increase invasion or plasma membrane localization of V-ATPases in MCF10a cells (Figs. 4 and 7). Understanding the molecular basis of these results will require additional studies of the functional interaction between the a3 and a4 isoforms in breast tumor cells.
To complement our analysis of the effect of a subunit knockdown on invasion by MCF10CA1a cells, we also tested the effect of overexpression of a subunit isoforms on the invasive potential of the parental MCF10a cell line. Importantly, overexpression of a3 but not the other a subunit isoforms dramatically increased invasion by MCF10a cells (Fig. 4). To the best of our knowledge, these are the first data demonstrating that elevated expression of a particular V-ATPase isoform can increase the invasion by tumor cells. Because a3 is known to localize V-ATPases to the plasma membrane of osteoclasts (13, 16–18), we expected that overexpression of a3 was increasing localization of V-ATPases to the plasma membrane of MCF10a cells. Immunocytochemical localization confirmed increased plasma membrane staining of V-ATPases in cells overexpressing a3 (Fig. 7). Consistent with this idea, MCF10CA1a cells express significantly higher levels of plasma membrane V-ATPase relative to parental cells (Fig. 6). These results suggest that elevated a3 expression may be responsible for elevated V-ATPase localization to the plasma membrane and that plasma membrane V-ATPase contributes to enhanced invasion by breast tumor cells.
Comparison of the images in Fig. 7 reveals that only overexpression of a3 leads to increased plasma membrane staining. By contrast, overexpression of a1 and a2 shows increased intracellular staining, whereas overexpression of a4 looks similar to cells expressing the empty vector. It should be noted that the staining observed in MCF10a cells in Fig. 6 appears somewhat more punctate than in the empty vector-transfected cells in Fig. 7, which appears more diffuse. This may be due to the use of different secondary antibodies in the experiments employed in Figs. 6 and 7, which may result in a different sensitivity of detection. Nevertheless, a comparison of the images in Fig. 7 employing the same conditions and antibodies supports the conclusion that overexpression of a3 leads to increased peripheral localization of V-ATPase.
There are several mechanisms by which overexpression of a3 and up-regulation of plasma membrane V-ATPases may lead to enhanced invasion by tumor cells. One possibility is that high a3 expression leads to an increase in total V-ATPase expression within MCF10a cells. Although somewhat brighter staining was observed for cells overexpressing a3 (Fig. 7A), no increase in A subunit protein levels was detected in these cells (Fig. 5). All panels in Fig. 7 were obtained under identical experimental conditions with identical exposure times. It is possible that overexpression of a3 may result in more assembled complexes that appear more finely punctate (and thus brighter) than the more diffuse pattern of the vector-transfected cells.
For successful metastasis, tumor cells must degrade the extracellular matrix and migrate out of the primary tumor as well as escape the circulation or lymphatic system to colonize new sites. For some tumor cells, this process of invasion relies on secretion of a class of pH-dependent proteases known as cathepsins (36). These proteases normally reside in lysosomes and, therefore, require a low pH for activity. It has been postulated that plasma membrane V-ATPases create an acidic extracellular environment that allows the activation of secreted cathepsins (24). Secreted cathepsins, once activated, may also function in the proteolytic activation of other proteases that participate in invasion, such as matrix metalloproteinases (36, 37). Neutralization of the acidic extracellular pH of tumors has been shown to prevent the formation of metastases in mouse models (38). Moreover, pharmacologic or genetic disruption of V-ATPase activity has been shown to reduce matrix metalloproteinase activity in pancreatic cancer cells (27). The data presented in this study are consistent with this hypothesis but do not rule out other possible roles for V-ATPases in invasion.
Another possibility by which V-ATPases may participate in invasion is though TGF-β signaling. TGF-β1 can induce epithelial-to-mesenchymal transition in renal epithelial cells as well as increased expression of a number of V-ATPase subunits, including B2, E, and c (39). TGF-β1 treatment also stimulates V-ATPase activity and plasma membrane localization in these cells, and inhibition of V-ATPase activity blocks the TGF-β1 induced epithelial-to-mesenchymal transition (39). Although the effect of TGF-β stimulation on a3 expression has not been evaluated, one possibility is that a3 overexpression promotes invasion in MCF10a cells by a similar mechanism as TGF-β stimulation.
Several recent studies have suggested that V-ATPases regulate trafficking of proteins involved in invasion. Treatment of metastatic cells with the V-ATPase inhibitor archazolid limited leading edge localization of the epidermal growth factor receptor and blocked endocytic activation of Rac1, a member of the Rho family of small GTPases with known functions in cell migration (40). Inhibition of V-ATPases with bafilomycin or through siRNA silencing prevented peripheral localization of Rab27 secretory vesicles and blocked heat shock protein 90α secretion (41). Inhibiting V-ATPases may prevent peripheral localization of molecules indirectly because low extracellular pH induces a redistribution of lysosomes toward the cell periphery (42).
In summary, the results of this work, in conjunction with other studies, indicate that V-ATPases participate in the invasion of metastatic cancer cells. Importantly, expression of the a3 isoform appears to promote both plasma membrane localization of V-ATPases and invasion in closely related breast cancer cell lines. This suggests that the V-ATPase, and the a3 isoform in particular, may be effective therapeutic targets in limiting the metastatic spread of breast cancer cells.
Acknowledgments
We thank Rachel Liberman, Laura Stransky, and Kristina Cotter for discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34478 (to M. F.).
- V-ATPase
- vacuolar proton-translocating adenosine triphosphatase.
REFERENCES
- 1. Nguyen D. X., Bos P. D., Massagué J. (2009) Metastasis. From dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 2. Spano D., Zollo M. (2012) Tumor microenvironment. A main actor in the metastasis process. Clin. Exp. Metastasis 29, 381–395 [DOI] [PubMed] [Google Scholar]
- 3. Gupta G. P., Massagué J. (2006) Cancer metastasis. Building a framework. Cell 127, 679–695 [DOI] [PubMed] [Google Scholar]
- 4. Martínez-Zaguilán R., Seftor E. A., Seftor R. E., Chu Y. W., Gillies R. J., Hendrix M. J. (1996) Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis 14, 176–186 [DOI] [PubMed] [Google Scholar]
- 5. Gillet L., Roger S., Besson P., Lecaille F., Gore J., Bougnoux P., Lalmanach G., Le Guennec J. Y. (2009) Voltage-gated sodium channel activity promotes cysteine cathepsin-dependent invasiveness and colony growth of human cancer cells. J. Biol. Chem. 284, 8680–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forgac M. (2007) Vacuolar ATPases. Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 7. Li S. C., Kane P. M. (2009) The yeast lysosome-like vacuole. Endpoint and crossroads. Biochim. Biophys. Acta 1793, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown D., Paunescu T. G., Breton S., Marshansky V. (2009) Regulation of the V-ATPase in kidney epithelial cells. Dual role in acid-base homeostasis and vesicle trafficking. J. Exp. Biol. 212, 1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Toei M., Forgac M. (2008) Analysis of the membrane topology of transmembrane segments in the C-terminal hydrophobic domain of the yeast vacuolar ATPase subunit a (Vph1p) by chemical modification. J. Biol. Chem. 283, 20696–20702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H. (2001) The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276, 47411–47420 [DOI] [PubMed] [Google Scholar]
- 11. Toei M., Saum R., Forgac M. (2010) Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morel N., Dedieu J. C., Philippe J. M. (2003) Specific sorting of the a1 isoform of the V-H+ATPase a subunit to nerve terminals where it associates with both synaptic vesicles and the presynaptic plasma membrane. J. Cell Sci. 116, 4751–4762 [DOI] [PubMed] [Google Scholar]
- 13. Toyomura T., Murata Y., Yamamoto A., Oka T., Sun-Wada G. H., Wada Y., Futai M. (2003) From lysosomes to the plasma membrane. Localization of vacuolar-type H+-ATPase with the a3 isoform during osteoclast differentiation. J. Biol. Chem. 278, 22023–22030 [DOI] [PubMed] [Google Scholar]
- 14. Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 15. Sun-Wada G. H., Toyomura T., Murata Y., Yamamoto A., Futai M., Wada Y. (2006) The a3 isoform of V-ATPase regulates insulin secretion from pancreatic β-cells. J. Cell Sci. 119, 4531–4540 [DOI] [PubMed] [Google Scholar]
- 16. Frattini A., Orchard P. J., Sobacchi C., Giliani S., Abinun M., Mattsson J. P., Keeling D. J., Andersson A. K., Wallbrandt P., Zecca L., Notarangelo L. D., Vezzoni P., Villa A. (2000) Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 25, 343–346 [DOI] [PubMed] [Google Scholar]
- 17. Toyomura T., Oka T., Yamaguchi C., Wada Y., Futai M. (2000) Three subunit a isoforms of mouse vacuolar H+-ATPase. Preferential expression of the a3 isoform during osteoclast differentiation. J. Biol. Chem. 275, 8760–8765 [DOI] [PubMed] [Google Scholar]
- 18. Hu Y., Nyman J., Muhonen P., Väänänen H. K., Laitala-Leinonen T. (2005) Inhibition of the osteoclast V-ATPase by small interfering RNAs. FEBS Lett. 579, 4937–4942 [DOI] [PubMed] [Google Scholar]
- 19. Smith A. N., Skaug J., Choate K. A., Nayir A., Bakkaloglu A., Ozen S., Hulton S. A., Sanjad S. A., Al-Sabban E. A., Lifton R. P., Scherer S. W., Karet F. E. (2000) Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 26, 71–75 [DOI] [PubMed] [Google Scholar]
- 20. Oka T., Murata Y., Namba M., Yoshimizu T., Toyomura T., Yamamoto A., Sun-Wada G. H., Hamasaki N., Wada Y., Futai M. (2001) a4, a unique kidney-specific isoform of mouse vacuolar H+-ATPase subunit a. J. Biol. Chem. 276, 40050–40054 [DOI] [PubMed] [Google Scholar]
- 21. Smith A. N., Finberg K. E., Wagner C. A., Lifton R. P., Devonald M. A., Su Y., Karet F. E. (2001) Molecular cloning and characterization of Atp6n1b. A novel fourth murine vacuolar H+-ATPase a-subunit gene. J. Biol. Chem. 276, 42382–42388 [DOI] [PubMed] [Google Scholar]
- 22. Pietrement C., Sun-Wada G. H., Silva N. D., McKee M., Marshansky V., Brown D., Futai M., Breton S. (2006) Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol. Reprod. 74, 185–194 [DOI] [PubMed] [Google Scholar]
- 23. Sennoune S. R., Bakunts K., Martínez G. M., Chua-Tuan J. L., Kebir Y., Attaya M. N., Martínez-Zaguilán R. (2004) Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential. Distribution and functional activity. Am. J. Physiol. Cell Physiol. 286, C1443–52 [DOI] [PubMed] [Google Scholar]
- 24. Hinton A., Sennoune S. R., Bond S., Fang M., Reuveni M., Sahagian G. G., Jay D., Martinez-Zaguilan R., Forgac M. (2009) Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells. J. Biol. Chem. 284, 16400–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soule H. D., Vazguez J., Long A., Albert S., Brennan M. (1973) A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 51, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 26. Cailleau R., Young R., Olivé M., Reeves W. J. (1974) Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 53, 661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chung C., Mader C. C., Schmitz J. C., Atladottir J., Fitchev P., Cornwell M. L., Koleske A. J., Crawford S. E., Gorelick F. (2011) The vacuolar-ATPase modulates matrix metalloproteinase isoforms in human pancreatic cancer. Lab. Invest. 91, 732–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishisho T., Hata K., Nakanishi M., Morita Y., Sun-Wada G. H., Wada Y., Yasui N., Yoneda T. (2011) The a3 isoform vacuolar type H+-ATPase promotes distant metastasis in the mouse B16 melanoma cells. Mol. Cancer Res. 9, 845–855 [DOI] [PubMed] [Google Scholar]
- 29. Michel V., Licon-Munoz Y., Trujillo K., Bisoffi M., Parra K. J. (2013) Inhibitors of vacuolar ATPase proton pumps inhibit human prostate cancer cell invasion and prostate-specific antigen expression and secretion. Int. J. Cancer 132, E1-E10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soule H. D., Maloney T. M., Wolman S. R., Peterson W. D., Jr., Brenz R., McGrath C. M., Russo J., Pauley R. J., Jones R. F., Brooks S. C. (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 [PubMed] [Google Scholar]
- 31. Santner S. J., Dawson P. J., Tait L., Soule H. D., Eliason J., Mohamed A. N., Wolman S. R., Heppner G. H., Miller F. R. (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65, 101–110 [DOI] [PubMed] [Google Scholar]
- 32. Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 33. Partridge J., Flaherty P. (2009) An in vitro FluoroBlok tumor invasion assay. J. Vis. Exp. 29, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dawson P. J., Wolman S. R., Tait L., Heppner G. H., Miller F. R. (1996) MCF10AT. A model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 148, 313–319 [PMC free article] [PubMed] [Google Scholar]
- 35. Rojas J. D., Sennoune S. R., Maiti D., Bakunts K., Reuveni M., Sanka S. C., Martinez G. M., Seftor E. A., Meininger C. J., Wu G., Wesson D. E., Hendrix M. J., Martínez-Zaguilán R. (2006) Vacuolar-type H+-ATPases at the plasma membrane regulate pH and cell migration in microvascular endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 291, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 36. Gocheva V., Joyce J. A. (2007) Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 6, 60–64 [DOI] [PubMed] [Google Scholar]
- 37. Gondi C. S., Rao J. S. (2013) Cathepsin B as a cancer target. Expert Opin. Ther. Targets 17, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robey I. F., Baggett B. K., Kirkpatrick N. D., Roe D. J., Dosescu J., Sloane B. F., Hashim A. I., Morse D. L., Raghunand N., Gatenby R. A., Gillies R. J. (2009). Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 69, 2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao X., Yang Q., Qin J., Zhao S., Li X., Fan J., Chen W., Zhou Y., Mao H., Yu X. (2012) V-ATPase promotes transforming growth factor-β-induced epithelial-mesenchymal transition of rat proximal tubular epithelial cells. Am. J. Physiol. Renal Physiol. 302, 1121–1132 [DOI] [PubMed] [Google Scholar]
- 40. Wiedmann R. M., von Schwarzenberg K., Palamidessi A., Schreiner L., Kubisch R., Liebl J., Schempp C., Trauner D., Vereb G., Zahler S., Wagner E., Müller R., Scita G., Vollmar A. M. (2012) The V-ATPase-inhibitor archazolid abrogates tumor metastasis via inhibition of endocytic activation of the Rho-GTPase Rac1. Cancer Res. 72, 5976–5987 [DOI] [PubMed] [Google Scholar]
- 41. Hendrix A., Sormunen R., Westbroek W., Lambein K., Denys H., Sys G., Braems G., Van den Broecke R., Cocquyt V., Gespach C., Bracke M., De Wever O. (2013) Vacuolar H+ ATPase expression and activity is required for Rab27B-dependent invasive growth and metastasis of breast cancer. Int. J. Cancer 133, 843–854 [DOI] [PubMed] [Google Scholar]
- 42. Glunde K., Guggino S. E., Solaiyappan M., Pathak A. P., Ichikawa Y., Bhujwalla Z. M. (2003) Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia 5, 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]